Abstract
Photoswitched tethered ligands (PTLs) can be used to remotely control protein function with light. We have studied the geometric and conformational factors that determine the efficacy of PTL gating in the ionotropic glutamate receptor iGluR6 using a family of photoiosomerizable MAG (maleimide-azobenzene-glutamate) PTLs that covalently attach to the clamshell ligand-binding domain. Experiments and molecular dynamics simulations of the modified proteins show that optical switching depends on 2 factors: (i) the relative occupancy of the binding pocket in the 2 photoisomers of MAG and (ii) the degree of clamshell closure that is possible given the disposition of the MAG linker. A synthesized short version of MAG turns the channel on in either the cis or trans state, depending on the point of attachment. This yin/yang optical control makes it possible for 1 wavelength of light to elicit action potentials in one set of neurons, while deexciting a second set of neurons in the same preparation, whereas a second wavelength has the opposite effect. The ability to generate opposite responses with a single PTL and 2 versions of a target channel, which can be expressed in different cell types, paves the way for engineering opponency in neurons that mediate opposing functions.
Keywords: glutamate receptor, ion channel, optics, photoswitch
A major challenge in biology is to develop new ways of determining how proteins function in cells and how their activity in cell circuits underlies the behavior of the organism. An attractive approach is to use light as both an input and output for interrogation and manipulation of the functional state of proteins. Although there has been significant progress in optical detection of protein function over the last 20 years, the development of optical remote control has accelerated recently. Much of the effort has been focused on cell signaling, with a particular emphasis on controlling the activity of ion channels. Several naturally photosensitive channels and pumps have been cloned and used in a variety of biological preparations (1). In parallel, 3 classes of chemical–biological approaches have been used to obtain optical control over biological signaling: (i) free photolabile “caged” ligands, (ii) photoisomerizable (photochromic) free ligands (2), and (iii) photoswitched tethered ligands (PTLs) (3–6). Of these, the most molecularly focused are the PTLs, which selectively attach to specific protein targets and exclusively endow them with sensitivity to light.
PTLs have been used in ion channels to conditionally present an agonist to an allosteric regulatory site, originally in the nicotinic acetylcholine receptor (7–10), and recently in a kainate receptor, iGluR6 (LiGluR) (11–13) or to conditionally present a blocker to the Shaker K+ channel (SPARK) (14, 15). PTLs contain the ligand (agonist/antagonist or blocker) at one end, a reactive group that attaches covalently to the protein at the other end, and a linker in the middle that contains a photoisomerizable moieity, such as azobenzene. The PTL is anchored in a site-directed manner to the protein of interest, usually at a cysteine that is introduced by mutagenesis, near the ligand-binding site. Two wavelengths of irradiation are used to isomerize the azobenzene back and forth between an extended trans state and a bent cis state, projecting the ligand at an angle and shortening the end-to-end distance by 0.35 nm or more, depending on the length of the PTL (26). Optical control is obtained when the ligand preferentially binds in one of the states.
The PTL blocker for the Shaker K+ channel, MAQ (maleimide-azobenzene-quaternary ammonium), was designed based on the structure of the homologous KcsA K+ channel (16) and on previous knowledge that a tethered quaternary ammonium compound dangling from a passive linker could permanently block the outer end of the channel when conjugated to a cysteine located close enough for the extended linker to allow the ligand to reach the pore (17). Insertion of an azobenzene into the linker enabled the PTL to be long enough for the QA to reach and block the pore in the trans state but not in the cis state, when the linker is shorter (14). This design is expected to apply generally for proteins in which there is a “line of sight” from the PTL anchoring position to the ligand-binding site.
The design is more complicated for controlling agonist binding to an allosteric regulatory domain. In a clamshell domain, in which ligand binding stabilizes a closed conformation to activate the protein, the cysteine attachment site is better placed outside of the conserved clamshell “mouth” to avoid altering residues whose interactions contribute to closed-state stability. This could mean a loss of line of sight from the anchoring site to the binding site as suggested for the first version of LiGluR and its PTL, maleimide azobenzene glutamate (MAG) (11, 12).
