Abstract
There is considerable interest in uncovering the pathway of amyloid formation because the toxic properties of amyloid likely stems from prefibril intermediates and not the fully formed fibrils. Using a recently invented method of collecting 2-dimensional infrared spectra and site-specific isotope labeling, we have measured the development of secondary structures for 6 residues during the aggregation process of the 37-residue polypeptide associated with type 2 diabetes, the human islet amyloid polypeptide (hIAPP). By monitoring the kinetics at 6 different labeled sites, we find that the peptides initially develop well-ordered structure in the region of the chain that is close to the ordered loop of the fibrils, followed by formation of the 2 parallel β-sheets with the N-terminal β-sheet likely forming before the C-terminal sheet. This experimental approach provides a detailed view of the aggregation pathway of hIAPP fibril formation as well as a general methodology for studying other amyloid forming proteins without the use of structure-perturbing labels.
Keywords: aggregation, amylin, fibers, human islet amyloid polypeptide, nucleation
More than 20 different diseases are associated with proteins that form insoluble amyloid fibers (1). In large quantities, organ function is disrupted by the formation of amyloid deposits, but for several amyloid diseases, there is evidence that the toxic entities are actually prefibril intermediates (2, 3). Although they have been the focus of numerous studies, details about these critical intermediates have been elusive, mostly because it is extraordinarily difficult to obtain structural and kinetic information for amyloid aggregation. The difficulty arises because high-resolution techniques do not have the time resolution required to track the structural changes, nor can they be easily applied to aggregating systems. Optical techniques that do have sufficient time resolution, such as circular dichroism spectroscopy, provide only a low-resolution view of structure. Other techniques, like electron spin resonance and fluorescence spectroscopy, require bulky labels that can perturb the structure and dynamics. Mechanistic information is vital to understand the mechanism of protein misfolding as well as to design inhibitors that subvert the pathway of amyloid formation. What is needed is a technique with sufficient time resolution to observe intermediates, provide residue-level structural information, is nonperturbing, and, ideally, can be used to test molecular dynamics simulations.
A technique that satisfies these criteria is 2D infrared (2D IR) spectroscopy when used with site-specific isotope labeling (4). We have recently demonstrated a technological approach for collecting 2D IR spectra that is particularly well-suited for studying amyloid formation (5). Our method uses a mid-IR pulse shaper to automate data collection, much like an NMR spectrometer, so that spectra can be collected quickly enough to monitor fibril kinetics on the fly. In this article, we combine this automated version of 2D IR spectroscopy with site-specific 13C18O-labeled peptides. Isotope labeling permits the kinetics of individual residues to be measured so that hIAPP fibril formation can be followed on a residue-by-residue basis. The resulting kinetics and secondary structure information provides a detailed pathway for the formation of the fibril backbone.
The deposition of amyloid fibrils in the islets of Langerhans of the pancreas is a common pathological feature of type 2 diabetes, and hIAPP is the major protein component of these amyloid deposits (6–9). The polypeptide hormone is stored with insulin in the secretory granules of the pancreatic β-islet cell and is secreted to the extracellular space in response to the same factors that cause the release of insulin. Synthetic amyloid aggregates are toxic to insulin-producing β-cells, supporting the argument that hIAPP fibril formation could play a role in the loss of β cell mass in type 2 diabetes. The extent of amyloid deposition appears to correlate with the severity of the disease, further highlighting a relationship between amyloid deposition in the pancreas and the progression of the latter stages of type 2 diabetes (8, 9).
To monitor the structures and kinetics of individual residues in hIAPP, we use 13C18O labeling of the backbone carbonyl groups to resolve single amide I stretches (10). Isotope labeling and IR spectroscopy have been used previously to study amyloids, which have provided insight into general aggregation mechanisms (11, 12). Most studies rely on small peptide fragments, which do not necessarily translate into the context of the full protein. For example, studies using small fragments of hIAPP led to the proposal that residues 20–29 formed the core of the cross-β-structure, but recent structural studies of full-length IAPP reveal that this is not the case (13). Furthermore, the kinetics of amyloid formation by short fragments of IAPP differs dramatically from that observed for the full-length hormone. For these reasons, we have chosen to isotope label and study the full 37-residue peptide.
