Abstract
The DNA replication–division cycle of eukaryotic cells is controlled by a complex network of regulatory proteins, called cyclin-dependent kinases, and their activators and inhibitors. Although comprehensive and accurate deterministic models of the control system are available for yeast cells, reliable stochastic simulations have not been carried out because the full reaction network has yet to be expressed in terms of elementary reaction steps. As a first step in this direction, we present a simplified version of the control system that is suitable for exact stochastic simulation of intrinsic noise caused by molecular fluctuations and extrinsic noise because of unequal division. The model is consistent with many characteristic features of noisy cell cycle progression in yeast populations, including the observation that mRNAs are present in very low abundance (≈1 mRNA molecule per cell for each expressed gene). For the control system to operate reliably at such low mRNA levels, some specific mRNAs in our model must have very short half-lives (<1 min). If these mRNA molecules are longer-lived (perhaps 2 min), then the intrinsic noise in our simulations is too large, and there must be some additional noise suppression mechanisms at work in cells.
Keywords: cyclin-dependent kinase, gene expression, network dynamics, stochastic model, mRNA turnover
To reproduce, a cell must make new copies of all of its components and then divide in half, partitioning every component more or less evenly to its two progeny, so that each newborn cell gets all of the machinery and information necessary to repeat the process. In particular, the genetic information of the cell must be precisely replicated and accurately partitioned at each division, so that each progeny cell gets one and only one copy of every chromosome. Furthermore, the DNA replication–division cycle must be coordinated with the growth cycle (the doubling of all other components of the cell) so that, on average, a cell lineage divides in half for each doubling of cell mass.
Progression through the eukaryotic cell cycle (Fig. 1A) is governed by a complex network of interacting genes and proteins that controls the activity of a family of cyclin-dependent protein kinases (CDKs) (1). When CDK activity is low, the cell is in a growing state (G1 phase) not yet committed to DNA replication and division. When CDK activity rises, the cell initiates DNA synthesis (S phase). G2 phase is the gap between the end of S phase and the beginning of M phase (mitosis = chromosome partitioning and nuclear division). CDK activity is destroyed as cells exit mitosis and undergo cell division. The newborn daughter cells are in G1 phase, with unreplicated DNA molecules and low CDK activity. They are also approximately half the size of their “mother” cell when she underwent division. The CDK control system is responsible for the alternation of DNA synthesis and mitosis and for the balance of growth and division.
Fig. 1.
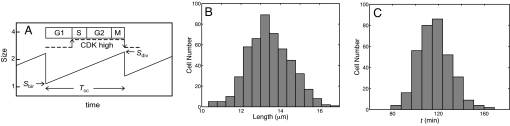
Cell cycle statistics. (A) Schematic illustration of a cell lineage undergoing growth and division. Each cell is born at a particular size (Sbir) and divides at some larger size (Sdiv). The birth size of a daughter cell is some fraction p of the division size of its mother, where p is a random number drawn from a normal distribution with mean 0.5 and 5% CV. The cell cycle time (Tcc) is the time between cell birth and division; it may also be called cell age at division. The interdivision period is divided into four phases, depending on the state of the chromosomal DNA: G1 (unreplicated chromosomes), S (replicating chromosomes), G2 (replicated chromosomes), M (dividing chromosomes). During G1 phase, CDK activity is low, whereas CDK activity is high during S + G2 + M phases. (B and C) Histograms for cell length (B) and age (C) in a sample of dividing fission yeast cells. Data were redrawn from Miyata et al. (23). Fission yeast cells are rod-shaped; they grow in length only at a fixed radius, so cell length is a proxy for cell size. For this yeast cell sample, mean size at division = 13.4 μm (CV = 7.5%), and mean age at division = 116 min (CV = 13.8%).
Although the cell division cycle is very precise and robust in many aspects, it is sloppy in other details (Fig. 1 B and C) (2). In a steady-state population of proliferating cells, although the distribution of cell size is time-independent and stable, the distribution of cell size at any particular stage of the cycle (e.g., at birth or at division) is quite variable. For example, for fission yeast cells, CVsize@division = (standard deviation of size at division)/(mean size at division) ≈ 7%. Fission yeast cells are even less fussy about the time they need to complete the cell cycle: CVage@division ≈ 14%. It is fair to say that a dividing cell takes pains to ensure that its DNA is accurately replicated and partitioned between its two progeny, but it is not particularly concerned with how long this process takes to do correctly, or (consequently) how large it is at division. Size fluctuations can be corrected in the next generation; indeed, in yeast cells, there is a strong negative correlation between size at birth and subsequent cell cycle time (3).
