Abstract
Asymmetric cell division is of fundamental importance in biology as it allows for the establishment of separate cell lineages during the development of multicellular organisms. Although microbial systems, including the yeast Saccharomyces cerevisiae, are excellent models of asymmetric cell division, this phenotype occurs in all cell divisions; consequently, models of lineage-specific segregation patterns in these systems do not exist. Here, we report the first example of lineage-specific asymmetric division in yeast. We used fluorescent tags to show that components of the yeast kinetochore, the protein complex that anchors chromosomes to the mitotic spindle, divide asymmetrically in a single postmeiotic lineage. This phenotype is not seen in vegetatively dividing haploid or diploid cells. This kinetochore asymmetry suggests a mechanism for the selective segregation of sister centromeres to daughter cells to establish different cell lineages or fates. These results provide a mechanistic link between lineage-defining asymmetry of metazoa with unicellular eukaryotes.
Keywords: meiosis, yeast spores
It is well known that the budding yeast, Saccharomyces cerevisiae, divides asymmetrically to produce phenotypically distinct mother and bud cells. This striking phenotype makes S. cerevisiae an excellent model of cellular polarity and asymmetric cell division (1). Mutational analysis combined with cell biology and physical studies have identified a number of the key regulators of this polarity (2–5). Some of these asymmetries such as mating-type switching are specific to yeast (6, 7), but others, such as the cell cycle control of polarity or aging, are likely conserved in metazoans (8, 9). A key feature of asymmetric yeast cell divisions is that they occur during each and every cell cycle. Hence, yeast are not thought to undergo repeated asymmetric divisions that are confined to a single lineage of cells, such as those seen in the development of metazoa or in stem cell niches (both patterns of asymmetry are illustrated in Fig. S1).
Unlike previous studies, here, we show that members of 4 separate kinetochore subcomplexes undergo asymmetric segregation specifically in a single yeast lineage. We used a simple and innovative strategy to track the segregation of fluorescently-tagged proteins from a spore to its progeny. Because we focus on spores that have lost the gene encoding the fluorescent protein during meiosis, the protein levels are unaffected by transcription. We observed the asymmetric phenotype only in the division of yeast spores immediately after meiosis and in the mother lineage derived from these spores, and not in vegetatively growing haploid or diploid cells. Because this asymmetry is evident for kinetochore proteins, which tether centromere sequences to the mitotic spindle, it has the potential to segregate centromeres, if not whole chromosomes, in a specific pattern. A nonrandom sister chromatid segregation pattern has long been proposed to occur in multiple organisms, including yeast (10, 11). Such a mechanism may allow the cell to avoid the accumulation of replication errors in a defined lineage (12), or may enable meiotic selection to drive the evolution of centromeric sequences (13, 14). The phenotype described here provides an avenue through which such a nonrandom pattern of chromatid segregation could be achieved and concurrently provides a new yeast model for the cellular asymmetry characterized in specific populations of metazoan cells, such as the asymmetric divisions reported in various stem cell lineages.
Results
Method to Track Nonencoded Protein in Yeast.
We reasoned that if a unique lineage does exist in yeast, it would be established during the emergence of a new strain. Under normal circumstances there are only 2 moments in the yeast life cycle when a new strain is created: when 2 haploid yeast cells mate to create a new diploid and during meiosis when a diploid cell produces 4 new haploid spores. We decided to study meiosis, because this is a likely instance in which molecular programs would be reset within the nucleus. In addition, the transition from diploid to haploid allowed us to develop a simple method to follow the segregation of protein without having to consider the confounding effects of transcription. In a diploid strain, we tagged the protein encoded by 1 allele of a gene with yellow fluorescent protein (YFP) and the other allele with cyan fluorescent protein (CFP). We next induced this diploid strain to enter meiosis and produce 4 haploid spores, each of which inherits only one of the tagged alleles, but both of the tagged proteins. In this way, we were able to observe the nonencoded protein inherited from the original diploid cell as it segregated from the spore to its daughter cells (Fig. 1). Differential levels of transcription in progeny cells could confound our results, but our method of quantitating nonencoded protein avoids this problem.
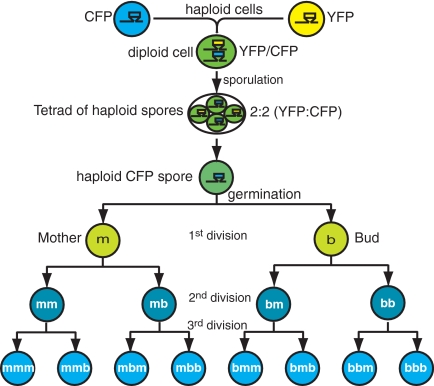
Fig. 1.