To elucidate the structural basis of optical switching of iGluR6, we tested 3 MAGs, including a newly synthesized short MAG0 [see supporting information (SI) Appendix], at a series of attachment sites surrounding the glutamate-binding pocket. We find that the cis state is the activating state for 1 or more of the MAGs at most of the sites. However, some combinations activated in trans. All-atom molecular dynamics (MD) simulations of iGluR6 with tethered MAG0 in explicit solvent, including an analysis of the occupancy of the glutamate-binding site and the degree of clamshell closure that is possible given the location of the linker, provided insight into the mechanism of photoswitched activation and the basis of its state dependence. Strikingly, 1 of the PTLs, MAG0, was found to activate in cis at some attachment sites but in trans at others. This could be used to drive 2 populations of neurons in the same preparation to fire in a complementary manner, paving the way toward engineering opponency into distinct subgroups of neurons.
Results
Strategies for Optical Control.
To endow a protein of interest with sensitivity to light, the simplest design is for a PTL whose ligand-binding pocket is within an unhindered line of sight to a nearby cysteine anchoring site, so that photoisomerization changes linker length, and only 1 of the lengths is compatible with ligand binding (Fig. 1 A and B). The design is more complicated for controlling agonist binding deep in a regulatory domain, especially in clamshell domains where it may not be possible to install the anchoring cysteine within a line of sight of the ligand-binding pocket. In the first light-gated version of iGluR6 (11), the PTL, maleimide azobenzene glutamate 1 (MAG1), was attached to the “lip” of the clamshell, facing away from the glutamate-binding site, and the cis state was the activating state, consistent with the notion that at this site the linker needs to turn a corner to point the ligand into the binding site (Fig. 1C). We explored the mechanism of optical switching of iGluR6 conjugated site-specifically with 3 variants of MAG, including a newly synthesized short MAG0 (Fig. 1F).
Fig. 1.
Models of state-dependent liganding by MAG and MAG structures. (A–D) Models of photoswitching of iGluR6 by MAG. (A–B) Line-of-sight and matched length. Two conceptual versions of how isomerization from trans (long) to cis (short) can impact whether the glutamate reaches the binding site and docks. In A, trans is too long and cannot dock, but cis is shorter and docks. In B, cis is too short, but trans is long enough to reach. (C) Turning the corner. Another difference between cis and trans, in that cis is bent and can turn a corner, whereas trans is straight and will point away from the attachment site. For an attachment site that is outside the binding pocket, as illustrated, the bent cis state is favored to orient the glutamate into the binding pocket. (D) Even when glutamate points into the binding pocket, certain linker geometry may interfere with clamshell closure. (E) Structures of photoisomerizable azobenzene in cis state under illumination at 380 nm and in trans state in the dark and at 500-nm illumination. (F) Three MAG variants contain a maleimide (M) at one end, the photoisomerizable azobenzene (A) and an allyl glutamate (G) agonist at the other end and differ only in linker length by number of glycines.
Functional Assays with a Family of MAGs at a Series of Attachment Sites.
In an initial screen to measure the activity of iGluR6 in response to free glutamate and optical switching of MAG, we took advantage of the channel's calcium permeability by performing calcium imaging in HEK293 cells. Sixteen cysteine mutants were examined around the glutamate binding site (Fig. 2A and Table S1). The sites were chosen to ring the binding pocket and lie on the surface of the protein where they would be expected to be accessible to MAG and where mutation to cysteine and conjugation of MAG would be expected to produce a minimal disruption to structure and function. Of the 16 sites tested, 11 yielded functional channels that showed glutamate-activated rises in internal calcium. Three MAG variants, MAG0, MAG1, and MAG2 (Fig. 1E), were tested at these sites. Six of the cysteine mutants showed optical responses to 1 or more of the MAGs. (Table S1). Interestingly, site 486 and 2 nearby sites in the upper lobe, 482 and 484, were found to be more strongly activated by trans-MAG0 than cis-Mag0.