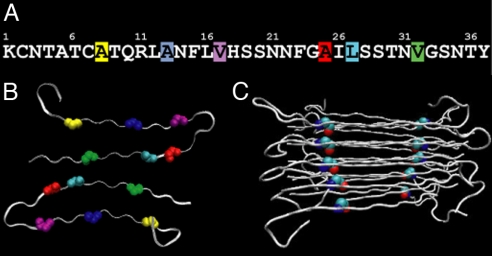
Six separate samples of hIAPP were prepared, each containing a single 13C18O-labeled residue. The positions of the 6 labeled residues are shown in Fig. 1 superimposed on the recent model of the hIAPP protofibril deduced from solid-state NMR (ssNMR) (13). hIAPP forms a cross-β-strand structure that is composed of 2 twisted columns of peptides. According to the ssNMR studies, each column is created from a stack of peptides that are folded into a structure containing 2 β-strands with an ordered loop between residues 18 and 27. Hydrogen bonding occurs along the stacks so that there are 4 β-sheets running in parallel along the lengths of the fibrils. The labeled residues are spaced along the peptide (Fig. 1) at positions chosen to probe key regions of the fibril. Because the peptides are stacked, each isotope label forms 2 columns, illustrated in Fig. 1C for Ala-25. The kinetics we report below reflects the arrangement of the isotopes into their respective columns.
Fig. 1.
Structure of hIAPP fibrils according to solid-state NMR. (A) Sequence of hIAPP with the 6 isotope-labeled residues highlighted: Ala-8 (yellow), Ala-13 (blue), Val-17 (purple), Ala-25 (red), Leu-27 (cyan), and Val-32 (green). (B) Cross-section through a fibril, illustrating that the fibrils are composed of 2 columns of peptides to create 4 β-sheets (that run in and out of the page). The labeled residues are highlighted. (C) Side view of the protofibril, illustrating how a peptide with a single labeled residue creates 2 columns of isotope labels that run along the β-sheets (shown here for Ala-25).
Results
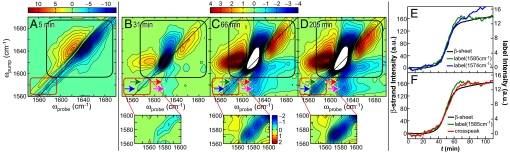
Shown in Fig. 2 are 4 2D IR spectra collected at successively later times during amyloid formation, starting from an initially unaggregated sample of hIAPP labeled at Ala-25. The 4 spectra are generated from many thousands of spectra that were collected [see supporting information (SI) Text]. The most prominent features in these spectra are between 1,617 and 1,670 cm−1 and are due to the unlabeled residues (inside the black squares in Fig. 2). The unlabeled peaks give information on the overall assembly kinetics of the amyloid, which were previously reported (5). At t = 5 min, the unlabeled features consists of 2 out-of-phase peaks at ωpump = 1,645 cm−1. In 2D IR spectra, vibrational modes create doublets in which the negative peak is located on the diagonal and can be interpreted much like a traditional infrared absorption peak, albeit with improved frequency resolution. The 2 broad peaks at t = 5 min are typical of random coil peptide structures with large structural distributions. The isotope-labeled features appear near 1,580 cm−1 (inside the red squares in Fig. 2), which provides information specific to the folding kinetics of Ala-25. At t = 5 min., the isotope-labeled absorption is very broad and weak, indicating that Ala-25 is conformationally disordered, which is consistent with the nature of the random coil state.
Fig. 2.
Representative 2D IR spectra and kinetics curves of hIAPP labeled at Ala-25. (A) The first 2D IR spectrum is at t = 5 min. (B–D) Difference 2D IR spectra at t = 31, 66, and 205 min, calculated by subtracting the t = 5-min spectrum. Black boxes surround the spectral features of the unlabeled portion of the peptide, whereas red boxes enclose the diagonal peaks of the isotope labeled Ala-25. Blue and green arrows highlight the 2 labeled features, whereas magenta and red arrows point to the cross-peak between the 13C18O Ala-25 and the unlabeled β-sheet. (E) Kinetics of the diagonal peaks of the unlabeled β-sheet at 1,617 cm−1 and the 2 label features (blue and green arrows). (F) Kinetics of the cross-peak are compared with the diagonal peaks. The left y axis is for the β-sheet, and the right y axis is for the other features.
To highlight the changes that occur in the 2D IR spectra during the aggregation processes, the remaining 2D IR spectra in Fig. 2 are plotted as difference spectra, calculated by subtracting the 2D IR spectrum in Fig. 2A from the others. As time progresses, the random coil doublet disappears, whereas a doublet grows in at ωpump = 1,617 cm−1. The 1,617-cm−1 features are the signature of β-strand amyloid growth. Concurrent with the growth of the β-strand, 2 isotope-labeled features appear at 1,574 and 1,585 cm−1. The lower-frequency peak eventually becomes more intense (Fig. 2 D and E). There do not appear to be cross-peaks between the 2 features, indicating that the coupling is either weak or nonexistent between the 2 modes (see Insets). Large cross-peaks grow in between the isotope labels and the unlabeled β-strand peak at 1,617 cm−1 (see arrows), indicating that Ala-25 is strongly coupled to the β-sheets. Thus, the cross-peaks measure the kinetics of Ala-25 folding into the β-sheet configuration of the amyloid fibril. Fig. 2F plots the intensity of the peaks for the unlabeled β-strand, Ala-25, and the cross-peak as a function of time. All 3 kinetic curves are sigmoidal, as is typical of amyloid kinetics, and all 3 have a time to half-maximum (t50) that is virtually identical, indicating that when Ala-25 becomes part of the ordered fibril, it assembles directly into a β-strand structure.