These correlations are evidence for a size control (or size threshold) operating at some critical transition in the cell cycle. Cells that are larger (or smaller) than average at birth take a shorter (or longer) time to grow to the threshold size for commitment to the DNA replication–division cycle and hence have a shorter (or longer) than average cycle time. In a classic article, Koch and Schaechter (4) worked out the implications of this idea of a size threshold for the G1/S transition [see supporting information (SI) Appendix]. For “sloppy size control” models under very general conditions, they showed that CVage@division ≈ 2 × CVsize@division. The Koch–Schaechter model demonstrated that much of the variability of cell cycle properties can be explained by inequities in the cell division process and a size threshold for the G1/S transition. They also assumed some variability in the time of progression through S, G2, and M phases. This variability is caused, presumably, by random fluctuations in the molecular machinery that coordinates and executes these events. Given the small size of a yeast cell (≈30 fL) and the low concentration of regulatory proteins (≈50 nM), the total number of molecules of each regulatory protein in a cell is limited (≈1,000), and intrinsic molecular fluctuations are not only inevitable but also large enough (CVmolecular noise ≈ N−1/2 ≈ 3%) to interact significantly with extrinsic fluctuations (e.g., CVunequal division ≈ 5%). Even more significant is the recent discovery that the average number of specific mRNA molecules per yeast cell is very small: 0.5–5 molecules per cell (Table 1). At such low numbers, molecular fluctuations must be very large indeed, and they are likely amplified many-fold in going from mRNA to protein (5). In the face of such potentially large fluctuations, it is hard to understand how the CDK control system can function with any reliability.
Table 1.
Numbers of molecules (per haploid yeast cell) for several cell cycle genes
| Gene | mRNA* | Protein |
|
|---|---|---|---|
| Ref. 24 | Ref. 25 | ||
| CDC28 | 2.2 | 6,700 | 6,000 |
| CLN2 | 1.2 | 1,300 | 1,000 |
| CLN3 | 1.1 | ND† | 110 |
| CLB2 | 1.1 | 340 | 500 |
| CLB5 | 0.9 | 520 | 390 |
| SWI5 | 0.8 | 690 | ND |
| MCM1 | 1.6 | 9,000 | ND |
| SIC1 | 1.9 | 770 | 100 |
| CDC14 | 1.0 | 8,500 | ND |
*mRNA data are from http://web.wi.mit.edu/young/expression/transcriptome.html.
†ND, not done.
It is our intention in this article to explore the relative contributions of intrinsic and extrinsic noise sources to variability in cell cycle properties of yeast and other eukaryotes. To be reliable, the model must capture the basic dynamic features of the CDK control system (Table 2), it must be formulated in terms of elementary chemical reactions so that the effects of molecular noise can be calculated accurately, and it must operate in the experimentally measured ranges of protein numbers and mRNA numbers. The model must have a mechanism for size regulation, and it must account for cell growth and sloppy cell division. By turning on and off the various sources of noise in the model, we will investigate the relative roles of protein fluctuations, mRNA fluctuations, and uneven division.
Table 2.
Principal molecular players in the CDK control system
| Cdk1 | A protein kinase, phosphorylates specific proteins involved in the initiation of DNA synthesis and mitosis. |
| Cyclin B | A partner of Cdk1, activates Cdk1 and directs its kinase activity to specific substrates. |
| APC | An E3 ubiquitin-conjugating enzyme, labels specific proteins for degradation by the proteasome. |
| Cdc20 | A partner of APC, directs its labeling activity to specific proteins. |
| Cdh1 | A partner of APC, similar to Cdc20 |
| Cdc14 | A protein phosphatase, reverses the phosphorylation reactions catalyzed by Cdk1 |
X, CycB–Cdk1; Y, Cdh1–APC; Z, Cdc20 and Cdc14.
There have been several earlier studies of noise effects on cell cycle progression. Starting from ordinary differential equation models of yeast cell cycle controls, Sveiczer et al. (6, 7) and Steuer (8) derived stochastic, Langevin-type equations that superimposed white noise on an underlying deterministic system. Mura and Csikasz-Nagy (9) took a different approach, by using Gillespie's algorithm to simulate stochastic evolution of Chen's deterministic model (10) of the budding yeast cell cycle. In all of these cases, although one may question the validity of the stochastic approach to compute intrinsic noise in the control system correctly, the authors drew interesting conclusions about certain mutant cells where stochastic fluctuations play important roles in their aberrant behavior. Zhang et al. (11), Braunewell and Bornholdt (12), Ge et al. (13), and Okabe and Sasai (14) have presented stochastic models of the yeast cell cycle based on a deterministic Boolean model from Li et al. (15). The main concern of all of these authors was the robustness of cell cycle progression in the presence of intrinsic and extrinsic sources of noise. None of them compared their models to observed statistics of cell cycle properties in wild-type or mutant cells.