A diagram of the double-fluorescent tag method. Two haploid cells containing a given gene fused to either one of CFP (blue) or YFP (yellow) fluorescent tags were created. These 2 haploids were mated together to form a diploid that contained the gene in question, tagged with heterozygous fluorescent markers (CFP/YFP, shown as green). After sporulation, the individual haploid spores only contained either the CFP or YFP tagged gene, but inherited both CFP and YFP fluorescent protein from the parental diploid. Because the spores divided to produce successive generations of progeny, the protein that is no longer genetically encoded was diluted away. In this case, we illustrate a CFP spore that loses its inherited YFP protein (i.e., becomes more blue) in successive generations.
Asymmetry of Mtw1.
We first examined MTW1, a homolog of the mammalian MIS12 gene, whose protein forms part of the conserved Mtw1/MIND kinetochore complex that promotes biorientation of the mitotic spindle (15, 16). Elegant studies have shown that Mtw1 protein levels can be assessed using fluorescent intensity of tagged protein (17). Using time-lapse fluorescence microscopy, we followed individual spores as they divided and were able to track the segregation of both nonencoded and total Mtw1 protein that localized to the kinetochore.
For each spore, we first quantitated the total amount of both yellow (YFP) and cyan (CFP)-tagged Mtw1 protein in the kinetochore, as the spore went through 3 rounds of cell division (Figs. 2 and 3 and Fig. S2). At the first division, a spore divides to give a mother (m) and bud (b) (Figs. 1 and 2). In the second round of division, each cell divides again into mother and bud: mm, mb (descended from the mother) and bm, bb (descended from the bud). In the third round of division, each of the 4 cells divides again into mother and bud: mmm, mmb, etc. Because all of the yeast kinetochores colocalize to a single locus in the nucleus, the tagged Mtw1 protein is visualized as a single fluorescent focus (actually 2 foci before mitosis). It was this fluorescence that was used to quantify the levels of the tagged kinetochore protein immediately after cytokinesis. The ratio of both nonencoded and total protein in the mother versus the bud (m/b) was calculated for 3 rounds of division to assess potential asymmetry.
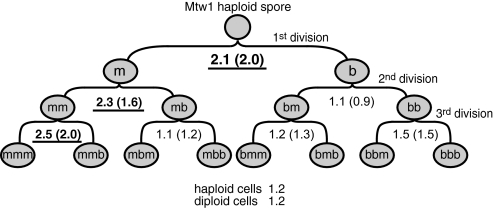
Fig. 2.
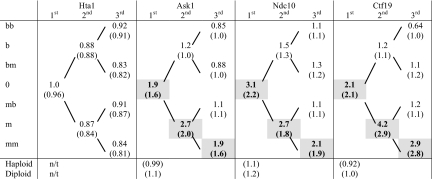
The distribution of Mtw1 protein in postmeiotic cell divisions. For each separate cell division, the ratio of nonencoded protein in the kinetochore of the mother versus the bud cell was calculated (m/b ratio). The mean value of these m/b ratios is shown for each division. The m/b ratios that show asymmetry in the mother lineage are underlined and in bold. The m/b ratios of the mother lineage (n = 88 separate cell divisions) versus the other lineages (n = 51 separate cell divisions) are significantly different (P = 8.8 × 10−6 using a 2-tailed t test with unequal variance). In parentheses, the same ratios are calculated using the measurements of the total protein in the kinetochore. Additionally, the mean ratio of distribution of protein between mother and bud in haploid and diploid cells is shown. The raw quantitative data are in Dataset S1, and the summary statistics are in Table S1.
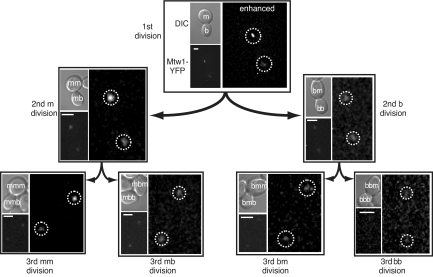
Fig. 3.
A spore derived from the diploid strain W7247, MTW1-YFP/MTW1-CFP, is shown dividing 3 times. The images are arranged as in Fig. 2. Each image shows a differential interference contrast (DIC) image (Upper) and below a fluorescent YFP image (YFP, the white scale bar is 2.5 μm). Right images in each example are contrast enhanced to illustrate the (a)symmetry of each division – the kinetochore foci are located inside the dashed circle. For clarity, only the encoded protein is shown, in this case YFP. The quantitative analysis of the fluorescence levels in this lineage is summarized in Dataset S1 (Mtw1 spore AM). More examples or Mtw1 asymmetry are provided in the Fig. S2.