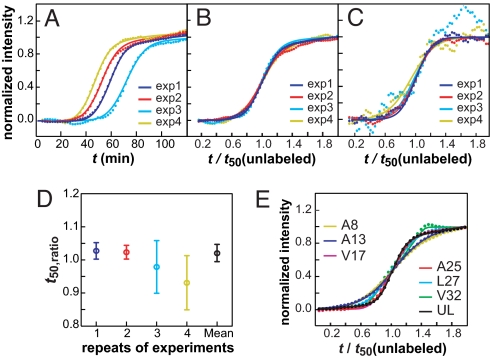
The kinetics of assembly were monitored by using 5 other labeled sites. The spectra are similar to those obtained for the Ala-25-labeled sample (Figs. S1 and S2), but the time at which the different residues are incorporated into β-sheets differs dramatically. Before presenting these results, we first describe our analysis procedure and the reproducibility of our experiments, because these are particularly important issues with regard to amyloid formation in this detailed studies of amyloid kinetics using 2D IR spectroscopy. It is well known that amyloid kinetics are not perfectly reproducible from one experiment to the next because they are extremely sensitive to conditions that are difficult to control, such as how rapidly the solution is mixed (14). Procedures for comparing separate datasets have been established by using fluorescence and CD spectroscopy (15). Our procedure is similar to these well-established methods but accounts for issues specific to infrared spectroscopy. First, we established that the unlabeled β-sheet feature near 1,617 cm−1 can be analyzed by using the same protocols as for kinetic measurements using ThT fluorescence. This task was accomplished by running fluorescence and FTIR kinetics experiments in parallel on hIAPP labeled at either Ala-13 or Leu-27. We found that the β-sheet feature in the FTIR spectrum matches the fluorescence trace nearly perfectly (see Fig. S3). This observation also confirms our expectation that isotope labeling does not affect the kinetics of the unlabeled β-sheet feature because it does not alter the peptide structure. Next, we investigated the reproducibility of our kinetics experiments. Shown in Fig. 3A are the kinetic traces of the unlabeled β-strand feature from 4 separate 2D IR experiments on the same sample of hIAPP labeled at Ala-25. The t50 times for these 4 experiments fall between 30 and 60 min. This range of times prohibits the averaging of successive 2D IR datasets, which is why our rapid-scan version of 2D IR spectroscopy is necessary to accomplish these experiments. Fortunately, it has been established from fluorescence studies that kinetic scans on wide-ranging time scales can be compared by scaling the time axes with their respective t50. The scaled kinetics match as long as the mechanism is unchanged (15). Following this procedure, we first established the value of t50 for each scan by fitting the kinetic traces associated with the spectral features due to the unlabeled 12C16O oscillators to a sigmoidal function (fits shown in Fig. 3A; function given by Eq. s1 in SI Text), which we then used to scale the time axes. On this scaled axis, the sigmoidal curves are quite similar (Fig. 3B). The similarity of the slopes indicate that all 4 experiments are monitoring the same aggregation mechanism, as is expected from identical samples, and confirms that the scaling procedure is valid for infrared spectroscopy. Having determined the scaling parameters from the unlabeled β-strand features, we then scale the time course of the spectral features associated with the labeled site by using the same parameters. For example, Fig. 3C shows the time course of the Ala-25 signal for the same 4 experiments after scaling with the t50 times used above, along with their sigmoidal fits. The good agreement confirms that the features from the unlabeled sites provide an internal standard, which allows us to objectively and reliably determine the appropriate scaling parameter. The signal-to-noise ratio of the label kinetics is lower than the unlabeled β-strand features because the label intensities, as expected, are ≈13 times weaker. To account for signal-to-noise ratio, we calculate the error in the ratio of the t50 times between the label and the β-strand features (Eqs. s2 to s4 in SI Text). The resulting ratios of t50 and their error bars are plotted in Fig. 3D. All 4 measurements give a similar ratio, with much smaller error bars associated with the less-noisy datasets, as expected. Because the error bars are related to the laser fluctuations, we combine them into a single data point by using a weighted average (Eqs. s5 and s6 in SI Text), which gives t50(label)/t50(unlabeled) ≡ t50,ratio = 1.02 ± 0.03 for Ala-25. Finally, we compare the scaled kinetics of the unlabeled β-sheet feature from all 7 samples (6 labeled peptides and 1 unlabeled peptide), in Fig. 3E. The slopes show slightly more variation (mostly because of Ala-8 and Ala-13) than for multiple runs of the same sample (Fig. 3B), which may be caused by small differences in HPLC purification of the peptides but is not large enough to indicate a significantly different aggregation mechanism (15, 16). The fiber morphologies are similar as measured by TEM (Fig. S4), which provide another check on the aggregation mechanism, because different mechanisms often give rise to different fibril structures (17). Additional experimental data and details on our fitting procedure are given in SI Text. The reproducibility of these control experiments leads us to conclude that the kinetics from individual 2D IR measurements can be compared by following the standard analysis methods of fluorescence experiments by using the unlabeled β-sheet feature as a reference. The β-sheet feature provides an internal standard, which allows one to deconvolve the normal run-to-run variations from the more interesting differences associated with the kinetics of assembly. The presence of such a built-in internal standard is an added bonus of the isotope-edited 2D IR technique.