The Model
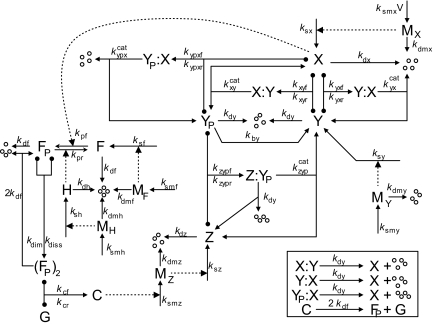
Our model of the CDK control system (Fig. 2) is based on earlier work by Sabouri-Ghomi et al. (16), who “unpacked” a phenomenological model from Tyson and Novak (17). The Tyson–Novak model is based on a bistable switch, created by the antagonism between the complex CycB–Cdk1 (called X) and the complex Cdh1–APC (called Y). Active Y catalyzes the degradation of CycB (thereby destroying X), whereas active X phosphorylates Cdh1 (thereby inactivating Y). In G1 phase, the switch is in the off position (i.e., X off and Y on), and it flips to the on position at the G1/S transition. Tyson and Novak modeled the phosphorylation and dephosphorylation of Y by nonelementary (Michaelis–Menten) rate laws. To prepare the Tyson–Novak model for stochastic simulations, Sabouri-Ghomi et al. (16) unpacked the Michaelis–Menten steps into elementary reactions (substrate + enzyme ↔ complex → enzyme + product), and their article should be consulted to see how this unpacking must be done to maintain bistability of the switch.
Fig. 2.
Molecular mechanism regulating the activity of cyclin B-dependent protein kinase. X, CycB–Cdk1; Y, Cdh1—APC; YP, phosphorylated (inactive) Y; Z, Cdc20 and Cdc14 (composite species); G, gene encoding Z; F, transcription factor controlling the expression of G; FP, phosphorylated form of F; H, enzyme that removes phosphate group from FP; (FP)2, dimeric form of F; C, Z gene bound to (FP)2 and actively transcribing MZ, the messenger RNA for Z; MX etc., messenger RNAs for all other primary gene products in the model; four small circles, products of protein and mRNA degradation reactions. A T-shaped arrow with balls on the cross-bar indicates a reversible binding reaction. (Inset) Additional degradation reactions necessary to maintain approximately constant concentrations of total Y and F proteins during the cell cycle. We assume that the complex YP–X has enough CDK activity to drive DNA synthesis but not mitosis; only the free form of X is sufficient to drive the cell into mitosis.
The Sabouri-Ghomi model deals only with the G1/S transition (X off → X on) and how it is triggered by an increase in cell size. To model a complete cell cycle, we need a mechanism for the M/G1 transition (X on → X off). Tyson and Novak (17) attribute this transition to the action of a pair of proteins, Cdc20 and Cdc14. High activity of X (CycB–Cdk1) in M phase promotes the synthesis and activation of Cdc20, which in turn promotes the activation of Cdc14 (a phosphatase). Cdc14 dephosphorylates and activates Cdh1–APC (Y). In the Tyson–Novak model, Cdc20 and Cdc14 are lumped together into a single variable Z, and the production rate of Z is given by a Hill function (another phenomenological, nonelementary rate law), depending on the activity of X. In the same spirit as Sabouri-Ghomi et al., we unpack the Hill function in terms of a transcription factor, F, that is phosphorylated by X and then forms a dimer (FP)2, that binds to and up-regulates expression of the gene encoding Z. Each step in the unpacked mechanism is assumed to be elementary (at the level of detail appropriate to this model), with kinetics given by the law of mass action.