Through this analysis we found that the kinetochore of the mother cells contains more Mtw1 protein than the bud, but only in the mother cell lineage directly descended from the spore. In the first division, the kinetochore of the m cell contains over twice as much nonencoded Mtw1 as that of the b cell, on average (m/b ratio of 2.1) (Fig. 2). In the second round of division, Mtw1 segregates asymmetrically from the m cell to the new mother (mm, with a m/b ratio of 2.3) but segregates symmetrically from the b cell to its daughters (m/b ratio of 0.9). In the third division, Mtw1 segregates preferentially from the mm cell to the third generation mother cell (mmm, with a m/b ratio of 2.5) while segregating evenly in all other divisions (m/b ratios of 1.1, 1.2 and 1.5). The mean m/b ratio of nonencoded Mtw1 segregation in the mother lineage is 2.2 (SEM = 0.22, n = 85), which is significantly different (P = 8.8 × 10−6 using a 2-tailed t test with unequal variance) from that of the nonmother lineage (mean m/b ratio of 1.1, SEM = 0.07, n = 46; see Table S1).
This asymmetry is not restricted to the nonencoded protein; when examining total Mtw1 protein (which includes both encoded and nonencoded) at the kinetochore in both the spores and their descendants, the same asymmetric segregation pattern is observed (ratios shown in parentheses in Fig. 2). By distinguishing nonencoded from total protein at the kinetochore, we show that this asymmetric phenotype is not due to mother-specific overexpression of the MTW1 gene in this lineage, but instead can be attributed to differential protein segregation. Although not all mother lineage divisions show asymmetric Mtw1 segregation, the frequency of asymmetric division in the mother lineage is much higher than the other lineages, summarized in Fig. S3. We note that the mother-lineage divisions that do not show Mtw1 asymmetry, instead segregate symmetrically (mean m/b ratio = 1.1). Because it has been found that there is an intrinsic asymmetry in the 4 spores of a tetrad (18), it is possible that not all spores in a tetrad show lineage-specific asymmetry. However, we cannot easily test this hypothesis because we are unable to analyze all 4 products of a single tetrad.
Finally, we examined the segregation of total Mtw1 protein at the kinetochore in vegetatively growing haploid and diploid cells and found that it is symmetrical (m/b ratios of 1.2) irrespective of whether the protein was tagged with either YFP or CFP (Fig. 2). Thus, the asymmetric Mtw1 segregation phenotype is specific to the postmeiotic mother lineage.
Symmetric Division of Histones.
Next, we tested whether the asymmetric segregation pattern of Mtw1 that was observed could be explained by nonspecific asymmetric segregation of all nuclear proteins in this pedigree of cells. It is known that, in yeast, damaged protein preferentially localizes to mother cells (19) and it is possible that this phenotype is more pronounced in postmeiotic cells than in haploid or diploid cells. It is also possible that the asymmetric Mtw1 phenotype is caused by some unforeseen postmeiotic effect caused by the fluorescent protein tags. To test these hypotheses, we repeated our experiments by differentially tagging the alleles that encode the H2A histone protein, Hta1. A diploid strain containing both HTA1-YFP and HTA1-CFP (W7912) was sporulated and the amounts of both nonencoded and total Hta1 in the nucleus of individual spores and their progeny were quantitated (Fig. 4). Hta1 protein (either nonencoded or total) segregated equally to the mother and bud at the first, second and third postmeiotic divisions (ratio of 0.96, 0.89 and 0.84 respectively, Table 1). The average m/b ratio for all divisions of the mother lineage (mean m/b ratio = 0.90) is not significantly different from those of the other lineages (mean m/b ratio = 0.88). These data indicate that the asymmetric segregation of Mtw1 is not a general feature of postmeiotic nuclear proteins in yeast. Additionally, two other proteins, TetR (the tetracycline repressor) and Rad52 (a central DNA repair protein), segregate equivalently in both mother and non-mother lineages (SI Methods).