Fig. 2.
Locations of MAG attachment and patch clamp current recordings of light responses. (A) Location of 16 residues around the glutamate-binding pocket (star) of the iGluR6 LBD that were individually mutated to cysteine to serve as MAG attachment sites. (B and C) Representative whole-cell patch clamp recordings of light responses in HEK293 cells expressing iGluR6 with cysteine substitution sites conjugated to MAG. Currents are elicited by near-saturating (300 μM) glutamate (blue bars) and by changes in wavelength from 380 to 500 nm during constant illumination (black traces). Time bars represent 25 s. (B) Two cysteine sites activated by the cis state of the longest MAG. (C) Three cysteine sites activated by the trans state of the shortest MAG, including the sites in B.
Five of the cysteine mutants were selected for further analysis by whole-cell patch clamping. Inward currents were measured in voltage clamp in response to exposures to glutamate and to illumination at 380 and 500 nm (Fig. 2 B and C). The patch clamp analysis confirmed the calcium imaging for all of the sites examined, with cis being more active than trans in all cases except for MAG0 at 482, 484, and 486, where the greater activation was in trans (Fig. 2 B and C).
Although comparing optical and glutamate responses reports which isomer activates more strongly, it does not reveal the degree of channel opening induced by either isomer. To determine this, we measured dose–response curves for inhibition of MAG activation by the competitive antagonist DNQX (Fig. 3). Responses to glutamate, 380-nm light, and 500-nm light were measured in the absence of DNQX, and then various concentrations of DNQX (ranging over 3 orders of magnitude, up to the millimolar solubility limit) were washed on, and photocurrents were measured under illumination at 380 and 500 nm (Fig. 3 A and B). The fractional currents, normalized to the glutamate response, were plotted against DNQX concentration (Fig. 3 C and D). As can be seen from this analysis (Fig. 4), some sites, such as 439, activated almost exclusively in 1 state for all 3 of the MAGs, whereas other sites, such as 482, were strongly and almost equally activated by both isomers for all 3 of the MAGs. Site 439 was the best of the cis activators and site 486 was the best of the trans activators, whereas MAG0 showed the biggest isomer preference (Figs. 2–4). iGluR6(439C)-MAG0 showed the biggest change in effective concentration that we observed of all of the sites, with an ≈120-fold-higher DNQX IC50 in cis than in trans (Fig. 3C).
Fig. 3.
DNQX titrations for cis-on and trans-on channels. (A and B) Current traces of iGluR6(439C)-MAG0 (A) and iGluR6(486C)-MAG0 (B) in response to switching from illumination at 380 and 500 nm in the presence of different concentrations of DNQX (μM). DNQX can be seen to reduce inward current at both wavelengths. p2/g2, cis photocurrent normalized to glutamate-evoked current; p3/g2, trans photocurrent normalized to glutamate-evoked current. Red bars indicate 300 μM glutamate. (C and D) DNQX titration curves of iGluR6(439C)-MAG0 (C) and iGluR6(486C)-MAG0 (D) in cis (purple symbols and fits) and trans (green symbols and fits). Currents are calculated as defined in A and B and plotted against log[DNQX]. Values are means of 3–6 experiments. Error bars are SEM.
Fig. 4.
Summary of patch clamp recordings of photoswitching by 3 MAGs at 5 attachment sites. Bar graph showing amplitudes of current responses evoked by the 3 MAGs at 5 attachment sites by illumination at 380 nm (cis) and 500 nm (trans). Photocurrents are calculated and normalized to current evoked by 300 μM glutamate, as in Fig. 3. Bars are means of 3–4 experiments. Error bars are SEM. Note that MAG0 activates more in trans at sites 482, 484, and 486, with relative potency of trans/cis = 486 > 484 > 482. Note also that all of the MAGs activate more in cis at 439.