The model of the CDK control system that we use here is given in full detail in Fig. 2. The mass action rate laws corresponding to this mechanism are written in full in Table S1 in the SI Appendix, and the rate constant values we employ are given in Table S2 in the SI Appendix. Details on how we carry out deterministic and stochastic simulations of the model are provided in the SI Appendix, with programs provided in Dataset S1, Dataset S2, and Dataset S3. Our full model has several additional features worth highlighting. (i) All variables are given as total numbers of molecules per cell, e.g., NX = total number of “free” (uncomplexed) molecules of X per cell. (ii) We take into account the continuous increase in cell volume during the cell cycle and the attendant dilution of molecular concentrations in the cell. (iii) We include synthesis and degradation of all proteins in the model, to maintain, where appropriate, the Tyson–Novak assumption of constant total concentrations of species like Y and F. Protein synthesis rates are proportional to cell volume, V(t), because the number of ribosomes in a cell increases as the cell grows. (iv) We include mRNAs for all primary gene products in the model. We have chosen rate constant values to give absolute numbers of protein and mRNA species in decent agreement with experimental measurements. Because thousands of copies of each protein species must be made from only ≈1 specific mRNA molecule per cell during a 100-min cell cycle, we must assume very fast translation rates (20–50 protein molecules per mRNA molecule per min). The polypeptide chain elongation rate in yeast cells is ≈10 aa per s (18), so it takes ≈1 min to make a molecule of cyclin B. With 10–20 ribosomes per mRNA, we can get close to the translation rates assumed in the model. (v) The synthesis rate of cyclin B mRNA (species MX in Fig. 2) is assumed to be proportional to cell size. This assumption makes the total amount of CycB (relative to the total amount of Cdh1) increase as the cell grows, which induces the G1/S transition when the cell reaches a threshold size. This is the basis of “size control” in the model.
Results
Deterministic Simulation.
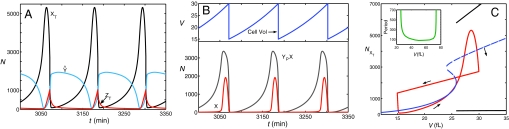
To establish a baseline, we first carry out a completely deterministic simulation (Fig. 3 A and B) of the model with precise division in half. There is no variability whatsoever (Table 3, row 1).
Fig. 3.
Deterministic simulations of the model. (A) Time courses for total amounts of cyclin B protein (XT, black line), Cdc20 protein (ZT, red line), and unphosphorylated Cdh1 protein (Ŷ blue line). (B) Time courses for free (uncomplexed) X (red line), YP–X (gray line), and cell volume (blue line). (C) The figure 8-shaped curve (red) is a cell cycle trajectory created by plotting, parametrically in t, the curves for XT(t) and V(t) from A and B, respectively. The small arrows indicate the direction of motion around this trajectory. Cell division is indicated by the abrupt jump from V = 29.8 to V = 14.9, which is triggered when Ŷ increases above 1,200 molecules. The S-shaped curve (blue) is a one-parameter bifurcation diagram (computed by using XPP-AUTO) for steady-state solutions of the molecular control system, treating cell size (V) as a bifurcation parameter. Solid line, stable steady state; dashed line, unstable steady state. The two black lines above and below the unstable steady-state curve indicate the presence of stable limit cycle oscillations for V > 25; the Upper (Lower) curve indicating the maximum (minimum) value of XT on the limit cycle for a particular value of V. (Inset) Period of limit cycle oscillations as a function of cell size. The control system generates oscillations by homoclinic (infinite period) bifurcations at V = 25 and V = 74.
Table 3.
Statistical properties of simulations
| Row | Type of noise | Cycle time, min |
Size at division, fL |
Size at birth, fL |
Avg. no. of Cdc20 mRNA molecules | Avg. no. of CycB protein molecules | |||
|---|---|---|---|---|---|---|---|---|---|
| Mean | CV, % | Mean | CV, % | Mean | CV, % | ||||
| 1 | Deterministic | 115.5 | 0 | 29.9 | 0 | 14.9 | 0 | 0.48 | 1,054 |
| 2 | Full stochastic | 115.8 | 13.8 | 29.2 | 8.4 | 14.6 | 9.8 | 0.52 | 1,062 |
| 3a* | Only extrinsic | 116.1 | 4.9 | 30.0 | 1.9 | 14.9 | 5.3 | 0.48 | 1,051 |
| 3b† | 116.1 | 5.1 | 29.9 | 2.0 | 14.9 | 5.3 | 0.46 | 1,037 | |
| 4 | Only intrinsic | 115.5 | 13.0 | 29.1 | 8.2 | 14.5 | 8.2 | 0.51 | 1,040 |
| 5‡ | Variance | (13.0)2 + (5.1)2 ≈ (13.8)2 | (8.2)2 + (2.0)2 ≈ (8.4)2 | (8.2)2 + (5.3)2 ≈ (9.8)2 | |||||
| 6a§ | τ1/2 = 120 s | 115.5 | 32.8 | 26.3 | 21.2 | 13.1 | 21.8 | 0.54 | 883 |
| 6b§ | τ1/2 = 60 s | 115.7 | 24.9 | 27.7 | 15.0 | 13.8 | 15.8 | 0.52 | 986 |
| 7a¶ | τX,Y = 12,12 s | 115.7 | 14.4 | 29.6 | 9.1 | 14.8 | 10.4 | 0.58 | 1,067 |
| 7b¶ | τX,Y = 12,120 s | 115.3 | 23.7 | 29.3 | 15.3 | 14.7 | 16.2 | 0.49 | 1,054 |
*Unequal division and strictly proportional distribution of protein and mRNA.