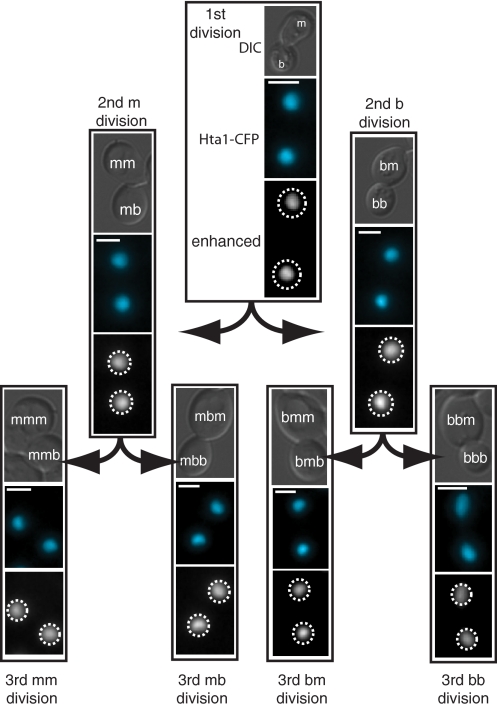
Fig. 4.
A spore derived from diploid strain W7912, HTA1-YFP/HTA1-CFP, is shown dividing 3 times; the images are arranged as in Fig. 3. The encoded CFP protein is shown, artificially colored to 480 nm. (Scale bar: 2.5 μm.) The dashed circles indicate the position of the nucleus in the contrast-enhanced images. The quantitative analysis of this lineage can be found in Dataset S1 (Hta1 spore 1-2).
Table 1.
Segregation of non-encoded (and total) protein during post-meiotic cell divisions
The mean m/b ratios of histone H2A (Hta1) and three kinetochore proteins (Ask1, Ndc10, Ctf19) are shown for post-meiotic cell divisions originating with the spore (0), as shown in Figure 2. The mean m/b ratios are shown both for non-encoded and, in parentheses, total protein. The asymmetric m/b ratios for the mother lineage are shaded and in bold. T-tests of the m/b ratios for nonencoded protein comparing the mother lineage against the other lineages are significantly different (P values of 4 × 10−4, 2 × 10−4 and 1 × 10−3 for Ask1, Ndc10 and Ctf19 respectively, two tailed, unequal variance). However, the segregation of histone Hta1 is equivalent in the mother and non-mother lineages (Hta1 gives a p values of 0.98, two tailed, unequal variance). Additionally, the mean m/b ratio of both haploid and diploid cells for Ask1, Ndc10 and Ctf19 are ≈1 (all cell divisions are combined). These values are in parentheses since they are measurements of total protein; n/t indicates ″not tested″. The raw quantitation data are in Dataset S1 together with a table of summary statistics, Table S1.
Asymmetry of 3 Other Kinetochore Proteins.
To explore whether the postmeiotic asymmetric segregation was unique to Mtw1, we examined 3 other kinetochore proteins, Ask1, Ndc10 and Ctf19. These proteins are each part of separate kinetochore complexes associated with either the centromeric DNA (Ndc10 and Ctf19) or the spindle microtubules (Ask1). We chose Ndc10 in particular because it is required to localize Mtw1 to the kinetochore (20). We sporulated 3 diploid strains containing differentially tagged Ask1, Ndc10 or Ctf19 (W7908, W7910 and W7909 respectively) and observed the spores through subsequent divisions. Like Mtw1, the nonencoded protein at the kinetochore segregates asymmetrically only in the pedigree of mother cells descended directly from the spore and not in the other lineages (Table 1 and Fig. S2). As before, total protein in the kinetochore segregated in the same asymmetric pattern as nonencoded protein. We saw no asymmetric segregation of these 3 proteins in either vegetatively growing haploid or diploid cells (Table 1). Consequently, the lineage-specific asymmetry phenotype appears to be a feature of the whole kinetochore and is not a unique feature of the Mtw1 protein.
Discussion
We have identified a unique pattern of asymmetric protein segregation in a single postmeiotic pedigree of S. cerevisiae, defined by the haploid spore and the mother lineage descended from it. Mother cells derived from a bud do not show this phenotype, nor do buds themselves. To the best of our knowledge, this is the first report of a phenotype in microorganisms that is passed down within, and defines, 1 single lineage of cells in a population (Fig. S1).
We find that 4 distinct kinetochore proteins—Ndc10, Ctf19, Mtw1, and Ask1—show the same asymmetric phenotype after meiosis. Ndc10, a member of the CBF3 complex, is part of the centromere-bound inner kinetochore (21). Ctf19 and Mtw1 are part of the COMA and MIND complexes respectively, which appear to bridge the inner and outer kinetochore (22); Ask1, a member of the DAM/DASH complex, is part of the outer kinetochore and interacts with the spindle microtubules (23). Because these 4 complexes span the kinetochore, it seems likely that the postmeiotic asymmetry is a general feature of the whole kinetochore. It is important to note that our results do not address the issue of whether or not the kinetochore complex itself is disassembled during S phase (17, 24, 25).