Whereas at 2 sites (482 and 486) the short MAG0 activated in trans and the longer MAGs activated in cis, as one might expect for a “reach” mechanism (Fig. 1B), at other sites (e.g., 439) the cis isomer activated well for all 3 of the MAGs (Fig. 5 and Table S1). This illustrated that a more sophisticated structural analysis is required to understand the mechanistic basis for photoactivation. Because the opposite behavior (cis activating vs. trans activating) observed for Mag0 at sites 439 and 486 is interesting from the point of view of engineering opponency into neuronal populations (see below), we decided to investigate further the structural basis for photoswitching by MAG0 using molecular dynamics simulations.
Fig. 5.
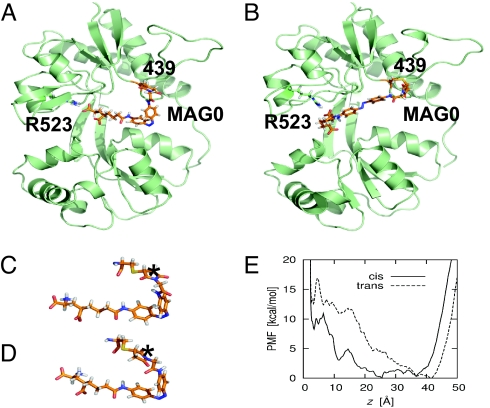
MD simulations of MAG0 docking in cis and trans. (A) MAG0 attached at site 439 and docked into an open iGluR6 cleft while in the cis configuration. (B) MAG0 docked in the trans configuration. (C and D) MAGs may covalently attach to a cysteine residue to yield 2 different R/S stereoisomers. MAG0 shown in C with (R)-configuration, and D with (S)-configuration at the attachment site. The asterisks indicate the asymmetric carbon in the succinimide ring. In A and B, MAG0 is shown with (R)-configuration at the attachment site. (E) Docking PMF for MAG0 with (R)-configuration at the attachment site restrained to either the cis or trans configuration at site 439. The order parameter for the PMF is the distance z between the guanidinium group of iGluR6 R523 and the α-carboxyl group of the MAG0 glutamate moiety. In our docking free energy calculations, the MAG0 glutamate is considered to be docked when z < 6 Å. All other docking PMFs are shown in SI Appendix.
MD Simulations of Ligand Docking and Clamshell Closure.
All-atom MD umbrella sampling simulations with explicit solvent were used to compute the “free energy landscape,” or “potential of mean force” (PMF), for docking a tethered MAG0 molecule into the iGluR6-binding site. The relative free energy between conformational states of MAG0, in this case docked vs. undocked, is a measure of the probability of finding MAG0 in those states (Eq. 1). The iGluR6 LBD was constrained to an open conformation, and the MAG0 azobenzene was constrained to either a cis or trans configuration. PMFs were computed for the 2 isomers at attachment sites 439 and 486. For each of the sites and isomers both the R/S stereoisomers of the bond between the maleimide of MAG and the cysteine were modeled. Consistent with the experimental observations, PMFs for 439C-MAG0 showed that cis MAG0 is more likely to bind than trans MAG0 (Fig. 5 A, B, and E, Fig. S1, and Table 1). However, PMFs for 486C-MAG0 also indicated that MAG0 is more likely to bind in cis (Fig. S1 and Table 1), demonstrating that additional factors must contribute to the state dependence of activation.
Table 1.