†Unequal division and binomial distribution of protein and mRNA molecules.
‡(CVrow4)2 + (CVrow3b)2 = (CVrow2)2.
§Varying half-life of constitutive mRNAs; compare with row 2, where τ1/2 = 12 s.
¶Half-lives of mRNAs for species X and Y, τ1/2 = 300 s for species MZ, MF, and MH.
In Fig. 3C we plot a one-parameter bifurcation diagram for the deterministic CDK control mechanism. For small values of V, the CDK control system has a unique stable steady state, corresponding to G1 phase of the cell cycle (X off, Y on). For V near 25 fL, the control system undergoes a homoclinic bifurcation to stable, large-amplitude, limit cycle solutions. [At very large size, V = 74 fL, the limit cycles are lost by another bifurcation to a stable M phase state (X on, Y off) of the cell cycle.] The narrow region of three steady states (23 fL < V < 25 fL) is created by the antagonistic relationship between CycB (X) and Cdh1 (Y). The limit cycle oscillations are driven by the negative feedback loop, whereby X up-regulates Z, which activates Y, which destroys X. The homoclinic bifurcation is a confluence of a saddle-node bifurcation of the antagonistic feedback loop with an infinite-period closed orbit of the negative-feedback loop.
In Fig. 3C we have superimposed on the bifurcation diagram a “cell cycle trajectory,” which is created by plotting (parametrically in t) the curve for XT(t) from Fig. 3A against the curve for V(t) from Fig. 3B. Fig. 3C shows how progression through the cell cycle in our model is related to the homoclinic bifurcation that separates the G1 stable steady state from the S + G2 + M stable limit cycle. The newborn cell (≈15 fL) is so small that the only stable attractor for the CDK control system is the G1 steady state. Hence, the newborn cell waits in G1 phase (with unreplicated DNA) until it grows large enough to surpass the homoclinic bifurcation point. At that point, the G1 stable steady state disappears (by coalescing with an unstable saddle point) and is replaced by a stable limit cycle. The CDK control system is attracted to the limit cycle (slowly at first, because this is a homoclinic bifurcation). As Cdh1 is inactivated, the level of CycB rises, and the cell enters S phase. At first, CycB is mostly associated with phosphorylated Cdh1 (YP–X), but eventually the CycB level swamps the supply of Cdh1. Free CycB then drives the cell into mitosis and activates the transcription factor (F) for Cdc20 (Z). As Z increases, it activates the phosphatase (Cdc14) that dephosphorylates Cdh1 (YP → Y). Active Cdh1 now destroys CycB, and this is the signal for the cell to divide. Cell division corresponds to the abrupt down-jump from V = 30 fL to 15 fL on the cell cycle trajectory.
The deterministic model exhibits the robust general properties that we expect of cell cycle progression. S phase and M phase alternate because the cell must enter S phase (the initial rise of XT after passing the homoclinic bifurcation point) before it can enter mitosis (when free X, uncomplexed with YP, finally accumulates). Then the cell must destroy X and return to G1 phase before it can initiate another round of DNA synthesis. Also, this mechanism automatically ensures that the cell cycle time is equal to the mass doubling time. Because the cell divides in half at the M/G1 transition, the time between successive G1/S transitions (passing the homoclinic bifurcation point, at the threshold cell size) must be exactly the time needed for cell size to increase 2-fold.
Fully Stochastic Simulation.
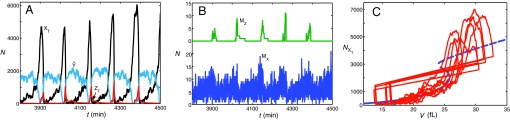
Next, we compute a fully stochastic simulation of the model with intrinsic noise at both protein and mRNA levels and extrinsic noise caused by uneven division and binomial distribution of protein and mRNA molecules to sister cells. A few typical cell cycle trajectories are illustrated in Fig. 4A, and their projections onto the deterministic bifurcation diagram are plotted in Fig. 4C. Although progression through the cell cycle is now quite noisy, it still maintains its association with the homoclinic bifurcation, on which depend the fundamental, robust characteristics of the cell cycle.
Fig. 4.