The function of the kinetochore asymmetry described here is not yet known. It is possible that this asymmetry is linked to that of the yeast spindle pole body, which is the yeast microtubule-organizing center (centrosome) that separates the kinetochores during cell division. The spindle pole body (SPB) proteins in yeast divide asymmetrically (26), the old spindle pole segregates to the bud, although this phenotype is not confined to a single lineage. This SPB asymmetric division is controlled by the cyclin dependent kinase Clb4-Cdc28 (27). In addition, a unique pattern of SPB asymmetry is observed during meiosis (18). Intriguingly, proteins in the equivalent metazoan centrosome are found to segregate asymmetrically in certain stem cell lineages (28). As in yeast, the function of this asymmetry remains elusive, however, it may provide an important structural reference for cell division (29). An important caveat to this idea is the remarkable finding that Drosophila centrosomes are not essential for mitosis or indeed most aspects of development (30). It will be interesting to determine whether the postmeiotic kinetochore segregation pattern that we have observed is linked to spindle pole body asymmetry.
Because the kinetochores are directly responsible for segregating chromatids during mitosis, a tempting hypothesis is that the asymmetric segregation is involved in nonrandom segregation of sister chromatids to daughter cells immediately after meiosis. For example, in metazoan stem cell lineages, the “immortal strand” hypothesis proposes that 1 strand of DNA from every chromosome is passed from 1 stem cell to the next by nonrandom chromatid segregation, thereby preventing stem cells from inheriting and accumulating DNA replication errors (12). This idea is controversial and has received experimental support in some adult stem cell lineages but not in others (31, 32). In yeast, there is conflicting evidence for the existence of nonrandom sister chromatid segregation (33, 34), but these studies do not, as we have here, examine postmeiotic yeast spores and their subsequent divisions.
An alternative view to whole-genome segregation is that only certain chromosomes segregate strands asymmetrically into a single lineage. Such a mechanism may allow the preservation of epigenetic differences at specific genomic loci within a defined cellular pedigree, and has been termed the “silent sister” hypothesis (32). Intriguingly, such a mechanism has received experimental support using a genetic readout in mammalian cells (35). The clearest way to test the immortal strand hypothesis in any system is to use DNA labeling to track strand segregation. However, this approach is difficult because cells that fail to divide (i.e., are quiescent) will retain label regardless of their DNA strand-segregation pattern. Further, such a strategy would fail to detect partial genome asymmetry.
Immortal chromosome strands in yeast seem unlikely, however, because of the occurrence of sister chromatid exchange (SCE). After DNA replication, homologous recombination between the 2 sister chromatids before mitosis would negate strand asymmetry because the recombined chromosome arms would be a mix of old and new DNA strands. Because SCE is estimated to be a frequent event in mitotic yeast (36, 37), it is unlikely that a whole chromosome strand would persist intact in yeast for multiple generations. One possibility is that SCE is inhibited in the postmeiotic yeast lineage to allow a chromatid DNA strand to remain distinct from its sister. To explore whether recombination is specifically inhibited in the postmeiotic mother lineage, we tested if a central homologous recombination protein, Rad52, forms foci in spores and their immediate descendents. Rad52 foci are indicative of double strand break repair by homologous recombination (38). An absence of these foci in the postmeiotic lineage suggests but does not prove an absence of SCE. However, we find that Rad52 foci are seen in both dividing spores and in the mother cells at the next division (“m” in Figs. 1 and 2; SI Methods), suggesting that recombination is active in this lineage of cells. Nevertheless, Rad52 foci may not be indicative of SCE and, indeed, SCE may be inhibited in the dividing spores.
An alternative view to “immortal strands” is that the kinetochore asymmetry found in the postmeiotic cells preferentially segregates only centromeric DNA to a single lineage. Because budding yeast centromere sequences are unidirectional, they have the potential to direct strand asymmetry. This strand distinction could be provided either by the centromere sequences themselves or by the direction of DNA synthesis through the centromere region. This latter idea has been proposed as a means to drive nonrandom chromatid segregation (11) and recent experiments demonstrate that such a mechanism exists in Escherichia coli (39). If a centromeric DNA or protein “mark” is established during meiosis, it could be bound or retained specifically by the asymmetrically dividing kinetochore, allowing cosegregation of centromere strands on the basis of their orientation (Fig. S4). For example, if kinetochores bind preferentially to a single strand of the centromere, this could facilitate strand discrimination. It should be possible to test this hypothesis using a dicentric chromosome in which both centromeres are aligned in the same orientation. Our prediction is that, in this case, an unusually low rate of dicentric chromosome breakage would be observed in the first division after meiosis and subsequently in the postmeiotic mother lineage.