MAG0 docking, LBD closure, and channel activation
| Variant | Gdock | 〈Δξ12〉 | Act(Δd) | Gclosure | Gtb |
|---|---|---|---|---|---|
| 439-cis-(R) | 9.4 | 0.9 | 0.04 | −8.3 | 1.1 |
| 439-trans-(R) | 13.7 | 1.4 | 0.008 | −5.6 | 8.1 |
| 439-cis-(S) | 6.6 | 1.6 | 0.004 | −5.0 | 1.6 |
| 439-trans-(S) | 10.1 | 6.3 | ≈0 | 1.1 | 11.2 |
| 486-cis-(R) | 10.1 | 2.4 | 0.0002 | −4.9 | 5.2 |
| 486-trans-(R) | 21.6 | 0.7 | 0.09 | −9.5 | 12.1 |
| 486-cis-(S) | 14.7 | 1.3 | 0.01 | −5.9 | 8.8 |
| 486-trans-(S) | 19.0 | 0.1 | 0.7 | −11.4 | 7.6 |
Gdock is the free energy of MAG0 docking calculated from the docking PMFs, in kcal/mol (Eq. 1 and SI Text). Effectiveness of activation [Act(Δd)] is estimated from the empirical relationship Eq. 2, with Δd being the average distance 〈Δξ12〉 estimated from MD (Materials and Methods and Fig. S3). Gclosure is the free energy of domain closure, estimated from the PMF for GluR2 LBD closure (18). Gtb = Gdock + Gclosure is the free energy of total binding.
An additional factor that needs to be considered to account for receptor activation is the amount of LBD closure (Eq. 2). The closure also increases the apparent agonist binding affinity. Comparing the extents of LBD closure for different conformations of 439C-MAG0 and 486C-MAG0 (Fig. S2) with (i) various GluR2 LBD ligand-complex crystal structures (Table 1 and Fig. S3), and (ii) free energies associated with GluR2 clamshell closure (18) (Table 1), suggested the relative activation between cis and trans forms (Eq. 3 and Table 2). In agreement with the experimental results, the cis form was estimated to activate more strongly for 439C-MAG0 and the trans form to activate more strongly for 486C-MAG0. Thus, one can account for the polarity of photoswitching of LiGluR by MAG by taking into account 2 factors: (i) the isomer dependence of docking of the glutamate end into the open LBD, as illustrated in Fig. 1 A–C and (ii) the ability of the LBD clamshell to close on the docked ligand, given the disposition of the linker (Fig. 1D), which has the effect that greater closure increases both channel activation and ligand-binding free energy.
Table 2.
Relative activation between cis and trans forms
| Variant | Relative ERA |
|---|---|
| 439-(R): cis/trans | 6.5 × 105 |
| 439-(S): cis/trans | ≈All cis |
| 486-(R): cis/trans | 244 |
| 486-(S): cis/trans | 0.002 |
Relative ERA predicted from Eq. 3 using the data from Table 1. In almost all cases shown in Table 1, the unfavorable docking free energy is mitigated by the favorable free energy of domain closure, which becomes more favorable with increasing domain closure. For attachment site 439, cis activates more than trans for both the (R) and (S) stereoisomer. For site 486, cis activates more than trans for (R), but trans activates more than cis for (S). Given equal proportions of (R) and (S) at site 486, trans would activate more than cis overall.
Yin/Yang Remote Control of Neuronal Activity with a Single Photoswitch.
Having found that a single MAG, MAG0, can activate from some sites in cis and other sites in trans, we explored the possibility of driving 2 subsets of neurons to fire in opposing manners within the same preparation with this 1 PTL. We chose the variants with the most dramatic preference for 1 state of MAG0, i.e., 439C for cis and 486C for trans (Figs. 3–5). Hippocampal neurons were transiently transfected with iGluR6(439C) and then later with iGluR6(486C). Because of the low (≈1–2%) efficiency of transfection, neurons that were successfully transfected expressed one or the other but not both of these cysteine versions of iGluR6 (Fig. 6A). We labeled the neurons with MAG0 and then carried out both optical and elelctrophysiological recordings of the transfected cells, which were identified by a coexpressed fluorescent protein. Calcium imaging showed that, as expected, illumination at 380 nm increased calcium in neurons expressing iGluR6(439C) and decreased it in neurons expressing iGluR6(486C), whereas illumination at 500 nm had the opposite effect (Fig. 6B). Untransfected neurons did not respond to the illumination. Whole-cell patch recordings under current clamp mode showed that short pulses of light at 375 nm elicited action potentials precisely and reproducibly in neurons expressing iGluR6(439C), whereas pulses of light at 488 nm elicited action potentials in neurons expressing iGluR6(486C) (Fig. 6C). Thus, our photoswitch MAG0 could provide complementary, opposing control of neural activity in subsets of neurons expressing either iGluR6(439C) or iGluR6(486C).