Stochastic simulations of the full model. These simulations include both intrinsic noise (fluctuations in the molecular regulatory system) and extrinsic noise (uneven cell division and binomial distribution of protein and mRNA molecules to daughter cells at division). (A) Time courses for total amounts of regulatory proteins, as in Fig. 3A. (B) Time courses for amounts of mRNA species, MZ (Upper) and MX (Lower). As described in the text, we assume that MZ has a half-life of 280 s, whereas MX (as well as all other mRNAs) has a much shorter half-life (12 s). (C) Series of stochastic cell cycle trajectories are superimposed on the bifurcation diagram of Fig. 3C.
The statistical properties of a large ensemble of simulated cells, summarized in Table 3 (row 2), reflect the characteristic sloppiness observed in populations of yeast cells (distributions of cycle time and sizes at division). The model is also in accord with typical average numbers of proteins and mRNAs per cell.
Relative Effects of Intrinsic and Extrinsic Noise.
In Table 3 we also report the statistical properties of the model in the case of extrinsic noise only (row 3) or intrinsic noise only (row 4). In row 3, we simulate the CDK control system deterministically but allow for uneven division (CVdiv = 5%). The statistics in this case are quite similar whether or not molecules are distributed binomially to sister cells at division. In row 4, we simulate the CDK control system stochastically, assuming that cells divide precisely in half and distribute proteins and mRNAs evenly to sister cells. By comparing the statistical properties in rows 2, 3, and 4 of Table 3, we see that for most indicators, Varintrinsic > Varextrinsic and Varfull ≈ Varintrinsic + Varextrinsic (Table 3, row 5); that is to say, the two sources of noise contribute independently to the observed variability of cell cycle properties, with intrinsic noise being the primary source of variability.
Role of mRNA Fluctuations in Intrinsic Noise Levels.
Next we investigate more closely how the measures of cell cycle variability depend on the turnover rate of mRNAs. According to the measurements of Holstege et al. (19), the mRNAs for cell cycle control genes in yeast cells have half-lives of 10–20 min. If we use such long half-lives for the mRNAs in our model, then the intrinsic noise of the model is unacceptably large. The results in Table 3, row 2, are computed for a half-life of MZ of ≈5 min (kdmz = 0.15 min−1) and all other mRNAs with a half-life of 0.2 min. We tried many different values for the half-life of these constitutive mRNAs. For much longer half-life (e.g., 1 or 2 min; Table 3, rows 6a and 6b), the intrinsic noise in the control system is much too large. To keep the noise at an acceptable level, it is necessary that the mRNAs for species X and Y turn over rapidly (half-life = 0.2 min), whereas species Z, F, and H can have longer-lived mRNAs (5 min) (see Table 3, row 7a).
Discussion and Conclusion
We have proposed a model of eukaryotic cell growth and division that realistically incorporates the major sources of variability in cell cycle progression. Intrinsic noise is reflected in a simple model of the CDK control system, based on a bistable switch between Cdh1–APC (a stabilizer of G1 phase of the cell cycle) and CycB–Cdk1 (the driving signal for S + G2 + M phases of the cell cycle). The switch from G1 phase into S phase is controlled by growth to a threshold cell size, and the switch from M phase back to G1 is controlled by a negative feedback loop involving the “mitotic exit” proteins, Cdc20 and Cdc14. The interactions of the CDK control system are given by elementary chemical reactions that can be simulated accurately by Gillespie's stochastic simulation algorithm. Rate constant values are assigned to give realistic variations in numbers of molecules of each species during the cell cycle. mRNAs are included in the model so that we may study the effects of low mRNA numbers per cell and of mRNA turnover.
The interdivision time of yeast cells is known to be very sensitive to birth size (3). Large newborn cells have unusually short cell cycle times, whereas small newborn cells are unusually “old” at division. Hence, a major source of variability in cell cycle times is the distribution of cell sizes of newborn cells. This distribution reflects the variability of division sizes of mother cells and the unevenness of the cell division process itself. Uneven division is extrinsic to molecular fluctuations of the cell cycle control system, but variability of size at division depends in large part on intrinsic noise in the control system. A second source of extrinsic noise is the unequal partitioning of protein and mRNA molecules to sister cells at division. This effect is modeled as a binomial distribution process.
