Abstract
Although regulation of sigma factors has been intensively investigated, anti-sigma factors have not been identified in oxygenic photosynthetic organisms. A previous study suggested that the sigma factor, SigE, of the cyanobacterium Synechocystis sp. PCC 6803, a positive regulator of sugar catabolism, is posttranslationally activated by light-to-dark transition. In the present study, we found that the H subunit of Mg-chelatase ChlH interacts with sigma factor SigE by yeast two-hybrid screening, and immunoprecipitation analysis revealed that ChlH associates with SigE in a light-dependent manner in vivo. We also found that Mg2+ promotes the interaction of SigE and ChlH and determines their localization in vitro. In vitro transcription analysis demonstrated that ChlH inhibits the transcription activity of SigE. Based on these results, we propose a model in which ChlH functions as an anti-sigma factor, transducing light signals to SigE in a process mediated by Mg2+.
Keywords: cyanobacteria, transcription
Recent investigations have revealed that metabolic enzymes play multiple roles as well as catalyzing metabolic reactions, particularly in transcriptional regulation (1). For example, GAPDH in mammalian cells acts as a coactivator of transcription from the histone H2B promoter during the S phase (2). In yeast mitochondria, Arg 5,6, an enzyme involved in arginine biosynthesis, directly binds to the promoter region of COX1 (encoding a subunit of cytochrome c oxidase) and activates its transcription (3). In Bacillus subtilis, glutamine synthetase interacts with TnrA, a global nitrogen transcription factor conserved among Bacillus species, inhibiting the access of TnrA to the promoters of its regulatory genes (4). In higher plants, Arabidopsis thaliana hexokinase1 (Hxk1) forms a complex in the nucleus with VHA-B1 (1 of the 3 expressed isoforms of the B-subunit of the V1 complex in V-ATPase) and RPT5B (1 of the 2 expressed isoforms of the 19S regulatory particle triple-A ATPase) and is essential for glucose-dependent transcriptional repression (5). Metabolic enzymes regulating transcription have been discovered in species from several different kingdoms.
Transcription in bacteria is initiated by recognition of promoter sequences by a sigma factor of an RNA polymerase. Sigma factors are regulated at transcriptional, translational, and posttranslational levels. The role of anti-sigma factors in posttranslational regulation has been intensively investigated (6). The first identification of a protein inhibiting a sigma factor was in the T4 bacteriophage (7). Subsequently, it has been found that uninfected Escherichia coli also contains similar mechanisms, such as FlgM, which inactivates σF to transcribe the structural genes necessary for the late stage of flagella biogenesis (8). It also is known that sigma factors of Gram-positive bacteria, such as B. subtilis, are regulated by anti-sigma factors necessary for the control of sporulation and stress responses (9). In the case of oxygenic photosynthetic organisms, yeast two-hybrid analyses have shown that the proteins SapG of the unicellular cyanobacterium Synechococcus sp. PCC 7002 and SibI of A. thaliana interact with the sigma factor SigG of Synechococcus sp. PCC 7002 and the plastid sigma factor SIG1 of A. thaliana, respectively, although the biochemical functions of these interactions remain unknown (10, 11). To date, no anti-sigma factor has been identified in oxygenic photosynthetic organisms.
Group-2 sigma factors are known as “primary-like” sigma factors, sharing similar promoter recognition with group 1 (or primary) sigma factors, but are not essential for cellular viability (12, 13). A non–nitrogen-fixing, unicellular cyanobacterium, Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803), contains 4 group 2 sigma factors (sigB–E) (14). A previous study revealed that SigE is a positive regulator of sugar catabolism, such as glycolysis, the oxidative pentose phosphate (OPP) pathway, and glycogen catabolism (15). Sugar catabolism in photosynthetic organisms, including cyanobacteria, is essential under heterotrophic and dark conditions (16). Metabolomic analysis with Synechocystis 6803 cells has shown that glucose is degraded mainly through the OPP pathway under heterotrophic conditions (17); consistently, the transcript levels of genes in the OPP pathway [zwf (slr1843), opcA (slr1734), gnd (sll0329), and tal (slr1793)] are increased 1 h after transition from light to dark conditions in a SigE-dependent manner (15). SigE levels are not increased by light-to-dark transition (18), however, suggesting that SigE activity is up-regulated at the posttranslational level under dark conditions. In this study, we show that the H subunit of the Mg-chelatase ChlH interacts with SigE and represses transcription by SigE, indicating that this metabolic enzyme functions as an anti-sigma factor, possibly transducing light/dark signals to the SigE.
Results
Determination of ChlH as a SigE-Binding Protein and the SigE–ChlH Interaction In Vitro and In Vivo.
Data from a large-scale protein–protein interaction analysis showed that SigE interacted with 4 proteins: ORFs sll1965, slr1055, slr1702, and ssl1707 (19). We performed yeast two-hybrid screening of 3.8 × 106 independent clones of a Synechocystis 6803 genomic library with Synechocystis 6803 SigE as bait, yielding positive clones including 27 ORFs [supporting information (SI) Table S1]. Combining the 2 results, only 1 ORF (slr1055) was included as a positive clone in both experiments. The ORF slr1055 encodes ChlH, the H subunit of Mg-chelatase. Mg-chelatase is an enzyme in the chlorophyll biosynthesis pathway that catalyzes the reaction from protoporphyrin IX (Proto) to Mg-protoporphyrin IX (Mg-Proto) by integrating an Mg2+ ion (20, 21). In plants, ChlH is a multifunctional protein with roles in plastid-to-nucleus and plant hormone signal transduction pathways (see below).