Fig. 3.
Summary of experiments using Ala-25 labeled peptides to test reproducibility and data analysis methods. (A) Kinetics of unlabeled β-strand features at 1,617 cm−1 for 4 independent experiments (dots), along with sigmoidal fits (solid line) by using Eq. s1. (B) Curves from A scaled by their respective t50 times. (C) Kinetics of Ala-25 feature at 1,585 cm−1, using the same t50 times as in B along with sigmoidal fits. (D) Scaled t50 times and standard errors extracted from fits of β-sheet and isotope kinetics shown in B and C. Colors are assigned in the same way for all 4 experiments. (E) Scaled kinetics of unlabeled β-sheet features from all 6 labeled and the unlabeled peptides.
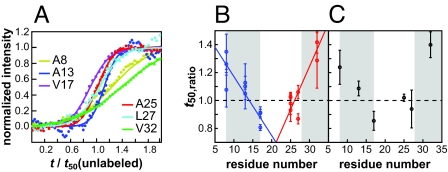
Having established the validity of our 2D IR approach, we now turn to comparing the relative times that the 6 labeled residues assemble into the fibril, which is the primary motivation for this study. Shown in Fig. 4A are scaled kinetic curves for each of the 6 isotope-labeled peaks. Simple visual inspection of these data reveals that these 6 residues exhibit strikingly different transition times; some residues transition sooner than the unlabeled β-strand feature (t50,ratio < 1), whereas others are later (t50,ratio > 1). The data shown in Fig. 4A are presented solely to visualize the differences in transition times. Like Ala-25, each labeled peptide was measured 2–4 times. The t50,ratio for the individual measurements are shown in Fig. 4B and the combined data point for each residue is plotted in Fig. 4C. The error bars in Fig. 4B become smaller when the data are averaged more before fitting, but averaging does not alter the mean values in Fig. 4C significantly (see Tables S1–S4 and Fig. S5).
Fig. 4.
Summary of the kinetics for all 6 labeled peptides. (A) Comparison of sample kinetic curves for the isotope-labeled peaks by using the same scaling factors as in Fig. 3E. (B) t50,ratio measured from the individual experiments. (C) Averages of the individual data points in B with weights inversely proportional to the square of individual errors. Error bars were determined with standard error described in Eqs. s4 and s7 in SI Text.
Of those residues that were measured, Val-17 is the first residue to transition and has a t50,ratio that is 10% sooner than the t50,ratio of the unlabeled residues (in all experiments performed, the Val-17 isotope label rises before the unlabeled β-sheet features). Val-32 is the last to transition with a t50,ratio that is 30% slower. On an absolute time scale, this 40% difference translates into a 20-min separation between the folding of Val-17 and Val-32 for a typical t50 folding time of 50 min. Leu-27 may also transition as early as V17, but it has the most uncertain t50,ratio of those residues measured.
The above experiments report on local structure through the changes in intensity and frequency of the isotope-labeled amide I band. There are 2 factors that affect the frequency and intensity of the bands in the 2D IR spectra: their environment and their coupling. To differentiate between the 2, we collected 2D IR spectra of Ala-13, Ala-25, and Leu-27 where only 25% of the peptides contain an isotope label (Fig. S6). Diluting the isotopes largely eliminates the effects of coupling between labeled residues. Coupling to the unlabeled residues is still present, but its influence on the label frequency is small because the frequency separation is large. We found that upon isotope dilution, the frequencies of these 3 residues lie within 5 cm−1 of each other (1,589–1,594 cm−1), indicating that their local electrostatic environments are comparable. Moreover, the isotope frequencies are higher when diluted, indicating that the coupling between isotope labels causes a negative frequency shift, which is consistent with the negative coupling constants predicted for β-sheets in amyloid fibril (12). These results emphasize that the isotope-labeled features that are the focus of this study arise from delocalized vibrational modes that involve the cooperative motions of several labeled amide I groups (18). Thus, according to these dilution experiments and the cross-peaks in the 2D IR spectra, we conclude that we are measuring β-sheet formation through the association of multiple peptides into columns of isotopically labeled residues. See also SI Text and Figs. S7–S9 for additional experimental data.