By studying the model under various circumstances, we reach a few general conclusions. Intrinsic and extrinsic sources of noise are mostly independent in their effects, and they both contribute significantly to the observed variability of cell cycle progression. Nonetheless, intrinsic molecular fluctuations in the control system are considerably noisier than extrinsic inequities in the division process. By far, the greatest source of intrinsic noise comes from low numbers of mRNA molecules reported for yeast cells, 0.5–5 mRNAs per expressed gene per cell (19). To limit the effects of mRNA fluctuations in our model, we have assumed that the turnover of cyclin B and Cdh1 mRNAs is very rapid. In this case, mRNA-directed protein synthesis is effectively time-averaged and not as noisy as might be expected. To fit our model to the typically observed variability of yeast cell cycles, we are compelled to assume that τ1/2 = 12 s for these two mRNAs, much shorter than any measured mRNA half-lives. Indeed, the rate of translation in yeast cells is ≈10 aa per s (18), so an mRNA molecule has to live at least 1 min to direct the synthesis of a protein such as cyclin B. It is hard to reconcile all of these facts, unless cells employ some other mechanism of noise suppression that is not evident in our simple model.
Fluctuations in gene expression have been studied by many authors (5, 20), who have shown that the CV for protein number (NP) is strongly dependent on fluctuations in mRNA number (NM), according to the elegant equation: (CVP)2 = <NP>−1 + τM/(τP + τM)<NM>−1, where <…> denotes “mean,” and τM and τP are the half-lives of mRNA and protein. In our case, <NP> ≈ 1,000 and <NM> ≈ 1, which explains why, to keep CVP < 10%, we must choose τM < τP/100. More sophisticated models of gene expression predict that protein noise may be reduced by periodic transcriptional “bursting” and mRNA “senescence” (20), but the effects are modest (2-fold, at best), so they cannot account for the noise suppression we seek.
In a recent article, most directly related to our work, Okabe and Sasai (14) studied a stochastic model of the yeast cell cycle, including extrinsic fluctuations at cell division and intrinsic fluctuations in gene activation and mRNA abundances. They assumed a 5-min half-life for mRNAs and very low abundances (10–100 molecules) of proteins. They found that intrinsic noise in mRNA levels is non-Poissonian and much larger than noise in protein levels, which they attributed to the role of checkpoints in governing progression through the cell cycle. It is difficult to compare our approach with theirs because we do not include checkpoints explicitly in our model, and they do not compute CVs for cell size and age at division. Hence, the mechanism of noise suppression in the cell cycle remains unresolved.
An accurate stochastic model of the eukaryotic cell cycle will be useful not only for addressing general questions of intrinsic and extrinsic variability, as in this article, but also for making contact with the growing body of experimental studies of specific proteins in single cells by quantitative flow cytometry (21) and fluorescence microscopy (22). For these purposes, the stochastic model of the CDK control system must be more detailed and extensive than the model provided here. The present model keeps track of only cyclin B-dependent kinase; a realistic model would require cyclin A-, cyclin D-, and cyclin E-dependent kinases as well. Our single G1 stabilizer, Cdh1, would need to be complemented by cyclin-dependent kinase inhibitors (CKIs) such as Sic1 (in yeast) or p21 and p27 (in mammalian cells). The mitotic exit network requires, in addition to Cdc20 and Cdc14, other crucial components, such as Net1 and Polo kinase. In many cell types, it is important to follow transcriptional regulation not only of Cdc20 but also of cyclins and CKIs.
We are working toward the goal of a more accurate stochastic model of the CDK control system, but it is no simple task to convert successful phenomenological models, e.g., Chen et al. (10) for the budding yeast cell cycle, into elementary reaction mechanisms supportive of accurate, reliable stochastic simulations. This article illustrates how a simple phenomenological model (17) with 3 variables and 18 parameters quickly expands into a stochastic model with 18 components and 36 rate constants. One shudders to think what might be required to unpack Chen's model, with 30+ dynamic variables and 100+ phenomenological kinetic constants. Nonetheless, the challenge is clear: realistic, accurate, stochastic models of cell cycle regulation are needed to confront quantitative measurements of specific regulatory proteins and mRNAs in single cells. We hope that this work shines some light on the issues involved and the way to progress.
Supplementary Material
Acknowledgments.
We thank Mohsen Sabouri-Ghomi for his preliminary work on the model studied here. This work was supported by the National Institutes of Health Grant 1 R01 GM078989.
Note Added in Proof:
Recently, Zenklusen et al. (26) have shown that the high-throughput measurements of mRNA abundances underestimate these numbers by 5-fold or more. Repeating our full stochastic simulations with 5-fold higher mRNA abundances, we get statistical properties similar to Table 3, row 7a, for τX = τY = 120 s. Although these mRNA half-lives are more reasonable, they are still 5- to 10-fold shorter than generally accepted values (19).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810034106/DCSupplemental.