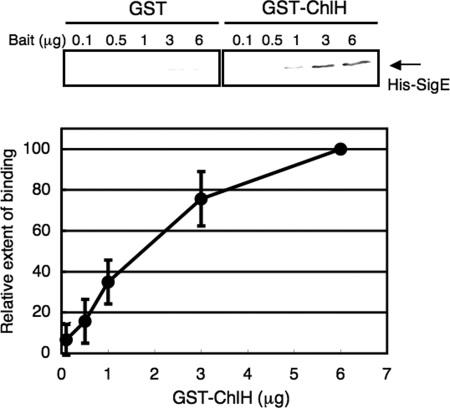
Subsequently, we purified histidine-tagged SigE (His-SigE) and GST-tagged ChlH (GST-ChlH) from E. coli (Fig. S1A). Using GST-ChlH or GST as bait proteins, the amounts of coprecipitated His-SigE were found to increase in a concentration-dependent manner with GST-ChlH, but not with GST (Fig. 1), indicating that ChlH interacts with His-SigE in vitro. Because yeast two-hybrid screening suggested that other proteins also might interact with SigE, we tested another protein, glutamine synthetase (GS), for an interaction with His-SigE. Using purified GST-tagged GS (GST-GS), we performed a GST-pulldown analysis; the results show that GST-GS interacts with His-SigE at background levels (Fig. S1B). In addition, we tested whether another sigma factor interacts with ChlH. GST-pulldown analysis revealed that purified histidine-tagged SigA (His-SigA) did not bind GST-ChlH (Fig. S1C).
Fig. 1.
Interaction between SigE and ChlH in vitro: GST-pulldown analysis. Various amounts of GST or GST-ChlH were bound to glutathione-sepharose 4B beads and mixed with His-SigE (1.0 μg), after which bead-bound proteins were subjected to immunoblotting with antiserum to SigE. Data represent mean ± SD of values from 3 independent experiments. The level is calibrated relative to the pull-down value at 6 μg GST-ChlH, which is set at 100%.
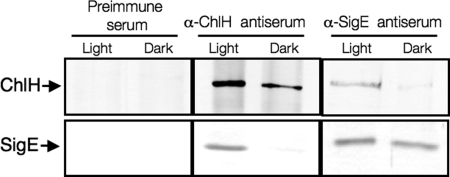
We then investigated the interaction between SigE and ChlH in vivo. To examine the effect of light conditions, we used proteins from cells grown under light or dark conditions for 1 h. SigE and ChlH proteins from light-grown cells were coimmunoprecipitated by rat anti-SigE antiserum or rabbit anti-ChlH antiserum, but neither was precipitated by rabbit preimmune antiserum (Fig. 2). Immunoprecipitation with anti-ChlH or SigE antiserum also revealed that SigE or ChlH proteins from dark-grown cells were not coimmunoprecipitated (Fig. 2).
Fig. 2.
Immunoprecipitation analysis. Proteins from GT Synechocystis 6803 cells grown in modified BG-11 (A750 = ≈2.0) under light or dark conditions (1 h) were mixed with 50 μL of rabbit preimmune antiserum, rabbit antiserum against ChlH, or rat antiserum against SigE. Proteins from the collected cells were extracted in PBS containing 1 mM MgCl2. Precipitated SigE or ChlH proteins were detected by immunoblotting with rabbit or rat SigE or rabbit ChlH antiserum.
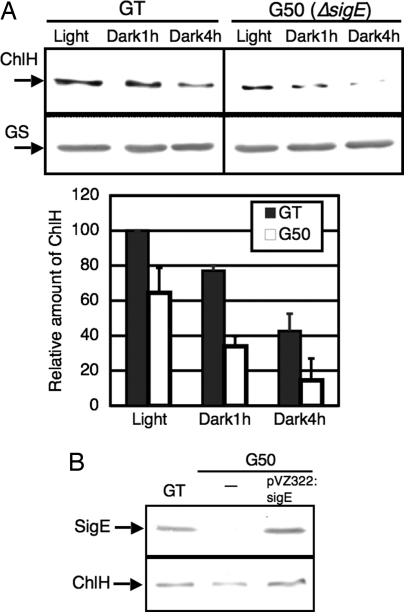
Next, we examined the protein levels of ChlH during light-to-dark transition. Immunoblotting with antiserum to ChlH revealed that ChlH protein levels decreased to about 80% and 40% of those under light conditions at 1 h and 4 h after light-to-dark transition, respectively (Fig. 3A). In addition, disruption of sigE, such as that in the sigE insertion mutant G50 (15), resulted in a 40% decrease in ChlH protein levels under light conditions (Fig. 3A). To confirm that the decreased protein levels of ChlH in G50 actually resulted from sigE deficiency, we performed a complementation experiment with a plasmid carrying the wild-type sigE gene, pVZ322:sigE (22). Immunoblotting revealed that wild-type sigE gene restored protein levels of ChlH (Fig. 3B).
Fig. 3.