Discussion
Amyloid formation is generally described as nucleation dependent process, although it is often observed that the concentration dependence of the kinetics is weaker than observed for classic nucleation dependent polymerization (15). If hIAPP fibers are created by the concerted formation of a nucleus in which the entire polypeptide adopts the structure found in the fibril, followed by the addition of monomers that template onto the ends of the fibers extremely quickly, we would expect to observe the same t50,ratio for all 6 labeled residues. Furthermore, all 6 labeled residues would have a t50,ratio = 1, because their apparent rates of assembly would be indistinguishable from the unlabeled residues. Thus, this model of amyloid formation predicts that the data in Fig. 4B should all fall on a line at t50,ratio = 1. However, a t test indicates that the t50,ratio for many of the residues are not equal and a goodness-of-fit test leads us to conclude that Ala-25 is the only residue that is likely to have t50,ratio = 1 (see Statistical Analysis in SI Text). Therefore, our results indicate that hIAPP clearly has a diverse and interesting free-energy landscape that is more complicated than simply an initial random coil ensemble that undergoes a concerted transition to form a perfect fiber-like nucleus that then elongates by the addition of monomers that quickly assume their final fold. Instead, the landscape must contain multiple barriers to cause slow folding of the monomers after their addition to the fiber ends. In principle, the shape and width of the unlabeled β-sheet transition should reflect the entire range of kinetics because all of the residues contribute to it. In fact, our previous 2D IR study of the unlabeled β-sheet feature hinted at complex kinetics (5). However, because 37 amino acids contribute to the unlabeled feature, many of which are strongly coupled, the kinetics of assembly for individual residues will be obscured by the others. The experiments reported here demonstrate that when individual residues are monitored by using isotope labeling, details of the folding and assembly process are resolved that are otherwise masked by overlapping spectral features.
Because the earliest residues to transition are in the middle of the peptide (Val-17, Ala-25, and Leu-27) and the last ones on the ends, one possible explanation for our data is that nucleation occurs near or at the loop, followed by folding down the N- and C-terminal β-sheets. Thus, it may be possible to describe the data in Fig. 4B by 2 lines, one that describes the rate of formation for the N-terminal β-strand and the other for the C-terminal β-strand. To test this hypothesis, we fit the data for residues 8, 13, and 17 to one line and 25, 27, and 32 to another. This fit cannot be rejected according to a goodness-of-fit test. Furthermore, according to an F test, it is between 22 and 167 times more likely that the data fits to this 2-line model rather than a simple nucleation model in which all of the residues have t50,ratio = 1 (see Statistical Analysis in SI Text).
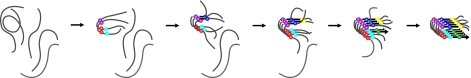
Shown in Fig. 5 is a schematic representation of this mechanism. In the first step, a series of peptides aggregate to create a nucleus in which the loop is formed, but the β-sheets are not. This nucleus serves as a template for elongation of the fiber by the addition of monomers to the loop at the ends of the fiber. Concomitant to elongation, the β-sheets form, with the N-terminal β-sheet folding in approximately half the time that it takes the C-terminal sheet to fold, according to the 2-line fit to the data. At this time, we cannot say exactly which residues are involved in the initial assembly step. Certainly Val-17 is involved because it is the first to aggregate, and Ala-25 and Leu-27 may be as well, because they have t50,ratios closest to Val-17. Because we do not yet have a spectroscopic signature for when the ordering of the loop occurs, we cannot say for certain that it is the loop itself that is the critical substructure. We suspect that the 2 columns of peptides grow at different rates, which would explain why formation of the C-terminal sheet, which resides at the interface of the 2 columns (see Fig. 1B), is slower than the outside sheets formed by the N-terminal region of the chain, so that the region containing Val-32 does not fully form a β-sheet until it is stabilized by the second column of peptides. Because the cross-peaks appear at the same rate as the isotope-labeled diagonal peaks, we believe that when the β-sheets form, they are in-register. If unaligned strands were to occur, followed by an ordering of the hydrogen bonds, as has been seen in amyloid fragment studies (11), then we would expect the cross-peaks to grow in before the isotope labeled peaks, because the isotope labels would be coupled to β-sheet normal modes even if they are not coupled among themselves. In principle, more sophisticated isotope labeling schemes could be used to test these hypotheses. Nonetheless, the results are both consistent with the nucleation mechanism obtained from standard biophysical approaches (15) although they provide residue-specific details that cannot be obtained otherwise.