Fig. 6.
Opposite polarity photoswitching of neuronal activity with same MAG at 2 different attachment sites. (A) Neurons in the same culture expressing either iGluR6(486C) or iGluR6(439C) are loaded with Fluo4-AM for calcium-imaging and labeled with MAG0. (B) Illumination with 380-nm light (purple bars) causes an increase in calcium (and consequently Fluo4-AM emission intensity) in cells expressing iGluR6(439C)-MAG0 and a decrease in cells expressing iGluR6(486C)-MAG0. When illuminated with 500-nm light (green bars), the opposite occurs. The scale bar for Fluo4-AM emission intensity is ΔF/F. (C) Action potentials can be precisely stimulated with brief pulses (2–7 ms) of moderately intense light (≈6 mW/mm2). Neurons expressing iGluR6(439C) and labeled with MAG0 are activated by 375-nm light (purple arrows), whereas neurons expressing iGluR6(486C) and labeled with MAG0 are activated by 488-nm light (green arrows).
Discussion
We experimentally tested 3 MAGs of different lengths at a series of attachment sites on the iGluR6 LBD. These sites were situated on the “lips” of the clamshell in a ring around the glutamate-binding pocket. From this analysis, we identified several sites with large gating responses to light. The efficacy and cis/trans dependence of the optical gating differed between attachment sites and the MAG variants. MD simulations revealed the mechanism of photoswitching by MAG, showing that channel activation depends on 2 factors: (ii) the fraction of rotamers of a cysteine-anchored MAG that enable the glutamate to orient in the binding site of the open LBD and (ii) the ability of the clamshell to close, given the location of the linker. Greater clamshell closure was shown to contribute in 2 ways: increasing the opening of the gate (19) and increasing the affinity for the glutamate (18). It should be noted that despite the fact that the maleimide linkage can result in unequal probabilities of R/S stereoisomers and that labeling may not reach saturation for all of the subunits, clear on/off effects are found in living cells. Thus, it seems that a kind of “binary filtering” may be going on by the allosteric channel.
A new short version of MAG0 emerged as the best photoswitch in 2 regards. First, MAG0 has the biggest change in effective concentration seen to date: ≈120-fold higher in cis than in trans, based on competition with the antagonist DNQX. Second, MAG0 has the unique property that it activates iGluR6 at some attachment sites in cis but at other sites in trans. When the best cis activator [iGluR6(439C)] and best trans activator [iGluR6(486C)] are expressed in different subsets of neurons within the same preparation, labeling with MAG0 endows the 2 sets of cells with opposite sensitivity to light. Neurons expressing iGluR6(439C) fire in response to pulses of 380-nm light and those expressing iGluR6(486C) fire in response to pulses of 500-nm light. The ability to use 1 PTL simultaneously to excite 1 subpopulation of neurons and deexcite another could pave the way for engineering opponency into distinct subgroups of neurons, such as, for example, ON and OFF retinal ganglion cells, which have overlapping receptive fields but opposite responses to light.
Materials and Methods
Synthesis of MAG0.
MAG synthesis and chemical analysis is described in SI Appendix.
Site-Directed Mutagenesis.
Cysteine point mutations were substituted outside of the glutamate-binding site of the iGluR6 LBD. The iGluR6 cDNA containing R621Q mutations (for high Ca2+ permeability) was obtained from K. M. Partin (Colorado State University, Fort Collins, CO). Cysteine point mutations were introduced into this clone by using the QuikChange site-directed mutagenesis kit (Stratagene). The forward and reverse oligonucleotide primer sequences of the cysteine point mutations are shown in Table S2. The following PCR profile was used: 1 cycle (95 °C for 30 s); 20 cycles (95 °C for 30 s, 55 °C for 1 min, 68 °C for 12 min); 1 cycle (68 °C for 12 min).
Cell Culture and Transfection.
