Abstract
Modelers of molecular signaling networks must cope with the combinatorial explosion of protein states generated by posttranslational modifications and complex formation. Rule-based models provide a powerful alternative to approaches that require explicit enumeration of all possible molecular species of a system. Such models consist of formal rules stipulating the (partial) contexts wherein specific protein–protein interactions occur. These contexts specify molecular patterns that are usually less detailed than molecular species. Yet, the execution of rule-based dynamics requires stochastic simulation, which can be very costly. It thus appears desirable to convert a rule-based model into a reduced system of differential equations by exploiting the granularity at which rules specify interactions. We present a formal (and automated) method for constructing a coarse-grained and self-consistent dynamical system aimed at molecular patterns that are distinguishable by the dynamics of the original system as posited by the rules. The method is formally sound and never requires the execution of the rule-based model. The coarse-grained variables do not depend on the values of the rate constants appearing in the rules, and typically form a system of greatly reduced dimension that can be amenable to numerical integration and further model reduction techniques.
Keywords: protein interaction networks, rule-based models, model reduction, distinguishability, information carriers
Molecular biology is spectacularly successful in disassembling cellular systems and anchoring cell-biological behaviors of staggering complexity in chemistry. This raises the challenge of reconstituting molecular systems formally, in pursuit of principles that would make their behavior more intelligible and their control more deliberate. This pursuit is as much driven by the practical need to cure disease as it reflects a desire for a theoretical perspective needed to understand the complexity of cellular phenotypes. In achieving such a perspective, we must deal with two broad problems.
First, we must be able to represent and analyze molecular interaction systems of combinatorial complexity. Although ubiquitous, such systems are perhaps most notorious in the context of cellular signaling. The posttranslational modification of proteins and their noncovalent association into transient complexes generate an astronomical number of possible molecular species that can relay signals (1). The question then becomes how to reason about system dynamics if we cannot possibly consider a differential equation for each chemical species that can appear in a system.
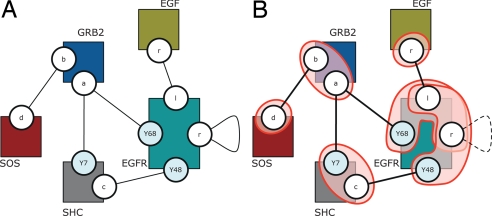
Second, understanding systems requires resisting the temptation of adopting the view of an outside observer. The outside view is indeed appropriate for the chemical analysis of a network, since the experimenter deliberately interacts in specific ways with the network to create measurable distinctions. Yet, the network, as a dynamical system, may not be capable of making these same distinctions. For example, an experimental technique might differentiate between SOS recruited to the membrane via GRB2 bound to SHC bound to the EGF receptor and SOS recruited via GRB2 bound to the EGF receptor directly. However, from the perspective of the EGF signaling system, such a difference might not be observable for lack of an endogenous interaction through which it could become consequential. The endogenous units of the dynamics may differ from the exogenous units of the analysis.
In an attempt at mitigating the first problem, analytical model reduction techniques eliminate variables on the basis of algebraic constraints such as conservation equations and quasi-steady-state conditions obtained mainly by exploiting separations of time and/or concentration scales (for example refs. 2 and 3). Numerical model reduction consists in integrating the kinetic rate equations of the full network and subsequently building a reduced model based on species that were observed to be significantly populated (4). Yet, all these techniques hinge on an explicit representation of the full network, which severely curtails their applicability to larger systems.
The past few years have seen the emergence of several approaches (5–8) that represent signaling systems in terms of rules stipulating conditions for specific interactions among proteins. These conditions typically specify (far) less than the full state of all proteins involved in an interaction. In this way, rules capture combinatorial complexity but avoid an explicit representation of the complete reaction network involving all possible molecular species. Yet, to explore the dynamics of a system of rules, such approaches must resort to stochastic simulations (6, 9, 10), whose event-based nature exacts a high computational cost. Ordinary differential equations (ODEs) would be highly useful for rapidly exploring system dynamics by numerical integration, but a flat-out expansion of rules into ODEs would, of course, fall victim to the combinatorial explosion. To nonetheless assemble ODEs from rules, a coarse-graining approach has been recently proposed (11–16). The idea is to convert a rule-based model into a reduced system of rate equations by identifying molecular patterns (sets of species) that act “independently” (16). We believe this approach to be promising, because it seems natural that a system described by rules might be characterized by dynamical units that are less specific than molecular species. We proceed in the same spirit, but differ significantly by seeking as variables those molecular patterns that establish the finest level of resolution at which the dynamics of the system is capable of making distinctions, thus rendering finer-grained patterns unwarranted. This we call internal coarse-graining. Moreover, our approach is formal, avoiding the limitations listed in ref. 16.
The next section surveys the language, Kappa (17), in which we cast rules of interaction. Kappa forms the basis of a substantive, formal, yet intuitive modeling framework (7, 9, 18, 19). Access to the Kappa modeling platform is provided at www.cellucidate.com.