(A) Amounts of ChlH proteins in the GT strain of Synechocystis 6803 and the sigE mutant (G50) under light and dark conditions. Total protein (5 μg) was subjected to immunoblotting. (Lower Panel) GS protein levels as a control. Data represent mean ± SD of values from 3 independent experiments. The level is calibrated relative to the value in the GT strain of Synechocystis 6803 under light conditions, which is set at 100%. (B) Amounts of ChlH proteins in GT, G50, and G50 carrying pVZ322:sigE. Cells were grown in modified BG-11 under light conditions, and total protein (5 μg) was subjected to immunoblotting.
Mg2+-Dependent Interaction Between SigE and ChlH.
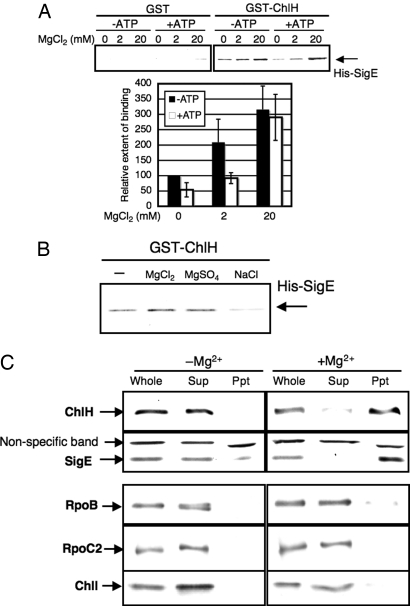
Because it has been suggested that Mg2+ and ATP associate with Synechocystis 6803 ChlH (23), we tested the effects of these cofactors on the SigE–ChlH interaction. GST-pulldown analysis revealed that the addition of MgCl2 strengthened the interaction between GST-ChlH and His-SigE (Fig. 4A). The amounts of coprecipitated His-SigE proteins in the presence of 2 or 20 mM MgCl2 were 2.1 or 3.2 times those in the absence of MgCl2, respectively (Fig. 4A). MgSO4 strengthened the interaction, but NaCl did not, indicating that Mg2+ specifically promoted the interaction (Fig. 4B). In contrast, the addition of ATP inhibited the interaction (Fig. 4A). The amount of His-SigE in the presence of 1 mM ATP was 54% of that in the absence of ATP. The inhibitory effect of ATP was eliminated by increasing the Mg2+ concentration (Fig. 4A). EDTA did not inhibit the interaction, suggesting that the inhibitory effect of ATP was not due to chelating Mg2+ (Fig. S2).
Fig. 4.
Mg2+-dependent interaction between SigE and ChlH. (A) Effect of Mg2+ and ATP on the SigE–ChlH interaction. GST or GST-ChlH (2.0 μg) bound to glutathione-sepharose 4B beads and mixed with His-SigE (1.0 μg) in the absence or presence of 2 or 20 mM MgCl2 and with or without 1 mM ATP. Precipitated proteins were detected by immunoblotting. Data represent mean ± SD of values from 4 independent experiments. The level is calibrated relative to the value obtained in the absence of both MgCl2 and ATP, which is set at 100%. (B) Specific effect of Mg2+ on the SigE–ChlH interaction. GST-ChlH (2.0 μg) bound to glutathione-sepharose 4B beads and mixed with His-SigE (1.0 μg) in the absence or presence of 10 mM MgCl2 or MgSO4 or 20 mM NaCl. Precipitated proteins were detected by immunoblotting. (C) Colocalization of ChlH and SigE depending on Mg2+ concentration. Cells were disrupted in the absence or presence of 10 mM MgCl2 (whole cell extract), and divided into soluble and membrane fractions (Sup and Ppt, respectively) by ultracentrifugation. Proteins (5 μg/lane) were detected by immunoblotting.
ChlH proteins of higher plants fractionate differently, depending on the concentration of Mg2+ in the lysis buffer (24, 25). Previous experiments have found that ChlH fractionates within the membrane fraction at Mg2+ concentrations exceeding 5 mM and within the soluble fraction at concentrations below 1 mM (24, 25). We carried out a similar experiment using glucose-tolerant (GT) cells of Synechocystis 6803 (the GT strain of Synechocystis 6803). GT Synechocystis 6803 cells were disrupted in lysis buffer with or without 10 mM MgCl2, and soluble and membrane fractions were separated by ultracentrifugation (see Materials and Methods). As is the case in higher plants, Synechocystis 6803 ChlH proteins were detected mostly within the membrane fraction in the presence of Mg2+, whereas they were detected in the soluble fraction in the absence of Mg2+ (Fig. 4C). Like the ChlH proteins, most SigE proteins were detected within the membrane fraction in the presence of Mg2+ or in the soluble fraction in the absence of Mg2+ (Fig. 4C). In contrast, 3 subunits of Mg-chelatase or RNA polymerase—ChlI, RpoB (beta subunit), and RpoC2 (beta prime subunit)—were detected within the soluble fraction irrespective of the presence or absence of Mg2+ (Fig. 4C). Disruption of sigE did not affect the localization of ChlH (Fig. S3).
ChlH Represses Transcription by RNA Polymerase Containing SigE In Vitro.