References
- 1.Tyson JJ, Novak B. Temporal organization of the cell cycle. Curr Biol. 2008;18:R759–R768. doi: 10.1016/j.cub.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyson JJ. The coordination of cell growth and division: Intentional or incidental? Bioessays. 1985;2:72–77. [Google Scholar]
- 3.Fantes PA. Control of cell size and cycle time in Schizosaccharomyces pombe. J Cell Sci. 1977;24:51–67. doi: 10.1242/jcs.24.1.51. [DOI] [PubMed] [Google Scholar]
- 4.Koch AL, Schaechter M. A model for statistics of the cell division process. J Gen Microbiol. 1962;29:435–454. doi: 10.1099/00221287-29-3-435. [DOI] [PubMed] [Google Scholar]
- 5.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sveiczer A, Csikasz-Nagy A, Gyorffy B, Tyson JJ, Novak B. Modeling the fission yeast cell cycle: Quantized cycle times in wee1−cdc25Δ mutant cells. Proc Natl Acad Sci USA. 2000;97:7865–7870. doi: 10.1073/pnas.97.14.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sveiczer A, Tyson JJ, Novak B. A stochastic, molecular model of the fission yeast cell cycle: Role of the nucleocytoplasmic ratio in cycle time regulation. Biophys Chem. 2001;92:1–15. doi: 10.1016/s0301-4622(01)00183-1. [DOI] [PubMed] [Google Scholar]
- 8.Steuer R. Effects of stochasticity in models of the cell cycle: From quantized cycle times to noise-induced oscillations. J Theor Biol. 2004;228:293–301. doi: 10.1016/j.jtbi.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Mura I, Csikasz-Nagy A. Stochastic petri net extension of a yeast cell cycle model. J Theor Biol. 2008;254:850–860. doi: 10.1016/j.jtbi.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Chen KC, et al. Integrative analysis of cell cycle control in budding yeast. Mol Biol Cell. 2004;15:3841–3862. doi: 10.1091/mbc.E03-11-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Stochastic model of yeast cell cycle network. Physica D. 2006;219:35–39. [Google Scholar]
- 12.Braunewell S, Bornholdt S. Superstability of the yeast cell cycle dynamics: Ensuring causality in the presence of biochemical stochasticity. J Theor Biol. 2007;245:638–643. doi: 10.1016/j.jtbi.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Ge H, Qian H, Qian M. Synchronized dynamics and nonequilibrium steady states in a stochastic cell cycle network. Math Biosci. 2008;211:132–152. doi: 10.1016/j.mbs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Okabe Y, Sasai M. Stable stochastic dynamics in yeast cell cycle. Biophys J. 2007;93:3451–3459. doi: 10.1529/biophysj.107.109991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li F, Long T, Lu Y, Ouyang Q, Tang C. The yeast cell cycle network is robustly designed. Proc Natl Acad Sci USA. 2004;101:4781–4786. doi: 10.1073/pnas.0305937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabouri-Ghomi M, Ciliberto A, Kar S, Novak B, Tyson JJ. Antagonism and bistability in protein interaction networks. J Theor Biol. 2008;250:209–218. doi: 10.1016/j.jtbi.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Tyson JJ, Novak B. Regulation of the eukaryotic cell cycle: Molecular antagonism, hysteresis and irreversible transitions. J Theor Biol. 2001;210:249–263. doi: 10.1006/jtbi.2001.2293. [DOI] [PubMed] [Google Scholar]
- 18.Von der Haar T. A quantitative estimation of the global translational activity in logarithmically growing yeast cells. BMC Syst Biol. 2008;2:87. doi: 10.1186/1752-0509-2-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 20.Pedraza JM, Paulsson J. Effects of molecular memory and bursting on fluctuations in gene expression. Science. 2008;319:339–343. doi: 10.1126/science.1144331. [DOI] [PubMed] [Google Scholar]
- 21.Darzynkiewicz Z, Crissman H, Jacobberger JW. Cytometry of the cell cycle: Cycling through history. Cytometry A. 2004;58:21–32. doi: 10.1002/cyto.a.20003. [DOI] [PubMed] [Google Scholar]
- 22.Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effect of molecular noise and size control on the variability in the budding yeast cell cycle. Nature. 2007;448:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- 23.Miyata H, Miyata M, Ito M. The cell cycle in the fission yeast, Schizosaccharomyces pombe. I. Relationship between cell size and cycle time. Cell Struct Funct. 1978;3:39–46. [Google Scholar]
- 24.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 25.Cross FR, Archambault V, Miller M, Klovstad M. Testing a mathematical model of the yeast cell cycle. Mol Biol Cell. 2002;13:52–70. doi: 10.1091/mbc.01-05-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.