Kappa: A Language for Molecular Biology
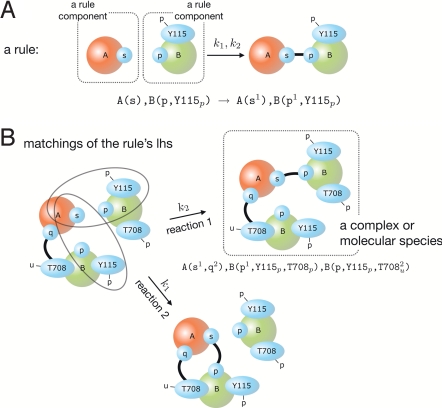
Kappa (17) is a formal language for defining agents (typically meant to represent proteins) as sets of sites that constitute abstract resources for interaction, as illustrated in Fig. 1 and extensively detailed in section 1 of supporting information (SI) Appendix. Sites can hold an internal state, as generated through posttranslational modifications, and engage in binding relations with sites of other agents. An association of proteins is a connected (site) graph, called a complex (of agents), as shown in the box of Fig. 1B. The nodes of the graph are agents, but the endpoints of edges are sites, which belong to agents. Although an agent can bear many connections, a site can bear only 1.
Fig. 1.
Rules and reactions in Kappa. (A) A rule captures a high-level mechanistic statement (empirical or hypothetical) about a protein–protein interaction in terms of a rewrite directive plus rate constant(s). The left-hand side (lhs) of the rule is a pattern of partially specified agents and represents the contextual information necessary for identifying reaction instances that proceed according to the rule. The right-hand side (rhs) expresses the actions that may occur when the conditions specified on the lhs are met in a reaction mixture of Kappa agents. A maximal connected subgraph on the lhs of a rule is called a rule component. (B) The rule in A matches a combination of agents in 2 distinct ways giving rise to 2 possible reactions with different outcomes. Note that because of their local nature, Kappa rules with >1 lhs component may apply in both a unimolecular and bimolecular situation. This is why such rules are given 2 rate constants, a first-order (k1) and a second-order (k2) constant. In a textual representation, agents are names followed by an interface of sites delimited by parentheses. Bonds are labeled by superscripts and internal states at a site by subscripts. In the graphical rendition, internal states are indicated as labeled barbs. See SI Appendix, section 1 and the section Kappa: A Language for Molecular Biology for more details.
Kappa is used to express tunable rules of interaction between proteins characterized by discrete modification and binding states. The idea of a rule, Fig. 1A, is to stipulate only the molecular context required for an interaction along with some rate constant(s). The left-hand side (lhs) of a rule is any site graph. Agents may mention a subset of their sites and omit states (SI Appendix, section 1.2). The right-hand side (rhs) exhibits the changes that occur when the lhs is matched (SI Appendix, section 1.4) in a mixture of agents. The difference between rhs and lhs is called the action of the rule. Sites mentioned on the lhs are said to be tested by the rule. Sites that are tested but not modified constitute the context of a rule's action. Because rules typically do not mention all of the sites and states of an agent, they keep combinatorial complexity implicit, obviating the need for eliminating it. A molecular species is a complex in which each agent occurs with a complete set of sites in definite states. We also refer to molecular species as ground-level objects. The complete set of sites defines the finest grain of resolution at which the state of an agent is known. Like rules, this set of sites can be updated to reflect new knowledge or hypotheses. Rules give rise to potentially numerous reaction instances [whose rate constants are related to the rate constant(s) of the rule]. These instances involve particular combinations of molecular species, each of which satisfies the context required for the rule to apply, see Fig. 1B and Fig. S4 in SI Appendix.
Kappa rules are both descriptions of mechanistic knowledge and executable instructions. In fact, we view Kappa as a programming language attuned to molecular signaling. Rules induce a stochastic dynamics on a mixture of agents, for which we implemented a general and efficient implicit-state version of the Doob–Gillespie algorithm (9). A Kappa model of a biological system is a concurrent computer program whose instructions are rules that asynchronously change the state of a shared store representing the reaction mixture on which the rules act. Computer programs are formal objects that can be analyzed statically. Static analysis assists in the discovery of behavioral properties of a program without running it, much like a system of differential equations can be analyzed without simulating it. Static analysis involves, for example, the inspection of causal dependencies among rules and an overapproximation of the molecular species reachable from an initial condition.
Kappa is closely related to BNGL (5), but differs from the latter in being a context-free grammar, that is, a language that expresses strictly local rules of action. The computational cost of checking whether a rule can apply to a given choice of reactants is bounded by the size of the rule's lhs and not by the reactants. This difference enables scalable simulation (9) and static analysis of the implied dynamical system (7), which plays a crucial role in the efficiency of the coarse-graining technique we describe here (see Remarks below and SI Appendix). The central role we attach to static analysis sets our framework apart from other rule-based approaches, such as BNGL (5) and “little b” (8), whose primary deliverable is the automated assembly of the full reaction network by generating all possible species and their reactions from a given set of rules. Yet, the combinatorial explosion inherent in molecular signaling makes such goals impractical and often impossible. In a pilot study of EGF signaling, we collated 71 rules representing mechanistic observations of pertinent protein–protein interactions. These rules would produce 1019 molecular species. Our current EGF model has grown to ≈350 rules. It thus appears more useful to forgo the expansion into an inscrutably large system of equations and, instead, apply static analysis techniques directly to the rule collection and explore the system with stochastic simulations that generate dynamical trajectories (6, 9, 10). Yet, such simulations are computationally expensive. This raises the question whether there is a system of ODEs that “corresponds” to a rule-based model, i.e. that constitutes its natural differential semantics.