It is possible that kinetochore asymmetry is important for the evolution of centromeres. Although the kinetochore is highly conserved, centromere sequences and their DNA binding proteins appear to be diverging rapidly in metazoa (13, 40). It is suggested that meiotic drive, which selects for specific products of meiosis, is responsible for this process during metazoan female meiosis (14, 40). Because female meiosis results in a single oocyte, as the other polar bodies degenerate, there is the potential to preferentially segregate specific alleles into the egg. Such meiotic drive is suggested to underlie the rapid divergence of centromeres and, by extension, drive speciation (41). Interestingly, recent sequencing studies from Saccharomyces paradoxus, a close relative of S. cerevisiae, show that the CDE II centromere sequences are undergoing unusually rapid divergence (42). This observation suggests that, like the centromeres of metazoa, yeast centromeres are also subject to drive. However, unlike female meioses, all 4 of the yeast meiotic products are viable. Hence, drive would need to be accomplished in another way. For example, if parental centromere strands are confined to a single lineage in postmeiotic yeast, then perhaps the kinetochore asymmetry described here enables a “postmeiotic drive” to select for retention of evolving centromeres sequences in the mother lineage. Centromeres whose sequences bind optimally to the coevolving kinetochore proteins would be preferentially retained in a single lineage derived from the spore, whereas evolutionarily less fit “old” centromeres would be distributed equally throughout the rest of the population.
Yeast has proven to be a useful model for a wide range of fundamental processes of multicellular organisms. Our results show that an additional unexpected phenotype—a stem cell-like, lineage-specific asymmetric cell division—is present and can be studied in this genetically tractable microorganism. Indeed, it should be possible to screen the yeast gene disruption library to identify the genes responsible for this lineage-specific asymmetry.
Materials and Methods
Strains and Media.
The yeast strains used in this study are listed in Table S2. The sporulation medium consisted of 2.5 g/L yeast extract, 1 g/L glucose, 20 g/L potassium acetate, 1.5% wt/vol bacto-agar, supplemented with 75 mg/L adenine, lhistidine, l-leucine, l-lysine, l-methionine, l-tryptophan and uracil adjusted to pH 7.0. Synthetic complete medium plus adenine (SC+Ad) consists of 1.72 g/L yeast nitrogen base, 5 g/L ammonium sulfate, 20 g/L glucose and supplemented with 20 mg/L, l-arginine, l-histidine, l-methionine, uracil; 30 mg/L l-isoleucine and l-tyrosine; 50 mg/L l-phenylalanine; 60 mg/L l-leucine; 120 mg/L adenine; 150 mg/L l-valine adjusted to pH 5.6. Haploid and diploid strains used for microscopy were grown overnight at 23 °C in SC+Ad medium and then prepared for microscopy.
Sporulation.
Diploid yeast were sporulated on solid sporulation medium for 3 days at 30 °C. Tetrads were then placed into liquid SC+Ad at 23 °C for 2–6 h to promote germination and breakage of the ascal wall. After brief vortexing, tetrads separated into individual spores, which were then prepared for microscopy.
Microscopy.
Two methods were used for sample preparation. In the first, equal volumes (1–2 μL) of 1.2% low melting point agarose made in SC+Ad (FMC Bioproducts) and yeast culture were mixed directly on a microscope slide (38). For the second, 2–3 μL of culture was mounted on an 1.4% low melting point agarose pad (43). Both methods yielded equivalent results. Cells were mounted on 76 × 25 mm plain glass microscope slides with a 22 × 22 mm no. 1.5 coverglass (Corning).