Fig. 5.
One hIAPP aggregation pathway that is consistent with our data. Labeled residues in register at each assembly stage are denoted with colored circles (purple, Val-17; red, Ala-25; cyan, Leu-27; blue, Ala-13; yellow, Ala-8; green, Val-32). Thick black arrows indicate the β-strand, whereas the portion of the peptide in random coil or ordered loop is shown in gray.
The folding mechanism is somewhat reminiscent of the zipper mechanism for β-hairpin folding. In the zipper mechanism, the β-turn forms first, followed by elongation of the hairpin toward the N and C termini by rapidly forming a series of hydrogen bonds (19, 20). However, we emphasize that the hydrogen bonding in amyloid β-hairpins is not the same; instead of hydrogen bonds linking the 2 halves of each peptide individually, hydrogen bonds in amyloids run along the fiber long axis to connect stacked hairpins. Thus, fundamentally different forces are likely to be responsible for loop formation in amyloid. Another very important point is that our data rule out the presence of large β-sheet structures during the lag phase. We estimate that not more than 5% of the peptide ensemble can reside in β-sheets with >4 strands, based on the background intensity at 1,617 cm−1, where large β-sheets absorb. β-Sheet aggregates of 3 strands or less could be present during the lag phase, because small β-sheets absorb near the random coil at 1,645 cm−1 (21).
Of the 6 residues measured, Ala-25 is particularly interesting because it is the only substitution that exhibits 2 isotope-labeled bands (see Fig. 2 and Figs. S1 and S2). Isotope dilution of Ala-25 causes both peaks to collapse into a single feature (Fig. S6). This finding indicates that Ala-25 contains 2 coupling constants of 3.5 and 15.5 cm−1. Two coupling constants indicate that there is a bimodal distribution of distances between adjacent Ala-25 residues. A bimodal distribution of distances could arise from 2 different populations of fibril structures or from the helical twist that breaks the symmetry between the 2 columns of peptides. The last possibility seems unlikely, because the twist is small over the delocalization length of a vibrational exciton (4–6 strands), and thus should be negligible. It seems more likely that there are 2 subpopulations of β-sheet structures and that the relative ratio of the 1,574- and 1,585-cm−1 features gives their relative population, which is consistent with the lack of a cross-peak between the 2 features. If true, then the fact that the 2 features have similar intensities up to 1.5 t50 indicates that the 2 subpopulations aggregate in tandem but that the subpopulation with the lower frequency isotope label (and, thus, smaller Ala-25 spacing) is eventually preferred (Fig. 2E). The argument for a common first phase of aggregation also agrees with fits to the data that predict nearly identical t50 times but a much larger contribution to a slower phase from the 1,574-cm−1 feature (also see Long-time kinetics in SI Text). Considering that Ala-25 is the only residue among the 6 that were labeled that exhibits 2 peaks and the only residue we measured that was assigned to random coil in the ssNMR structure, it appears that the ordered loop is the most sensitive region of the structure to this bimodal distribution.
At this time, we do not know why the initial ordering occurs at or near the loop. However, some insight may be gained by considering our results in the context of the well-known correlation between variations in the primary sequence of IAPP and the prevalence of islet amyloid in different species (22). For example, rodents do not form islet amyloid even though they produce IAPP. The sequence of rat IAPP differs from that of hIAPP at 6 positions; residues 18, 23, 25, 26, 28, and 29. Four of these residues flank the ordered loop and are located close to the amino acids that we postulate are the nucleation sites for β-sheet formation. It is also worth noting that modification of residues 24 and 26 by N-methylation leads to a nonamyloidogenic variant of human IAPP, as does a single mutation from Ile-26 to proline (23, 24). Interestingly substitutions outside of the 20–29 region but within the region we have identified as being involved with nucleation also profoundly affected amyloid formation (25). These studies highlight the putative importance of the region(s) we have identified as being critical for nucleation. Our results have important implications for inhibitor development because they provide the information required to design inhibitors that target the critical nucleation site. Along these lines, insulin and its isolated B-chain are known to be among the most potent inhibitors of amyloid formation by IAPP, and they are thought to interact predominantly within residues 7–19 of IAPP (26). Thus, rat and mouse IAPP might have evolved to prohibit amyloid formation by mutating the first 2 sites in the aggregation pathway: Val-17 and Ala-25.