From Rules to ODEs
Using a rule-based (as opposed to a reaction-based) model amounts to acknowledging that molecular species may not always be meaningful units of the dynamics. Such units should lump together species that cannot be distinguished by the dynamics arising from a given system of rules (see section 4, especially 4.2, of the SI Appendix). Moreover, the lumping must be self-consistent, meaning that the contribution of each rule to the rate of production or consumption of any unit should only depend on other units. In the following, we introduce 2 key properties that a suitable set of coarse-grained dynamical units—referred to as fragments (to be properly defined later)—should satisfy.
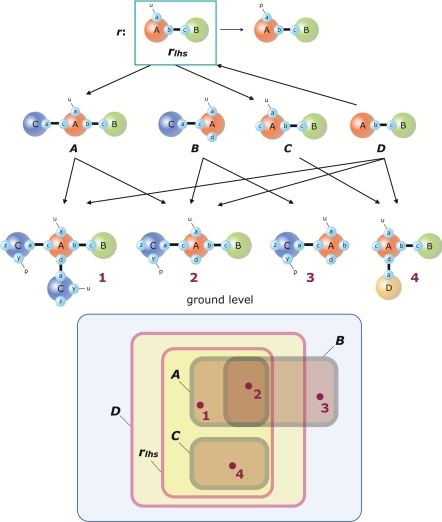
Property 1 (“No Overlap”).
No fragment properly overlaps a lhs component of a rule on a modified site. This property is defining of fragments and is key (but not enough) for expressing the rate function of a fragment in terms of fragments. The reasoning is illustrated in Fig. 2. The rule r at the top consumes those species that match its lhs component rlhs. We can think of a pattern X in terms of its extension X◇, which is the set of species that match X, accounting for the many ways in which any such species might match X (think symmetries). The extension rlhs◇ of rlhs is shown schematically at the bottom of Fig. 2 as a yellow area within the blue area standing for the set of all molecular species implied by the rules of a system and an initial condition. Fig. 2 provides assistance for reasoning about the suitability of a few sample patterns as potential fragments in light of Property 1. Consider pattern B. Although B does not itself match rlhs, some ground-level instances of B do, such as species 2. Thus, B◇ (properly) intersects rlhs◇, which makes it impossible to express the contribution of the unimolecular rule r to the consumption rate of B in terms of B alone. Rather, we would have to know at any time the fraction of molecular species that occurs in the intersection of B◇ with rlhs◇, which is a property that requires knowing the complete reaction mixture at any time. By contrast, A◇ is entirely contained within rlhs◇. As a consequence, the firing of rule r will consume the pattern A at a rate proportional to its concentration [A], defined at t = 0 by the number of embeddings of A in the reaction mixture. There is no need to know the reaction mixture for any subsequent time. The case of C is analogous to that of A.
Fig. 2.
Rules and fragments. The figure provides assistance in establishing criteria that define fragments, as detailed in the section From Rules to ODEs. The top row depicts a (unimolecular) rule whose lhs component is rlhs. The third row from top shows fully specified molecular species (ground-level objects), numbered 1 to 4. The second row depicts various patterns, A to D. Arrows indicate embedding relations of one pattern (graph) into another (see SI Appendix, section 1.4). The rectangles at the bottom provide a schematic of relationships between sets of molecular species that match the patterns A–D and rlhs. Note that D embeds into rlhs; its matching instances are therefore a superset of those of rlhs. Also, D does not overlap with rlhs on a site that r modifies. Hence r has no effect on D.
It is possible to refine B into B′ by adding context, such that B′◇ ⊂ rlhs◇. For example, connecting agent A at site b to agent B at c yields B′ ≡ C (a1), A (au, c1, d, b2), B (c2) with B′◇ = B◇ ∩ A◇. Thus, as far as rule r is concerned, patterns A, C, and B′ are fragment candidates by virtue of their extensions being inside rlhs◇. However, other rules in the system may further constrain these potential fragments. Indeed, our procedure to construct fragments depends on all rules of a given system.
Property 2 (“Orthogonality”).
Fragments must partition (in the extension sense) anything that is contained within a fragment, which we refer to as a subfragment. We show later that any lhs component of a rule is a subfragment (Property 1 clarifies this only for particular components). The rate equation for a fragment affected by a rule of molecularity >1 (i.e. a rule with 2 or more lhs components) gets a contribution consisting of a monomial involving several fragments. Consider, for example, a rule of type Z, Z′ → Z*, Z′, which modifies the lhs component Z into Z*. Consider further a particular fragment A that is a refinement of Z and is thus consumed by the rule (A◇ ⊂ Z◇). The consumption rate of A will be proportional to [A] [Z′]. If only 1 fragment, say ℬ, matches the lhs component Z′, then [Z′] = [ℬ]. However, there may be several fragments ℬi that match Z′, in which case [Z′] should be the sum over all [ℬi]. The only problem is that the ℬi might have ground-level extensions ℬi◇ that overlap, causing the naive sum to overcount. Thus, there must be a set of fragments that partitions Z′◇, so that [Z′] can be expressed as a sum of orthogonal fragments. Property 2 does more, however: It guarantees that the concentration of any subfragment can be expressed in terms of fragment concentrations. This will be needed down the road. Properties 1 and 2 jointly ensure a self-consistent coarse-grained system whose dynamics is sound. Soundness means that computing the ground-level dynamics and then coarse-graining yields the same result as coarse-graining at the outset and then running the coarse-grained dynamics.