Time-lapse fluorescence microscopy was performed with a Zeiss Axioplan II microscope using a 100×, 1.4 NA, oil-immersion differential interference contrast (DIC) objective (Carl Zeiss). Images were captured with an Orca-ER CCD camera (Hamamatsu). To minimize photobleaching, a 10% neutral density filter was used to reduce the exposure of samples to fluorescent light from a 100W mercury arc lamp (Osram). Fluorescently-tagged proteins were visualized using the following fluorescent filter sets: YFP, CFP and RFP (catalog nos. 41028, 31044v2, and 41002c respectively; Chroma). A stack of vertical images (z stack, 0.3–0.5 μm apart) was collected for each cell, at each time point. Images were typically taken at 60–90 min intervals to minimize photo-bleaching. Fluorescence image acquisition times for the tagged proteins used in this study were 1 second. To keep the microscope in focus for the duration of the experiment, a low concentration (<100 per slide) of 1-μm diameter polystyrene red fluorescent beads (catalog no. F13083; Molecular Probes) was included in the agar, and 1 bead was used to reset the plane of focus at each time point, using the autofocus module of Openlab imaging software (Improvision). A motorized microscope stage was used to allow the imaging of several cells on a single slide (BioPrecision2 with linear encoder, Ludl Electronic Products). In this way, images of multiple distinct cells were captured at regular intervals.
To capture later cell divisions in a given lineage, DIC images alone were taken for the first 5–6 h. After this time, fluorescent images were captured as described above. This allowed for the detection of a fluorescent signal at the later time points.
Elimination of Diploid Cells.
Because sporulation of diploid yeast is rarely 100% efficient and spores were not purified, there was a risk that diploid cells would be mistakenly analyzed. To avoid this problem, spores were identified in 4 different ways: first by morphology, because the spores have a distinctive small spherical appearance. Second, the spore wall often has a higher level of autofluorescence in the CFP and RFP channels than the normal yeast cell wall, a phenotype that is easily distinguished in fluorescent images. Third, haploid mother cells typically have sequential axial bud sites adjacent to each other, which contrast with diploids, which bud sequentially from opposite ends of the cell [although the W303 genetic background contains a mutant bud4 allele, which partially disrupts this haploid budding pattern (44)]. Finally, as a definitive mark of ploidy for the Mtw1 experiments, an array of tetracycline operator sequences located on chromosome III was used. This array is bound by the Tet-repressor, which is tagged with monomeric red fluorescent protein (TetR-mRFP). Fluorescence microscopy reveals either 2 RFP foci in diploids, or 1 focus indicative of haploids.
Image Analysis.
To reconstruct a 3-dimensional fluorescent image of the cells, the stack of vertically separated fluorescent images (z-stack) was deconvolved using Volocity software (Improvision). The resulting volume was used to calculate the amount of fluorescence (in relative units) for each kinetochore or nucleus using Volocity software (further description of microscopy methods and the controls undertaken to ensure consistent measurement of fluorescence intensity are in SI Methods and Fig. S5 and Fig. S6).
Statistics.
For each protein studied, the ratios of protein in the mother versus the bud at each division (m/b ratio) between all of the mother and non-mother lineages were compared using standard t tests. P values of 0.05 or less are considered to indicate significance. See Table S1 for summary statistics and Dataset S1 for raw quantitation data.
Supplementary Material
Acknowledgments.
We thank Argiris Efstratiadis and Rudy Leibel for motivating our interest in cellular asymmetry; Phil Heiter (University of British Columbia, Vancouver), Trisha Davis (University of Washington, Seattle), Kerry Bloom (University of North Carolina, Chapel Hill, NC), Michael Lisby (University of Copenhagen, Copenhagen), Kara Bernstein, Rebecca Burgess, Bob Reid, Qi Feng, and Jackie Barlow (Columbia University, New York) provided strains and/or helpful advice and suggestions on this manuscript. This work was supported by the Russ Berrie Diabetes Foundation and National Institutes of Health Grants GM50237 and GM67055.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811248106/DCSupplemental.