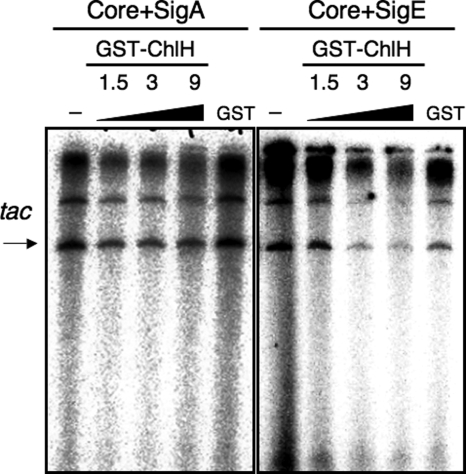
We performed in vitro transcription analysis to examine the posttranslational regulation of SigE by ChlH. Fig. 5 shows that transcription by RNA polymerase containing the group-1 sigma factor SigA was not affected by the addition of GST-ChlH; however, transcription by RNA polymerase containing SigE was repressed in the presence of GST-ChlH in a ChlH concentration–dependent manner. To exclude a possible effect of GST, we purified histidine-tagged ChlH (His-ChlH) from E. coli (Fig. S4A). In vitro transcription analysis revealed that His-ChlH also inhibited transcription by RNA polymerase containing SigE, but not transcription by RNA polymerase containing SigA (Fig. S4B). Although we tested the effects of increased concentrations of Mg2+ (from 3 mM to 20 mM), transcription itself was abolished in the presence of 20 mM MgCl2; thus, we could not examine the effect of Mg2+ on in vitro transcription (Fig. S4C). We also found that an excess amount of GST-ChlH completely abolished the transcription by RNA polymerase containing SigE (Fig. S4D).
Fig. 5.
ChlH repressed the transcriptional activity of SigE in vitro. An in vitro transcription analysis was performed by mixing purified His-SigA or His-SigE (1.5 pmol) with GST-ChlH (0, 1.5, 3, or 9 pmol) or GST (9 pmol), after which the native core of RNA polymerase (1 pmol) and 0.3 pmol of template DNA pKK223–3 was added. The resultant mRNAs were resolved by 5% urea-PAGE, and radioactivity was detected using a BAS-1000 image analyzer.
Discussion
Our findings demonstrate that the H subunit of Mg-chelatase, ChlH, interacts with the sigma factor SigE and represses transcriptional activity in vitro, and that this interaction is controlled by light conditions in vivo. It has been shown that Mg2+ concentrations in the stroma of spinach chloroplasts are altered by light-dark transition (26). The free Mg2+ concentration is about 0.5 mM under dark conditions and increases to about 2 mM during illumination. Although changes in the in vivo concentrations of free Mg2+ were not investigated in Synechocystis 6803, our preliminary analysis with a fluorescence probe mag-fura 2 (Invitrogen) revealed that free Mg2+ concentration in a Synechocystis cell is about 1 mM under light conditions (data not shown). Further analysis is needed to determine the changes of Mg2+ concentration by light-to-dark transition.
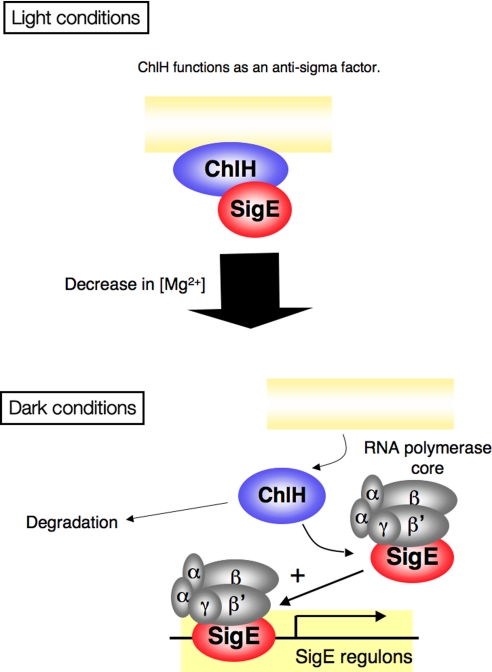
Based on these data, we propose a probable model for SigE regulation by ChlH (Fig. 6). Under light conditions, SigE interacts with ChlH, which is anchored to plasma or thylakoid membranes (Fig. 4C). During transition from light to dark, the decreased Mg2+ concentration causes ChlH to dissociate from the membrane, and the interaction between SigE and ChlH is abolished. Then free SigE is able to associate with the RNA polymerase core and activate transcription of SigE regulons (Fig. 6). In this model, ChlH transduces light signals by Mg2+, not by Proto or Mg-Proto, and regulates SigE at the posttranslational level. This model is consistent with the data showing that ChlH is a negative regulator of SigE transcription (Fig. 5). This model also is consistent with a previous study demonstrating increased expression of OPP pathway genes by light-to-dark transition in a SigE-dependent manner (15).
Fig. 6.
Probable model for the regulation of SigE by ChlH. Under light conditions, ChlH is membrane-associated and interacts with SigE, leading to repression of transcription by RNA polymerase interacting with SigE. When Proto binds to ChlH, ChlH is released from SigE and functions as a Mg-chelatase. During the transition from light to dark, ChlH dissociates from the membrane, and concomitantly, SigE dissociates from ChlH due to the decreasing Mg2+ concentration. As a result, free SigE can complex with the RNA polymerase core, resulting in activation of the transcription of SigE regulons.