Note that the (possibly infinite) set of molecular species is always a trivial set of fragments enjoying Properties 1 and 2, but typically far from optimizing our criterion of “dynamical distinguishability.” We can do much better without ever touching the ineffable ground-level network of species. As we show next, by proceeding directly from the rules, we construct dynamical units whose boundaries are carved out by the actions available to the system.
Constructing Coarse-Grained Fragments
In this section we implement Properties 1 and 2 by defining syntactical criteria with which we scan all rules in a model to determine which agents and sites belong to a fragment. As a test case, we apply these criteria to a rule-based model of a small section of early events in epidermal growth factor (EGF) signaling as adapted from ref. 20. These events include the binding of EGF (agent E) to the receptor (R), the subsequent dimerization of the receptor, and the eventual recruitment of SOS (O). The model consists of 39 rules r01–r39, listed in section 5.1 of SI Appendix. We write separate rules for binding and unbinding actions, because unbinding typically occurs under less-restrictive contexts than binding. The names of agent sites were chosen fairly arbitrarily. The biological accuracy of the published models from which we obtained the rules might be outdated, because knowledge about EGF signaling mechanisms keeps changing rapidly. Our goal here is not a particular biological insight, but a procedure of general interest. Together, the 39 rules of our test case imply 356 possible distinct molecular species. We shall see, however, that based on these rules of interaction, the system can only make 38 internal distinctions. Differential equations in these 38 variables self-consistently describe the dynamics of the system. It is very convenient to use a special map as a canvas for laying out which sites and bindings must appear together in a fragment. In ref. 7, we called this map the contact map (CM), Fig. 3A. The CM is generated automatically from a rule-based model and provides a summary of attainable interactions. The CM is a graph whose nodes are the agents that appear in the model. Recall that agents are sets of sites. These sites are the endpoints of edges representing possible binding interactions. Certain sites are colored to indicate that their internal state can be modified.
Fig. 3.
The contact map. (A) The contact map is a graph whose nodes are the agents in the model and whose edges are possible bonds between sites. Filled circles indicate sites with modifications of state. The contact map is a fine-grained version of what is known as a protein–protein interaction (PPI) map, in that its edges end in sites of agents and not just agents. (B) The annotated contact map (ACM) after decoration induced by the directives Cov1–Cov3 and Edg1.
Syntactical Criteria for Annotating the Contact Map.
We shall need the notion of a parsimonious covering, or covering for short. A covering C of a set S is a set of subsets of S, called classes, such that (i) no class is empty, (ii) no class is a subset of another class, and (iii) the union of all classes yields S. A covering differs from a partition in that the elements of a covering need not be pairwise disjoint.
In preparation for building fragments, we first annotate the CM with 2 types of information obtained by applying the syntactical criteria listed below. (i) For each agent type A, we define a covering C(A) of the set of its sites. (ii) For each edge in the CM, we define its type as either “solid” or “soft.” In a second step, we assemble fragments based on the annotated CM (ACM).
The following syntactical criteria determine valid coverings for an agent and the type of a bond. We follow up with some explanatory remarks.
Cov1 (backward closure).
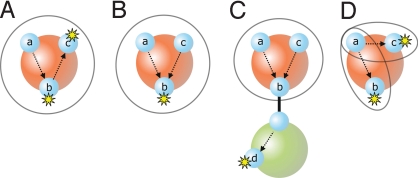
If a rule tests a site a in an agent A and modifies a site b in the same agent, any class in C(A) that contains b must also contain a (e.g. Fig. 4 A and B).
Fig. 4.
Examples illustrating the syntactical criteria Cov1 and Cov2 for determining classes in the covering of an agent. See section 3 of the SI Appendix for further details.
Cov2 (relay).
If a rule tests a site a in an agent A, and A is connected by some path through a site b to an agent that is modified, any class in C(A) that contains b must also contain a (e.g. Fig. 4C).
Cov3 (witness).
For each agent in an unmodified lhs component, there must be a class in the agent's covering that contains all of the sites tested by the rule.
Edg1.
A bond is solid if it occurs on the lhs of a rule that tests anything other than that bond.
Syntactical criteria Cov1–Cov3 and Edg1 implement Properties 1 and 2. To see this, define an overlap between 2 patterns X and Y as the set of agents and sites both mention along with a mutually compatible state. The overlap, if it exists, can be used as an instruction for gluing the patterns together, see section 2 of SI Appendix. Our discussion of Fig. 2 suggests that if a pattern has an overlap with a component on the lhs of a rule, and the overlap contains a site modified by the action of the rule, the pattern must be glued to the lhs component to become a fragment as far as that rule is concerned. Hence, a fragment A either has no overlap with the sites that are modified by the rule, or it contains a whole lhs component (SI Appendix, section 2). The same process—glue on overlap—is repeated for each rule and all agents, starting out with each site in its own class. Cov1 and Cov2 simply keep track of which sites of an agent must be mentioned together in a fragment as a result of this repeated glue-on-overlap. Cov3 takes care of the orthogonality property in the special case of a component required for an action but not modified by it (a “witness”).