In principle, some of the information reported here could have been obtained by using FTIR spectroscopy, but in practice, it is difficult to observe the isotope label against the background. Carbonyl isotope labels appear more prominently in 2D IR spectra than background solvent modes due to a nonlinear nature that enhances strong absorbers over weaker ones. Another advantage is that our rapid-scan technology can be used to suppress scatter by phase-cycling the pulses. Scatter is a huge problem with heterogeneous samples like amyloids and membranes. However, the real advantages of 2D IR spectroscopy remain to be discovered through experiments that focus on 2D line shapes and cross-peaks, which have the potential to time-resolve solvent expulsion and identify the secondary structures of intermediates, among other properties.
Conclusions
In summary, we have used isotope labeling and technological advances in 2D IR spectroscopy to gain residue-specific kinetic information about the aggregation pathway of hIAPP. We found that the amyloid fibrils associated with type 2 diabetes follow a multistep pathway involving a series of intermediate, largely β-sheet structures. Because at each point in time, our data provides the relative population of each residue in its final structure, it should be possible to test kinetic models for fibril formation by using these data. The methodology presented here is not limited to IAPP, nor is it limited to amyloid formation in homogenous solution. Membrane peptide interactions have been postulated to play an important role in amyloid formation (27), and our methodology is well suited to investigate such systems. It is also applicable to a wide range of proteins now that it is possible to synthesize large polypeptides with unnatural or labeled amino acids (28). Furthermore, 2D IR spectra can be simulated from X-ray, NMR, or molecular dynamics structures. Thus, our methodology has the potential to test and link disparate studies that together hold the promise of providing a comprehensive understanding of the elusive mechanism of amyloid fiber formation.
Materials and Methods
Six labeled peptide samples and 1 unlabeled peptide sample were prepared by using standard Fmoc chemistry and HPLC purification protocols. The peptide was denatured in d-HFIP and dried. Amyloid formation was initiated by redissolving in 20 mM deuterated potassium phosphate buffer at pH 7 to a peptide concentration of 1 mM. Fibrils produced by using this protocol have the same structure as those formed by using other methods (Fig. S4). TEM images of an aliquot removed from the sample immediately after initiation showed predominantly ≈15-nm spherical oligomers with trace numbers of fibers. Automated 2D-IR spectroscopy was used to follow aggregation by rapidly scanning a 4-cycle pulse sequence. There is ≈2-min dead time between initiation of the aggregation and the collection of the first 2D IR spectrum. Details of the data-collection procedure and analysis are given in SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Robert Tycko for providing the coordinates of the solid-state NMR structure of the IAPP fibril. D.P.R. appreciates helpful discussions with Dr. Andisheh Abedini (Harvard Medical School, Boston, MA) and Mr. Peter Marek (Stony Brook University). This work was supported by National Institutes of Health Grants GM 070941 and GM 078114 (to D.P.R.) and DK 79895 (to M.T.Z.). M.T.Z. also acknowledges the Sloan Foundation for support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805957106/DCSupplemental.
References
- 1.Sipe JD. Amyloidosis. Crit Rev Clin Lab Sci. 1994;31:325–354. doi: 10.3109/10408369409084679. [DOI] [PubMed] [Google Scholar]
- 2.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 3.Silveira JR, et al. The most infectious prion protein particles. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukherjee P, Kass I, Arkin I, Zanni MT. Picosecond dynamics of a membrane protein revealed by 2D IR. Proc Natl Acad Sci USA. 2006;103(10):3528–3533. doi: 10.1073/pnas.0508833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strasfeld DB, Ling YL, Shim S-H, Zanni MT. Tracking fibril formation in human islet amyloid polypeptide with automated 2D-IR spectroscopy. J Am Chem Soc. 2008;130(21):6698–6699. doi: 10.1021/ja801483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westermark P, et al. Amyloid fibrils in human insulinoma and islets of Langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. Proc Natl Acad Sci USA. 1987;84:3881–3885. doi: 10.1073/pnas.84.11.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper GJS, et al. Purification and characterization of a peptide from amyloid-rich pancreases of type 2 diabetic patients. Proc Natl Acad Sci USA. 1987;84:8628–8632. doi: 10.1073/pnas.84.23.8628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenzo A, Razzaboni B, Weir GC, Yankner BA. Pancreatic islet cell toxicity of amylin associated with type II diabetes mellitus. Nature. 1994;368:756–760. doi: 10.1038/368756a0. [DOI] [PubMed] [Google Scholar]