ChlH is known as a multifunctional protein. Shepherd et al. (27) reported that ChlH of Synechocystis 6803 accelerates the catalytic activity of ChlM, which catalyzes the conversion of Mg-Proto to Mg-protoporphyrin IX monomethyl ester. In higher plants, the chlH mutant gun5 cannot transduce signals from plastid to nucleus in A. thaliana (28). In A. thaliana, ChlH also functions as a receptor of the plant hormone abscisic acid (ABA) and as a positive regulator of ABA signal transduction (29). We propose a novel function of ChlH as an anti-sigma factor, repressing the transcription of SigE via a protein–protein interaction. In higher plants, ChlH and several sigma factors are localized within the plastid; however, phylogenetic analysis suggests that sigma factors in eukaryotes are derived from group-1 sigma factors of ancient cyanobacteria (30); therefore, regulation of a sigma factor by ChlH may be restricted to several cyanobacterial strains. Nevertheless, a recent study revealed that transcription within chloroplasts was regulated by Mg-Proto level, indicating the significance of tetrapyrrole metabolism for transcription within plastids (31). Thus, future studies may reveal that plastids also have regulatory mechanisms regulating sigma factors via protein–protein interactions with other proteins, including metabolic enzymes, similar to the repression of SigE by ChlH in cyanobacteria.
Materials and Methods
Bacterial Strains and Culture Conditions.
The GT strain of Synechocystis sp. PCC 6803, isolated by Williams (32), was grown in BG-110 liquid medium with 5 mM NH4Cl (buffered with 20 mM Hepes-KOH; pH 8.0), designated modified BG-11 medium. Liquid cultures were bubbled with 2% (vol/vol) CO2 in air at 30 °C under continuous white light (ca. 70 μmol photons m−2 s−1) (33). For plate cultures, BG-11 medium (17.5 mM NaNO3 and 20 mM Hepes-KOH; pH 8.0) was solidified using 1.5% (wt/vol) agar (Nissui) and incubated in air containing 2% (vol/vol) CO2 at 30 °C under continuous white light (ca. 70 μmol photons m−2 s−1). Growth and cell densities were measured at A750 with a Beckman DU640 spectrophotometer.
Yeast Two-Hybrid Analysis.
The full-length sigE gene (sll1689) of Synechocystis 6803 was amplified by PCR using Pfu polymerase (Clontech) and the specific primers 5′-GAGGGCGCGCCATGAGCGATATGTCTTCC-3′ and 5′-CGGTATCTATAACCAACCTTTGAG-3′, and then subcloned into the pAS2–1-AscI bait vector constructed by Sato et al. (19). The details of the yeast two-hybrid screening procedure were as described by Sato et al. (19).
Affinity Purification of GST-ChlH and His-SigE.
A region of the Synechocystis 6803 genome encoding ChlH was amplified by PCR using KOD polymerase (Toyobo) and the specific primers 5′-CGGGATCCGGATGTTTACTAACGTCAAGTC-3′ and 5′-ACTCGAGTTTATTCAACCCCTTCAATG-3′, digested with BamHI and XhoI (Takara), and inserted into the BamHI-XhoI sites of pGEX5X-2 (Amersham Pharmacia Bioscience). The resultant plasmid was designated pGEXChlH. Full-length sigE was amplified using KOD polymerase (Toyobo) and the specific primers 5′-CCGCATGCATGAGCGATATGTCTTCC-3′ and 5′-CCGATATCCTATAACCAACCTTTGAG-3′, digested with SphI and EcoRV (Takara), and inserted into the SphI-SmaI sites of pQE80L (Qiagen). The constructed plasmids encoding GST-ChlH or His-SigE were introduced separately into E. coli BL21 Codon Plus cells (Stratagene) by transformation. Expression was induced by the addition of 1 mM isopropyl-β-D-thiogalactopyranoside (Wako) to 1 L of LB medium, and the cells were cultured overnight at 27 °C. The cells were then collected by centrifugation and lysed in 30 mL of lysis buffer [40 mM Tris-HCl (pH 8.0), 5% glycerol, 5 mM EDTA, and 4.5% Triton X-100] by sonication (Branson Sonifier 450). The soluble fraction and the insoluble fraction that contained recombinant protein were divided by centrifugation of the cell lysate at 17,400 × g for 20 min at 4 °C. For purification of GST-ChlH, 800 μL of glutathione-sepharose 4B (GE Healthcare) was added to soluble fraction and gently mixed for 1 h at 4 °C. After centrifugation at 300 × g, resin incubated with GST-ChlH was washed 5 times with PBS containing 0.1% Triton X-100 and eluted 3 times with 400 μL of GST elution buffer [50 mM Tris-HCl (pH 8.0) and 10 mM reduced glutathione]. For purification of His-SigE, the insoluble fraction was washed with cell lysis buffer, suspended in sterilized water, and solubilized by the addition of half a volume of a solution containing 8 M urea, 50 mM Tris-HCl (pH 8.0), and 10 mM DTT and incubated for 1 h at 37 °C. The solubilized His-SigE proteins were dialyzed against His-binding buffer [50 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 0.1% Triton X-100] and purified with HIS-Select resin (Sigma). Protein concentration was determined using a Bio-Rad protein assay.
GST-Pulldown Analysis.