HEK293 cells were maintained in DMEM with 5% FBS on polyl-lysine-coated glass coverslips at ≈3 × 106 cells per milliliter and transiently cotransfected with various iGluR6 plasmids and EYFP at a ratio of 9:1 by using Lipofectamine 2000 (Invitrogen). Calcium imaging or patch clamping was performed 12–48 h after transfection.
Dissociated postnatal rat hippocampal neurons (P0-P5) were prepared and transfected as described previously (13). Serial transfections of iGluR6(486C) and iGluR6(439C) were performed identically but separated by 24 h.
Ca2+ Imaging and Patch Clamp Recordings.
Ca2+ imaging and patch clamp recordings were as described in SI Appendix.
MD Simulations and Analysis.
The atomic model for the iGluR6 LBD was constructed from the X-ray crystal structure PDB ID 1S50 (20). All simulations were performed by using the program CHARMM (21) with the all-atom potential energy function PARAM27 for proteins (22) and the TIP3P potential energy function for water (23). The software package Antechamber (24) was used to obtain GAFF (generalized amber force field) parameters (25) for MAG0 adapted for use in CHARMM. Additional details are described in SI Appendix.
The total effective receptor activation (ERA) is predicted from the probability of finding the tethered ligand in the binding site when the clamshell is open (“ligand docking”), times the probability of the LBD to adopt a closed conformation in the presence of a bound ligand (“domain closure”), times the effectiveness of receptor activation once the LBD has reached its maximum closure. The free energy of ligand docking onto an open clamshell, Gdock, is obtained as follows:
 |
where kB is Boltzmann's constant, T is temperature, and docking is considered to occur at z < 6 Å, where z is the distance between the guanidinium group of iGluR6 R523 and the α-carboxyl group of the MAG0 glutamate moiety. The PMF(z) of the tethered ligand is calculated with the LBD constrained to remain open. The free energy associated with the closure of the clamshell onto a docked ligand, Gclosure, is taken from the PMF for GluR2 LBD closure previously computed with all-atom MD simulations (18). The total binding free energy is Gtb = Gdock + Gclosure. The effectiveness of receptor activation depends also on the extent of LBD closure. This is predicted by using an empirical relationship derived from the change in the clamshell distance Δd taken from available crystal structures relative to the glutamate-bound crystal structure and the observed peak current for GluR2:
where Δd must be >0 (Fig. S3). Domain closure is measured in terms of the 2-dimensional order parameter (ξ1, ξ2), where ξ1 is the distance between the center-of-mass of residues 517–519 (Lobe 1) and 689–690 (Lobe 2), and ξ2 is the distance between residues 439–441 (Lobe 1) and 721–722 (Lobe 2). Δξ12 is the difference in cleft closure for the closed LBD relative to the crystal structure of GluR6 bound to glutamate, where ξ12 = (ξ1+ξ2)/2. 〈Δξ12〉 was calculated from the final 200 ps of a 1-ns MD trajectory. In our analysis, 〈Δξ12〉 is substituted for Δd above.
It follows that, for a given construct, the total effective receptor activation (ERA) is predicted from a combination of Act (Δd) times the probability of finding the tethered ligand in the binding site times the probability of finding the clamshell open. The ERA for 2 constructs (cis and trans) are compared as follows:
 |
where Gtb = Gdock + Gclosure.
Supplementary Material
Acknowledgments.
We thank K. M. Partin for the iGluR6 cDNA, T. Machen for guidance on calcium imaging, and Harald Janovjak for helpful discussion. This work was supported by Human Frontiers Science Program Grant RGP23-2005 and National Institutes of Health (NIH) Nanomedicine Development Center for the Optical Control of Biological Function Grant PN2 EY018241 as well as postdoctoral fellowships from the Japan Society for the Promotion of Science and the Institute of Tokyo Vascular Disease (to R.N.) and the Generalitat de Catalunya (Nanotechnology Program), Ministerio de Educación y Ciencia (Spain) and the Human Frontiers Science Program (to P.G.). D.T. thanks Novartis and Roche Biosciences for support. A.Y.L and B.R were supported by NIH Grant GM-62342.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811899106/DCSupplemental.