- 9.Kahn SE, Andrikopoulos S, Verchere CB. Islet amyloid: A long recognized but underappreciated pathological feature of type II diabetes. Diabetes. 1999;48:241–246. doi: 10.2337/diabetes.48.2.241. [DOI] [PubMed] [Google Scholar]
- 10.Torres J, Briggs JAG, Arkin IT. Multiple site-specific infrared dichroism of CD3-zeta, a transmembrane helix bundle. J Mol Biol. 2002;316(2):365–374. doi: 10.1006/jmbi.2001.5267. [DOI] [PubMed] [Google Scholar]
- 11.Petty SA, Decatur SM. Intersheet rearrangement of polypeptides during nucleation of beta-sheet aggregates. Proc Natl Acad Sci USA. 2005;102(40):14272–14277. doi: 10.1073/pnas.0502804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. Two-dimensional infrared spectra of isotopically diluted amyloid fibrils from Ab40. Proc Natl Acad Sci USA. 2008;105(22):7720–7725. doi: 10.1073/pnas.0802993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307(5707):262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 15.Padrick SB, Miranker AD. Islet amyloid: Phase partitioning and secondary nucleation are central to the mechanism of fibrillogenesis. Biochemistry. 2002;41(14):4694–4703. doi: 10.1021/bi0160462. [DOI] [PubMed] [Google Scholar]
- 16.Konarkowska B, Aitken JF, Kistler J, Zhang SP, Cooper GJS. The aggregation potential of human amylin determines its cytotoxicity towards islet beta-cells. FEBS J. 2006;273(15):3614–3624. doi: 10.1111/j.1742-4658.2006.05367.x. [DOI] [PubMed] [Google Scholar]
- 17.Goldsbury C, et al. Amyloid fibril formation from full-length and fragments of amylin. J Struct Biol. 2000;130(2–3):352–362. doi: 10.1006/jsbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- 18.Chung HS, Tokmakoff A. Visualization and characterization of the infrared active amide I vibrations of proteins. J Phys Chem B. 2006;110(6):2888–2898. doi: 10.1021/jp053956a. [DOI] [PubMed] [Google Scholar]
- 19.Munoz V, Henry ER, Hofrichter J, Eaton WA. A statistical mechanical model for beta-hairpin kinetics. Proc Natl Acad Sci USA. 1998;95(11):5872–5879. doi: 10.1073/pnas.95.11.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munoz V, et al. Folding and aggregation kinetics of a beta-hairpin. Biochemistry. 2006;45(23):7023–7035. doi: 10.1021/bi052556a. [DOI] [PubMed] [Google Scholar]
- 21.Hahn S, Kim SS, Lee C, Cho M. Characteristic two-dimensional IR spectroscopic features of antiparallel and parallel beta-sheet polypeptides: Simulation studies. J Chem Phys. 2005;123(8) doi: 10.1063/1.1997151. 084905. [DOI] [PubMed] [Google Scholar]
- 22.Westermark P, Engstrom U, Johnson KH, Westermark GT, Betsholtz C. Islet amyloid polypeptide: Pinpointing amino acid residues linked to amyloid fibril formation. Proc Natl Acad Sci USA. 1990;87:5036–5040. doi: 10.1073/pnas.87.13.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan LM, Tatarek-Nossol M, Velkova A, Kazantzis A, Kapurniotu Design of a mimic of nonamyloidogenic and bioactive human islet amyloid polypeptide (IAPP) as a nanomolar affinity inhibitor of IAPP cytotoxic fibrillogenesis. Proc Natl Acad Sci USA. 2006;103:2046–2051. doi: 10.1073/pnas.0507471103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abedini A, Meng F, Raleigh DP. A single-point mutation converts the highly amyloidogenic human islet amyloid polypeptide into a potent fibrillization inhibitor. J Am Chem Soc. 2007;129:11300–11301. doi: 10.1021/ja072157y. [DOI] [PubMed] [Google Scholar]
- 25.Abedini A, Raleigh DP. Destabilization of human IAPP amyloid fibrils by proline mutations outside of the putative amyloidogenic domain: Is there a critical amyloidogenic domain in human IAPP. J Mol Biol. 2006;355:274–281. doi: 10.1016/j.jmb.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 26.Gilead S, Wolfenson H, Gazit E. Molecular mapping of the recognition interface between the islet amyloid polypeptide and insulin. Angew Chem Int Ed. 2006;45:6476–6480. doi: 10.1002/anie.200602034. [DOI] [PubMed] [Google Scholar]
- 27.Engel MFM, et al. Membrane damage by human islet amyloid polypeptide through fibril growth at the membrane. Proc Natl Acad Sci USA. 2008;105(16):6033–6038. doi: 10.1073/pnas.0708354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dawson PE, Kent SBH. Synthesis of native proteins by chemical ligation. Annu Rev Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.