Purified GST or GST-ChlH was bound to 20 μL of glutathione-sepharose 4B for 30 min at room temperature, mixed with His-SigE in 500 μL of Hepes-binding buffer [50 mM Hepes-KOH (pH 8.0), 5% glycerol, and 0.1% Triton X-100] with MgCl2 at the concentrations indicated in the figure legends, and incubated for 1 h at room temperature. The resin was washed twice with 150 μL of Hepes-binding buffer, suspended in 50 μL of SDS sample buffer [250 mM Tris-HCl (pH 6.8), 20% sucrose, 20% 2-mercaptoethanol, 8% SDS, and 0.04% bromophenol blue], and then heated for 5 min at 96 °C. The released proteins were then subjected to SDS-PAGE and detected by immunoblot analysis with antiserum to SigE (15).
Rabbit ChlH Antiserum and Rat SigE Antiserum Production and Immunoblot Analysis.
Antisera to ChlH or SigE were produced commercially by Tanpaku Seisei Kogyo. Antiserum was raised in a rabbit using 2.0 mg of purified GST-ChlH and in rats using 1.5 mg of purified His-SigE. Immunoblotting with antiserum to ChlH or SigE was performed as described previously (12).
Immunoprecipitation Analysis.
The GT strain of Synechocystis 6803 cells was grown in modified BG-11 (A750 = ≈2.0) and collected by centrifugation (8,000 × g for 5 min). Cells from 0.5-L cultures were resuspended in 1.5 mL of PBS with 1 mM MgCl2, 2 mM PMSF, and 1 tablet of Complete Mini, EDTA-free (Roche), and then disrupted by sonication (Branson). After centrifugation at 17,400 × g for 10 min at 4 °C, the protein concentration of the supernatant was estimated using a BCA protein assay kit (Pierce), and the supernatant was used as a cell extract. Anti-SigE or preimmune antiserum from rat (50 μlL) was mixed with 400 μL of protein G sepharose (GE Healthcare) and 500 μL of PBS. After 30 min of shaking at room temperature, the resins incubated with antibodies were washed 3 times with 500 μL of PBS. Then resins were suspended in 500 μL of PBS, and 0.5 mg/mL of disuccinimidyl suberate (DSS; Pierce) was added to cross-link antibodies to the resins. After 40–50 min of mixing at room temperature, the resins were washed 5 times with 500 μL of glycine elution buffer (0.1 M glycine-HCl; pH 2.7) and 4 times with PBS. Then aliquots of cell extracts containing about 30 mg of total protein were added to the resin, and the mixture was incubated at 4 °C for 1 h. Resins were subsequently washed 3 times with 500 μL of PBS containing 1 mM MgCl2. Immunoprecipitated proteins were eluted with 150 μL of glycine elution buffer. Proteins were detected by immunoblotting with rabbit SigE antiserum (15) or with ChlH.
For immunoprecipitation with anti-ChlH antiserum, 50 μL of anti-ChlH or preimmune antiserum from rabbit was mixed with 200 μL of protein G sepharose (GE Healthcare) and 500 μL of PBS containing 1 mM MgCl2 and then cross-linked as described above. Cell extracts containing ≈25 mg of total protein (prepared in PBS with 1 mM MgCl2) were added to the resin, and the mixture was incubated at 4 °C for 1 h. Resins were then washed with 500 μL of PBS containing 1 mM MgCl2, and immunoprecipitated proteins were eluted with 200 μL of glycine elution buffer. The proteins were detected by immunoblotting with antiserum against SigE produced by rats or antiserum against ChlH produced by a rabbit.
Fractionation of Soluble and Membrane Fractions.
Cells of mid-exponential– phase cultures of the GT strain of Synechocystis 6803 (A750, 0.5–0.7) grown in 120 mL of modified BG-11 medium were collected by centrifugation at 6,600 × g for 4 min and then resuspended in 15 mL of Hepes-KOH (pH 8.0) with or without 10 mM MgCl2. After disruption of cells by sonication, undisrupted cells were removed by centrifugation at 17,400 × g for 20 min; the resulting supernatant was designated the whole cell extract. Then soluble and membrane fractions were divided by ultracentrifugation at 100,000 × g for 1 h. The protein concentrations of each fraction were estimated using a BCA protein assay kit (Pierce).
Affinity Purification of His-ChlH, GST-GS, and His-ChlI.
pGEXChlH was digested by BamHI and XhoI, and the resultant fragment was cloned into pQE82L (Qiagen) digested by BamHI and SalI (Toyobo). The constructed plasmid encoding His-ChlH was introduced into E. coli BL21 Codon Plus (Stratagene) by transformation. The expression of His-ChlH was induced by the addition of 1 mM isopropyl-β-D-thiogalactopyranoside (Wako) to 1 L of LB medium, after which the cells were cultured overnight at 27 °C. The cells were then collected by centrifugation, lysed in 30 mL of His-binding buffer by sonication (Branson Sonifier 450), and centrifuged at 17,400 × g for 20 min at 4 °C. The soluble fraction was gently mixed with 800 μL of HIS-Select for 1 h at 4 °C. After centrifugation at 300 × g, resin incubated with His-ChlH was washed 5 times with His-binding buffer containing 5 mM imidazol and eluted with 400 μL of His-binding buffer containing 500 mM imidazol.