References
- 1.Zhang F, Aravanis AM, Adamantidis A, de Lecea L, Deisseroth K. Circuit-breakers: Optical technologies for probing neural signals and systems. Nat Rev Neurosci. 2007;8:577–581. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- 2.Volgraf M, et al. Reversibly caged glutamate: A photochromic agonist of ionotropic glutamate receptors. J Am Chem Soc. 2007;129:260–261. doi: 10.1021/ja067269o. [DOI] [PubMed] [Google Scholar]
- 3.Miesenböck G, Kevrekidis IG. Optical imaging and control of genetically designated neurons in functioning circuits. Annu Rev Neurosci. 2005;28:533–563. doi: 10.1146/annurev.neuro.28.051804.101610. [DOI] [PubMed] [Google Scholar]
- 4.Gorostiza P, Isacoff EY. Optical switches and triggers for the manipulation of ion channels and pores. Mol Biosyst. 2007;3:686–704. doi: 10.1039/b710287a. [DOI] [PubMed] [Google Scholar]
- 5.Gorostiza P, Isacoff EY. Nanoengineering ion channels for optical control. Physiology. 2008;23:238–247. doi: 10.1152/physiol.00018.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorostiza P, Isacoff EY. Optical switches for remote and noninvasive control of cell signaling. Science. 2008;322:395–399. doi: 10.1126/science.1166022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartels E, Wassermann NH, Erlanger BF. Photochromic activators of the acetylcholine receptor. Proc Natl Acad Sci USA. 1971;68:1820–1823. doi: 10.1073/pnas.68.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lester HA, Krouse ME, Nass MM, Wassermann NH, Erlanger BF. A covalently bound photoisomerizable agonist: Comparison with reversibly bound agonists at electrophorus electroplaques. J Gen Physiol. 1980a;75:207–232. doi: 10.1085/jgp.75.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lester HA, et al. Electrophysiological experiments with photoisomerizable cholinergic compounds: Review and progress report. Ann NY Acad Sci. 1980;346:475–490. doi: 10.1111/j.1749-6632.1980.tb22118.x. [DOI] [PubMed] [Google Scholar]
- 10.Chabala LD, Lester HA. Activation of acetylcholine receptor channels by covalently bound agonists in cultured rat myoballs. J Physiol. 1986;379:83–108. doi: 10.1113/jphysiol.1986.sp016242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volgraf M, et al. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Bio. 2006;2:47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorostiza P, et al. Mechanisms of photoswitch conjugation and light activation of an ionotropic glutamate receptor. Proc Natl Acad Sci USA. 2007;104:10865–10870. doi: 10.1073/pnas.0701274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szobota S, et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–545. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Banghart M, Borges K, Isacoff EY, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers JJ, Banghart MR, Trauner D, Kramer RH. Light-induced depolarization of neurons using a modified Shaker K(+) channel and a molecular photoswitch. J Neurophysiol. 2006;96:2792–2796. doi: 10.1152/jn.00318.2006. [DOI] [PubMed] [Google Scholar]
- 16.Doyle DA, et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 17.Blaustein RO, Cole PA, Williams C, Miller C. Tethered blockers as molecular ‘tape measures’ for a voltage-gated K+ channel. Nat Struct Biol. 2000;7:309–311. doi: 10.1038/74076. [DOI] [PubMed] [Google Scholar]
- 18.Lau AY, Roux B. The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure (London) 2007;15:1203–1214. doi: 10.1016/j.str.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 20.Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: Molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 21.Brooks BR, et al. Comparison of simple potential functions for simulating liquid water. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 22.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Chem Physiol. 1983;79:926–935. [Google Scholar]
- 24.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graphics Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general AMBER force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 26.Kumar GS, Neckers DC. Photochemistry of azobenzene-containing polymers. Chem Rev. 1989;89:1915–1925. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.