The glue-on-overlap process can pull bonds into a fragment (see B′ in the discussion of Property 1). However, not all bonds are conduits of control between the parts, say X and Y, they connect. Suppose that the only time a bond appears on the lhs of a rule is in a so-called pure dissociation rule that tests nothing except the existence of the bond that is to be broken. No rule modifying X or Y depends on that bond (or the bond would figure in a rule other than the dissociation rule). As a consequence, the fragments containing X can stop short of including all possible states of Y and vice versa. The fragments containing X only need to specify whether or not X is connected to Y, but they do not need to specify Y itself. (And vice versa.) The directive Edg1 defines those bonds that carry constraints as solid. All bonds not characterized by Edg1 can be chosen as solid or soft, and we can choose to have smaller or larger covering classes (provided they satisfy Cov1–Cov3); the fragmentation is sound either way. However, soft bonds make for smaller fragments (see next section). Our policy is to obtain small fragments by choosing covering classes that are as small as possible and considering bonds to be soft when they appear only on the rhs of a rule (bonds that are only formed) and/or on the lhs of a pure dissociation.
Fragment Assembly.
To define fragments, it is convenient to extend the notion of complex with bond stubs. An agent with a bond stub is written A (aB@b), which means that A's site a is bound to B's b, without, however, including agent B in the complex.
Given an ACM, a fragment ℱ is a complex such that: Each agent has a set of sites that is a class, every site has an internal state if any, every site has a binding state—either free, bound, or stubbed, every stub must correspond to a soft bond in the ACM, and every bond is solid. A subfragment is a complex that embeds in a fragment.
To obtain a fragment, one starts with an agent and a site. The ACM then determines which further sites to add and which binding states (stubbed or not) are appropriate. When there is nothing more to add, one has a fragment.
As an example of this growth process, consider agent R in our rule set. According to the ACM in Fig. 3B, we have a choice between 2 classes. Suppose we choose class {l, r, Y48}. Next, we assign a state to each site in that class. For example, all sites are free, and Y48 is unphosphorylated. This yields fragment R (Y48u, l, r), which is ℱ34 in the complete list for our example (SI Appendix, section 2.3). Alternatively, we might choose Y48 to be phosophorylated (fragment ℱ15). Yet, if we choose Y48 to be also bound, then the solid link in the ACM forces agent S into the fragment, along with its site c as the link's endpoint. In turn, c forces inclusion of the class to which it belongs, {c, Y7}. Now we need to assign states to c and Y7 in agent S. For example, S (Y7p, c1), R (Y48p1, l, r), which is fragment ℱ04. A further fragment is obtained by considering site r in agent R to be bound. Site r can bind to another R agent, but the link is soft. A soft link at r does not force the inclusion of another instance of R. Instead, the bound state is only indicated with its type: S (Y7p, c1), R (Y48p1, l, rR@r). This fragment, however, does not show up in our list. Given our set of rules, the state in which R is dimerized at site r cannot occur if the ligand-binding site l is empty. Such a fragment is automatically eliminated from the list because a separate reachable state analysis (next section) recognizes it as inaccessible. Fragments as defined above enjoy the following properties:
Q1.
No fragment strictly overlaps with a rule component on a modified site.
Q2.
Any lhs component is contained in a fragment (i.e., is a subfragment).
Q3.
The concentration of any subfragment can be expressed as a linear combination of fragment concentrations (Eq. 17 in SI Appendix).
Q4.
Fragments are closed under rule actions.
Q1 is Property 1 (no overlap), whereas Q2 and Q3 imply Property 2 (orthogonality). Q4 means that fragments form a network of reactions (like species).
Q1 follows from Cov1 and 2 and Edg1; Q2 follows from Cov3 and Edg1 for nonmodified rule components, and Cov1 and 2 and Edg1 for modified ones; Q3 follows from the exhaustivity of the growth procedure for fragments, as does Q4.
Q1–3 ensure a sound translation from rules into an ODE system for fragments, as sketched next.
Assembling the Dynamical System for Fragments.