A region of glnA encoding GS was amplified by PCR using KOD polymerase (Toyobo) and the primers 5′-CATAGATCTTAATGGCCAGAACCCCCCA-3′ and 5′-GGCTCGAGCCTTAGCAGTCGTAGTACAAG-3′, digested by BglII and XhoI, and subcloned into pGEX5X-2 (Amersham Pharmacia Bioscience) digested by BamHI and XhoI. Expression and purification of GST-GS were performed as GST-ChlH. A region of chlI encoding the I subunit of Mg-chelatase was similarly amplified and cloned into a pQE82L vector with the primers 5′-CATAGATCTATGACTGCCACCCTTGCC-3′ and 5′-AAGATATCTTAAGCTTCATCGACAAC-3′. Expression and purification of His-ChlI were performed as His-ChlH. Rabbit antisera to GS and ChlI was produced commercially by Tanpaku Seisei Kogyo.
In Vitro Transcription Analysis.
His-SigA was expressed from an expression vector constructed previously (34) and purified from insoluble fractions as described for His-SigE (see above). In vitro transcription analysis was performed as described by Imashimizu et al. (35) with some modifications. Purified His-SigA or His-SigE (1.5 pmol) was mixed with GST-ChlH or His-ChlH in 10 μL of transcription buffer [50 mM Hepes-KOH (pH 8.0), 3 mM MgCl2, 1 mM DTT, 50 mM potassium glutamate, and 25 μg/mL BSA], and the mixture was incubated for 10 min at 30 °C. Then the native core of the RNA polymerase (1 pmol) of the thermophilic cyanobacteria Thermosynechococcus elongatus BP-1 was added to the mixture and incubated for 20 min at 30 °C. Subsequently, 25 μL of transcription buffer containing 0.3 pmol of template DNA pKK223–3 (GE Healthcare) was mixed with 10 μL of RNA polymerase mixture and then incubated for 20 min at 30 °C. RNA synthesis was initiated by the addition of a 15-μL prewarmed substrate mixture containing 160 μM each of ATP, GTP, and CTP as well as 50 μM UTP, 100 μg/mL of heparin, and 2 μCi [α-32P] of UTP (MP Bio Japan). After incubation for 5 min at 30 °C, the reaction was quenched by the addition of 50 μL of stop solution [40 mM EDTA-NaOH (pH 8.0) and 300 mg/mL of E. coli tRNA]. After ethanol precipitation, transcripts were separated by PAGE on a gel containing 5% polyacrylamide and 8 M urea, after which radioactivity was analyzed using a BAS1000 image analyzer (Fujifilm).
Supplementary Material
Acknowledgments.
We thank Drs. Neil C. Hunter, Alison Smith, Ayumi Tanaka, Nobuyoshi Mochizuki, Kintake Sonoike, and Tatsuru Masuda for helpful discussions. This work was supported by a Grant-in-Aid for Creative Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (16GS0304, to K.T.); a Grant-in-Aid for JSPS Fellows from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T.O.); and a Grant-in-Aid for Scientific Research for Plant Graduate Students from the Nara Institute of Science and Technology, supported by The Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T.O.).
Footnotes
The authors declare no conflicts of interest.
This article is a PNAS Direct Submission. C.E.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810040106/DCSupplemental.
References
- 1.Bhardwaj A, Wilkinson MF. A metabolic enzyme doing double duty as a transcription factor. BioEssays. 2005;27:467–471. doi: 10.1002/bies.20232. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Roeder RG, Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 3.Hall DA, et al . Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- 4.Wary LV, Jr, Zalieckas JM, Fisher SH. Bacillus subtilis glutamine synthetase controls gene expression through a protein–protein interaction with transcription factor TnrA. Cell. 2001;107:427–435. doi: 10.1016/s0092-8674(01)00572-4. [DOI] [PubMed] [Google Scholar]
- 5.Cho Y-H, Yoo S-D, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127:579–589. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 6.Helmann JD. Anti-sigma factors. Curr Opin Microbiol. 1999;2:135–141. doi: 10.1016/S1369-5274(99)80024-1. [DOI] [PubMed] [Google Scholar]
- 7.Minakhin L, Severinov K. Transcription regulation by bacteriophage T4 AsiA. Protein Expression Purif. 2005;41:1–8. doi: 10.1016/j.pep.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Chilcott KF, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudner DZ, Losick R. Morphological coupling in development: Lessons from prokaryotes. Dev Cell. 2001;1:733–742. doi: 10.1016/s1534-5807(01)00094-6. [DOI] [PubMed] [Google Scholar]
- 10.Inoue-Sakamoto K, et al. Group 3 sigma factors in the marine cyanobacterium Synechococcus sp. strain PCC 7002 are required for growth at low temperature. J Gen Appl Microbiol. 2007;53:89–104. doi: 10.2323/jgam.53.89. [DOI] [PubMed] [Google Scholar]
- 11.Morikawa K, Shiina T, Murakami S, Toyoshima Y. Novel nuclear-encoded proteins interacting with a plastid sigma factor, Sig1, in Arabidopsis thaliana. FEBS Lett. 2002;514:300–304. doi: 10.1016/s0014-5793(02)02388-8. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. Heterogeneity of the principal sigma factor in Escherichia coli: The rpoS gene product, sigma 38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli. Proc Natl Acad Sci U S A. 1993;90:3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goto-Seki A, Shirokane M, Masuda S, Tanaka K, Takahashi H. Specificity cross-talk among group 1 and group 2 sigma factors in the cyanobacterium Synechococcus sp. PCC7942: In vitro specificity and a phylogenetic analysis. Mol Microbiol. 1999;34:473–448. doi: 10.1046/j.1365-2958.1999.01608.x. [DOI] [PubMed] [Google Scholar]