References
- 1.Fischer JA. Molecular motors and developmental asymmetry. Curr Opin Genet Dev. 2000;10:489–496. doi: 10.1016/s0959-437x(00)00117-9. [DOI] [PubMed] [Google Scholar]
- 2.Chant J, Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- 3.Drees BL, et al. A protein interaction map for cell polarity development. J Cell Biol. 2001;154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pringle JR, et al. Establishment of cell polarity in yeast. Cold Spring Harb Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- 5.Lew DJ. Yeast polarity: Negative feedback shifts the focus. Curr Biol. 2005;15:R994–996. doi: 10.1016/j.cub.2005.11.048. [DOI] [PubMed] [Google Scholar]
- 6.Long RM, et al. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 7.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 8.Irazoqui JE, Lew DJ. Polarity establishment in yeast. J Cell Sci. 2004;117:2169–2171. doi: 10.1242/jcs.00953. [DOI] [PubMed] [Google Scholar]
- 9.McMurray MA, Gottschling DE. Aging and genetic instability in yeast. Curr Opin Microbiol. 2004;7:673–679. doi: 10.1016/j.mib.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Bell CD. Is mitotic chromatid segregation random? Histol Histopathol. 2005;20:1313–1320. doi: 10.14670/HH-20.1313. [DOI] [PubMed] [Google Scholar]
- 11.Lew DJ, Burke DJ, Dutta A. The immortal strand hypothesis: How could it work? Cell. 2008;133:21–23. doi: 10.1016/j.cell.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 13.Henikoff S, Ahmad K, Malik HS. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 14.Pardo-Manuel de Villena F, Sapienza C. Female meiosis drives karyotypic evolution in mammals. Genetics. 2001;159:1179–1189. doi: 10.1093/genetics/159.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:4931–4946. doi: 10.1091/mbc.E03-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinsky BA, Tatsutani SY, Collins KA, Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev Cell. 2003;5:735–745. doi: 10.1016/s1534-5807(03)00322-8. [DOI] [PubMed] [Google Scholar]
- 17.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taxis C, et al. Spore number control and breeding in Saccharomyces cerevisiae: A key role for a self-organizing system. J Cell Biol. 2005;171:627–640. doi: 10.1083/jcb.200507168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 20.Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- 21.Bouck D, Bloom K. The role of centromere-binding factor 3 (CBF3) in spindle stability, cytokinesis, and kinetochore attachment. Biochem Cell Biol. 2005;83:696–702. doi: 10.1139/o05-161. [DOI] [PubMed] [Google Scholar]
- 22.De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheeseman IM, et al. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mythreye K, Bloom KS. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J Cell Biol. 2003;160:833–843. doi: 10.1083/jcb.200211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira G, Tanaka TU, Nasmyth K, Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- 28.Yamashita YM, Fuller MT. Asymmetric centrosome behavior and the mechanisms of stem cell division. J Cell Biol. 2008;180:261–266. doi: 10.1083/jcb.200707083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusch J, Liakopoulos D, Barral Y. Spindle asymmetry: A compass for the cell. Trends Cell Biol. 2003;13:562–569. doi: 10.1016/j.tcb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Basto R, et al. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 31.Rando TA. The immortal strand hypothesis: Segregation and reconstruction. Cell. 2007;129:1239–1243. doi: 10.1016/j.cell.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 32.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 33.Williamson DH, Fennell DJ. In: Molecular Genetics in Yeast, Alfred Benson Symposium. von Wettstein D, Friis J, Kielland-Brandt M, Stenderup M, editors. Copenhagen: Munksgaard; 1981. pp. 89–107. [Google Scholar]
- 34.Neff MW, Burke DJ. Random segregation of chromatids at mitosis in Saccharomyces cerevisiae. Genetics. 1991;127:463–473. doi: 10.1093/genetics/127.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armakolas A, Klar AJ. Cell type regulates selective segregation of mouse chromosome 7 DNA strands in mitosis. Science. 2006;311:1146–1149. doi: 10.1126/science.1120519. [DOI] [PubMed] [Google Scholar]
- 36.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez-Barrera S, Cortes-Ledesma F, Wellinger RE, Aguilera A. Equal sister chromatid exchange is a major mechanism of double-strand break repair in yeast. Mol Cell. 2003;11:1661–1671. doi: 10.1016/s1097-2765(03)00183-7. [DOI] [PubMed] [Google Scholar]
- 38.Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 39.White MA, Eykelenboom JK, Lopez-Vernaza MA, Wilson E, Leach DR. Non-random segregation of sister chromosomes in Escherichia coli. Nature. 2008;455:1248–1250. doi: 10.1038/nature07282. [DOI] [PubMed] [Google Scholar]
- 40.Copenhaver GP. Who's driving the centromere? J Biol. 2004;3:17. doi: 10.1186/jbiol15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henikoff S, Malik HS. Centromeres: Selfish drivers. Nature. 2002;417:227. doi: 10.1038/417227a. [DOI] [PubMed] [Google Scholar]
- 42.Bensasson D, Zarowiecki M, Burt A, Koufopanou V. Rapid evolution of yeast centromeres in the absence of drive. Genetics. 2008;178:2161–2167. doi: 10.1534/genetics.107.083980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hailey DW, Davis TN, Muller EG. Fluorescence resonance energy transfer using color variants of green fluorescent protein. Methods Enzymol. 2002;351:34–49. doi: 10.1016/s0076-6879(02)51840-1. [DOI] [PubMed] [Google Scholar]
- 44.Voth WP, Olsen AE, Sbia M, Freedman KH, Stillman DJ. ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot Cell. 2005;4:1018–1028. doi: 10.1128/EC.4.6.1018-1028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.