The dynamical system for fragments is constructed by deriving mass action terms for the consumption and production of fragments from rules. We only sketch the reasoning here and provide a detailed account in section 6.4 of SI Appendix. Consider, for example, a rule of the form Z,Z′ → Z Z′, which binds 2 complexes Z and Z′. Based on this rule, the differential equation d[ℱi]/dt for each fragment ℱi that matches Z obtains a consumption term −γ[ℱi][Z′], where [Z′] is expressed as a sum of concentrations of orthogonal fragments using Q2 and 3. The factor γ depends on the rate constant of the rule and the number of ways that Z embeds into ℱi. On the production side, the kinetic terms depend on the bond type in the ACM. Consider, for example, a solid bond. A kinetic term γ[ℱi][ℱj] is generated for the differential equation d[ℱk]/dt of every fragment ℱk that matches Z
Z′, which binds 2 complexes Z and Z′. Based on this rule, the differential equation d[ℱi]/dt for each fragment ℱi that matches Z obtains a consumption term −γ[ℱi][Z′], where [Z′] is expressed as a sum of concentrations of orthogonal fragments using Q2 and 3. The factor γ depends on the rate constant of the rule and the number of ways that Z embeds into ℱi. On the production side, the kinetic terms depend on the bond type in the ACM. Consider, for example, a solid bond. A kinetic term γ[ℱi][ℱj] is generated for the differential equation d[ℱk]/dt of every fragment ℱk that matches Z Z′, where ℱi and ℱj are fragments matching Z and Z′, respectively, subject to the constraint that the match of ℱk is the disjoint sum of the embeddings of Z and Z′ into their respective fragments. If the bond in Z
Z′, where ℱi and ℱj are fragments matching Z and Z′, respectively, subject to the constraint that the match of ℱk is the disjoint sum of the embeddings of Z and Z′ into their respective fragments. If the bond in Z Z′ is soft and corresponds to a ··· A.a
Z′ is soft and corresponds to a ··· A.a b.B ···, one can replace Z
b.B ···, one can replace Z Z′ with ZB@b,Z′A@a, because there is no information in Z′A@a affecting ZB@b. Every fragment ℱk matching ZB@b gains a production term γ[ℱi][Z′], where ℱi matches Z and is related to the ℱk matching ZB@b. A similar argument applies to fragments that match Z′A@a.
Z′ with ZB@b,Z′A@a, because there is no information in Z′A@a affecting ZB@b. Every fragment ℱk matching ZB@b gains a production term γ[ℱi][Z′], where ℱi matches Z and is related to the ℱk matching ZB@b. A similar argument applies to fragments that match Z′A@a.
The dissociation of a solid bond Z Z′ will give rise to a piece Z (and also Z′) that embeds into a fragment ℱ. To determine the contribution of the dissociation rule to the rate of production of ℱ, we need the concentration of ℱ
Z′ will give rise to a piece Z (and also Z′) that embeds into a fragment ℱ. To determine the contribution of the dissociation rule to the rate of production of ℱ, we need the concentration of ℱ Z′. However, ℱ
Z′. However, ℱ Z′ is not itself a fragment but, rather, a subfragment. This is why, for our method to result in a closed system of equations, we must be able to express the concentration of a subfragment in terms of fragments (see Q3 and Property 2).
Z′ is not itself a fragment but, rather, a subfragment. This is why, for our method to result in a closed system of equations, we must be able to express the concentration of a subfragment in terms of fragments (see Q3 and Property 2).
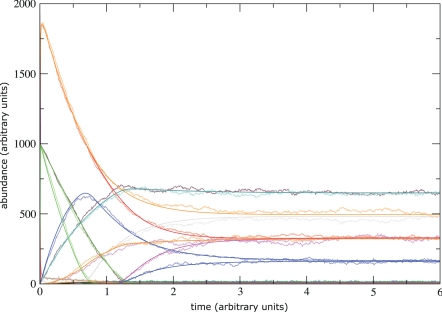
Fig. 5 was obtained by running a microscopic stochastic simulation of the early EGF test system, driven by rules r01–r39 while reporting the concentrations of fragments ℱ01–ℱ38. This stands as a proxy for the numerical integration of the deterministic ground-level system of 356 ODEs and the subsequent lumping of species into our 38 fragments. As a comparison, the smooth curves result from the direct numerical integration of the automatically generated ODE system for fragments.
Fig. 5.
Comparison between microscopic dynamics and fragment dynamics. Wiggly curves: The microscopic dynamics of the early EGFR example is executed with a Doob–Gillespie simulation (9) while reporting the coarse-grained fragment concentrations. This serves as a proxy for the deterministic microscopic dynamics. Steady curves: The output of the deterministic fragment dynamics. Still, many fragments (and many more molecular species) only acquire tiny concentration values, causing far fewer than 38 curves to be discernible by eye in this plot.
Remarks
Reachability.
Underlying several steps of our procedure is a very fast overapproximation α of the set of reachable species, deploying the framework of abstract interpretation (21) as described in ref. 19. This overapproximation comes into play at 3 junctures (i) The contact map reports edges and site modifications only if they are reachable by α. (ii) Fragments that are not reachable by α are discarded. (iii) The procedure for compressing rules (see below) makes use of α. In ref. 19, we characterize those special situations for which α is exact. (The present EGF example is such a case.)
Making Rules Concise.
Because fragment construction proceeds by inspecting the structure of rules, it is important that rules be concise, in the sense of avoiding redundant contextual conditions (tests) on their lhs. However, what classifies as redundant depends on the remaining rules in the model. Because rules record empirical observations or hypotheses, they tend to be crafted in isolation. Consider, for the sake of illustration, rule r02 expressing the binding of ligand to receptor: R (l, r), E (r) → R (l1, r), E (r1). The rule mentions 2 sites, l and r, of the receptor R. Site l is the ligand (EGF)-binding site, whose state is modified by the action of r02, whereas r is the site at which the receptor dimerizes (as described in r03). Rule r02 asserts that binding of E (EGF) to R requires not just a free l, but also a free r. Given the other rules of the model, there is no reachable state of the reaction mixture in which R could dimerize before binding E. Hence, in the context of the remaining 38 rules of this model, asking for site r to be free is a redundant condition for the firing of rule r02, because a free l implies a free r. Without removing such redundancies, fragments would be more numerous and bloated by fictitious dependencies. To reduce the extent to which this happens, we preprocess a rule system with an automatic compression that removes unnecessary contextual specifications. This technique rests on the reachability overapproximation referred to in the previous paragraph. In section 5.2 of the SI Appendix, we list the 39 compressed rules cr01–cr39 from which the 38 fragments were derived.