- 14.Kaneko T, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803, II: Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 15.Osanai T, et al. Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp. PCC 6803 by the group 2 σ factor SigE. J Biol Chem. 2005;280:30653–30659. doi: 10.1074/jbc.M505043200. [DOI] [PubMed] [Google Scholar]
- 16.Osanai T, Azuma M, Tanaka K. Sugar catabolism regulated by light and nitrogen status in the cyanobacterium Synechocystis sp. PCC 6803. Photochem Photobiol Sci. 2007;6:508–514. doi: 10.1039/b616219n. [DOI] [PubMed] [Google Scholar]
- 17.Yang C, Hua Q, Shimizu K. Integration of the information from gene expression and metabolic fluxes of the regulatory mechanisms in Synechocystis. Appl Microbiol Biotechnol. 2002;58:813–822. doi: 10.1007/s00253-002-0949-0. [DOI] [PubMed] [Google Scholar]
- 18.Imamura S, et al. Antagonistic dark/light-induced SigB/SigD, group 2 sigma factors, expression through redox potential and their roles in cyanobacteria. FEBS Lett. 2003;554:357–362. doi: 10.1016/s0014-5793(03)01188-8. [DOI] [PubMed] [Google Scholar]
- 19.Sato S, et al. A large-scale protein–protein interaction analysis in Synechocystis sp. PCC 6803. DNA Res. 2007;14:207–216. doi: 10.1093/dnares/dsm021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papenbrock J, Grimm B. Regulatory network of tetrapyrrole biosynthesis: Studies of intracellular signaling involved in metabolic and developmental control of plastids. Planta. 2001;213:667–681. doi: 10.1007/s004250100593. [DOI] [PubMed] [Google Scholar]
- 21.Reid JD, Hunter CN. Current understanding of the function of magnesium chelatase. Biochem Soc Trans. 2002;30:643–645. doi: 10.1042/bst0300643. [DOI] [PubMed] [Google Scholar]
- 22.Osanai T, et al. Nitrogen induction of sugar catabolic gene expression in Synechocystis sp. PCC 6803. DNA Res. 2006;13:185–195. doi: 10.1093/dnares/dsl010. [DOI] [PubMed] [Google Scholar]
- 23.Karger GA, Reid JD, Hunter CN. Characterization of the binding of deuteroporphyrin IX to the magnesium chelatase H subunit and spectroscopic properties of the complex. Biochemistry. 2001;40:9291–9299. doi: 10.1021/bi010562a. [DOI] [PubMed] [Google Scholar]
- 24.Gibson LC, et al. A putative Mg chelatase subunit from Arabidopsis thaliana cv C24: Sequence and transcript analysis of the gene, import of the protein into chloroplasts, and in situ localization of the transcript and protein. Plant Physiol. 1996;111:61–71. doi: 10.1104/pp.111.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakayama M, et al. Cloning and expression of the soybean chlH gene encoding a subunit of Mg-chelatase and localization of the Mg2+ concentration–dependent ChlH protein within the chloroplast. Plant Cell Physiol. 1998;39:275–284. doi: 10.1093/oxfordjournals.pcp.a029368. [DOI] [PubMed] [Google Scholar]
- 26.Ishijima S, Uchibori A, Takagi H, Maki R, Ohnishi M. Light-induced increase in free Mg2+ concentration in spinach chloroplast: Measurement of free Mg2+ by using a fluorescent probe and necessity of stromal alkalinization. Arch Biochem Biophys. 2003;412:126–132. doi: 10.1016/s0003-9861(03)00038-9. [DOI] [PubMed] [Google Scholar]
- 27.Shepherd M, McLean S, Hunter CN. Kinetic basis for linking the first two enzymes of chlorophyll biosynthesis. FEBS J. 2005;272:4532–4539. doi: 10.1111/j.1742-4658.2005.04873.x. [DOI] [PubMed] [Google Scholar]
- 28.Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci U S A. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen Y-Y, et al. The Mg-chelatase H subunit is an abscisic acid receptor. Nature. 2006;443:823–826. doi: 10.1038/nature05176. [DOI] [PubMed] [Google Scholar]
- 30.Minoda A, et al. Microarray profiling of plastid gene expression in a unicellular red alga, Cyanidioschyzon merolae. Plant Mol Biol. 2005;59:375–385. doi: 10.1007/s11103-005-0182-1. [DOI] [PubMed] [Google Scholar]
- 31.Ankele E, Kindgren P, Pesquet E, Strand A. In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell. 2007;19:1964–1979. doi: 10.1105/tpc.106.048744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams JGK. Construction of specific mutations in the Photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 1988;167:766–778. [Google Scholar]
- 33.Rippka R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 34.Seki A, et al. Light-responsive transcriptional regulation of the suf promoters involved in cyanobacterium Synechocystis sp. PCC 6803 Fe-S cluster biogenesis. FEBS Lett. 2006;580:5044–5048. doi: 10.1016/j.febslet.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 35.Imashimizu M, Hanaoka M, Seki A, Murakami KS, Tanaka K. The cyanobacterial principal sigma factor region 1.1 is involved in DNA binding in the free form and in transcription activity as a holoenzyme. FEBS Lett. 2006;580:3439–3444. doi: 10.1016/j.febslet.2006.05.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.