Role of Rate Constants.
All ground-level reactions into which a rule expands inherit its rate constant (after accounting for possible symmetry reductions upon expansion). Beyond any specific values of rate constants, rules themselves already imply a notion of kinetic distinguishability. For example, our toy model of early EGF events posits that the phosphorylated EGFR receptor (R) binds the protein SHC (S), which would read as R (Y48p), S (c) → R (Y48p1), S (c1). Yet, such a rule does not appear in the model. Rather, the same binding action between R and S is found in 2 rules r24 and r28 that differ in their contexts. Rule r24, R (Y48p), S (c, Y7u) → R (Y48p1),S (c1, Y7u), specifies that site Y7 of S must be unphosphorylated and free, whereas rule r28, R (Y48p), S (c, Y7p1),G (a1, b) → R (Y48p2),S (c2, Y7p1),G (a1, b), specifies that Y7 of S is phosphorylated and bound to G. The only reason to warrant such a distinction is an actual or hypothesized difference in the rate constants for the 2 contexts. Hence, regardless of the specific values of rate constants, positing 2 rules with different contexts for the same action affects the construction of fragments. The precise values of the rate constants of rules enter the ODEs for fragments, but they do not affect the fragments themselves, because the latter are based on the distinguishability of control flows shaped by rule contexts.
Limitations.
For our coarse-graining procedure to be well defined, rules must have unique ground-level molecularity, i.e., a rule with 2 lhs components must apply only to disjoint reactants (unlike in Fig. 1). Rules whose arity does not always match the arity of their ground-level instances (molecularity mismatches) can give rise to polymerization and result in an infinite number of fragments.
Multiple occurrences of the same agent in a rule do not constitute a problem; neither does the production of an agent. The destruction of an agent poses no theoretical problem either but is costly in terms of fragment numbers—as is the BNGL “.” (dot) operator.
We do not claim that our method generates a smallest set of fragments or that it is unique. In particular, our method carries a deliberate bias by defining fragments as connected patterns. As a consequence of our construction via an annotated contact map, fragments are closed under the operational semantics of Kappa, i.e., rules convert fragments into fragments (Q4). This allows us to conveniently picture a reaction network at the level of fragments. However, this is not necessary for sound coarse-graining, and alternatives remain to be explored.
We are mathematically certain that any information lost by our coarse-graining is not distinguishable by the microscopic dynamics. However, we cannot prove that all information retained in our fragments is distinguishable. One reason is that rule compression (see above) is, in general, an approximation.
Prior Art.
Our method differs from prior approaches in several aspects. First, our method is formal, which makes its properties more transparent and amenable to proof. It suffers from none of the limitations listed in ref. 16, as far as deterministic dynamics is concerned. Second, our approach focuses on interaction-based distinguishability rather than “independence.” In section 4 of SI Appendix, we provide some thoughts on independence and distinguishability that are conceptually useful for appreciating our stance but not needed for grasping our method. The similarity between the approach sketched in ref. 16 and our present work ends at directive Cov1, because control flows across bindings are treated differently. In section 8 of SI Appendix, we compare the outcome of our method with the manual procedure described in ref. 14.
Conclusions
Rule-based representations have been recently proposed to address the dynamics of combinatorial systems for which an expansion into the full reaction network is virtually impossible (5–7). It would be highly useful to construct a deterministic projection of rule-based dynamics for several reasons. On the practical side, rule-based models require stochastic simulations, which can be very time consuming. Although stochastic kinetics can provide insights not accessible from deterministic rate equations, the latter are useful for calibration, analysis, and judicious simplification. On the conceptual side, many of the molecular species that are, in principle, attainable by a given system seem unlikely to play a significant dynamical role, because they either are too improbable, or the dynamics of the system cannot differentiate them. The latter is already implicit in the use of rules, which specify patterns of interactions, rather than reactions between fully detailed molecular species.
We have presented a formal method for automatically generating a dynamical system of coarse-grained variables from a given set of rules, guided by a criterion of distinguishability. The method is exact in the sense that coarse-graining first and then integrating the fragment ODEs is equivalent to first integrating the network ODEs at the level of molecular species and then coarse-graining. The fact that the ground system is oftentimes ineffable because of combinatorial blow-up is of no consequence, because these patterns are constructed directly from the rules.
Our running test case was a limited model of early events in EGFR signaling (21), consisting of 39 rules that generate 356 molecular species. Our method yielded a dynamical system of 38 fragments. A pilot study on a larger section of the EGFR system (19), comprising 71 rules potentially expanding into 18,051,984,143,555,729,567 molecular species, yields 175,988 fragments, which reconnects the system to the realm of feasible ODEs.
In particular cases, fragments become independent units. (A necessary condition being that the coverings of all agents are partitions.) We call such systems “tileable.” In section 4.1 of SI Appendix, we provide a connection between tileability and invertibility. Although exact, our coarse-graining is not invertible, in general.
It might be biologically insightful to attempt a sensitivity analysis of the fragmentation process, to determine which rules, when changed, have the biggest impact on the nature and number of fragments. Can highly consequential rules be guessed from the annotated contact map? Issues like these suggest that internal coarse-graining is not only of practical use but of theoretical import for understanding the informational architecture of molecular signaling systems.
Supplementary Material
Footnotes
Conflict of interest statement: W.F. is a member of the Board of Directors of Plectix BioSystems Inc., a company that develops the Kappa modeling platform used in this research. J.F., V.D., J.K., and R.H. are consultants for Plectix BioSystems Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809908106/DCSupplemental.
References
- 1.Hlavacek WS, et al. Rules for modeling signal-transduction systems. Science STKE. 2006;344:re6. doi: 10.1126/stke.3442006re6. [DOI] [PubMed] [Google Scholar]
- 2.Krüger R, Heinrich R. Model reduction and analysis of robustness for the Wnt/β-catenin signal transduction pathway. Genome Inform. 2004;15:138–148. [PubMed] [Google Scholar]
- 3.Ciliberto A, Capuani F, Tyson JJ. Modeling networks of coupled enzymatic reactions using the total quasi-steady state approximation. PLoS Comput Biol. 2007;3:e45. doi: 10.1371/journal.pcbi.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faeder JR, Blinov ML, Goldstein B, Hlavacek WS. Combinatorial complexity and dynamical restriction of network flows in signal transduction. IEE Syst Biol. 2005;2:5–15. doi: 10.1049/sb:20045031. [DOI] [PubMed] [Google Scholar]
- 5.Blinov ML, Faeder JR, Hlavacek WS. BioNetGen: Software for rule-based modeling of signal transduction based on the interactions of molecular domains. Bioinformatics. 2004;20:3289–3292. doi: 10.1093/bioinformatics/bth378. [DOI] [PubMed] [Google Scholar]
- 6.Lok L, Brent R. Automatic generation of cellular reaction networks with Moleculizer 1.0. Nat Biotechnol. 2005;23:131–136. doi: 10.1038/nbt1054. [DOI] [PubMed] [Google Scholar]
- 7.Danos V, Feret J, Fontana W, Harmer R, Krivine J. Lecture Notes in Computer Science. Vol 4703. Lisboa, Portugal: Springer; 2007. Rule-based modelling of cellular signalling; pp. 17–41. [Google Scholar]
- 8.Mallavarapu A, Thomson M, Ullian B, Gunawardena J. Programming with models: Modularity and abstraction provide powerful capabilities for systems biology. J R Soc Interface. 2008 doi: 10.1098/rsif.2008.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danos V, Feret J, Fontana W, Krivine J. Lecture Notes in Computer Science. Vol 4807. Berlin: Springer; 2007. Scalable simulation of cellular signalling networks; pp. 139–157. [Google Scholar]
- 10.Yang J, Monine MI, Faeder JR, Hlavacek WS. Kinetic Monte Carlo method for rule-based modeling of biochemical networks. Phys Rev E. 2008;78 doi: 10.1103/PhysRevE.78.031910. 031910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borisov NM, Markevich NI, Hoek JB, Kholodenko BN. Trading the micro-world of combinatorial complexity for the macro-world of protein interaction domains. BioSystems. 2006;83:152–166. doi: 10.1016/j.biosystems.2005.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conzelmann H, Saez-Rodriguez J, Sauter T, Kholodenko BN, Gilles ED. A domain-oriented approach to the reduction of combinatorial complexity in signal transduction networks. BMC Bioinformatics. 2006;7:34. doi: 10.1186/1471-2105-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koschorreck M, Conzelmann H, Ebert S, Ederer M, Gilles ED. Reduced modeling of signal transduction–a modular approach. BMC Bioinformatics. 2007;13:336. doi: 10.1186/1471-2105-8-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conzelmann H, Fey D, Gilles ED. Exact model reduction of combinatorial reaction networks. BMC Systems Biology. 2008;2:78. doi: 10.1186/1752-0509-2-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conzelmann H. Stuttgart, Germany: Institut für Systemdynamik der Universität Stuttgart; 2008. PhD Thesis. [Google Scholar]
- 16.Borisov NM, Chistopolsky AS, Faeder JR, Kholodenko BN. Domain-oriented reduction of rule-based network models. IET Syst Biol. 2008;2:342–351. doi: 10.1049/iet-syb:20070081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danos V, Laneve C. Formal molecular biology. Theor Comput Sci. 2004;325:69–110. [Google Scholar]
- 18.Danos V, Feret J, Fontana W, Harmer R, Krivine J. Lecture Notes in Bioinformatics. Vol 5054. Cambridge, UK: Springer; 2008. Rule-based modelling, symmetries, refinements; pp. 103–122. [Google Scholar]
- 19.Danos V, Feret J, Fontana W, Krivine J. Lecture Notes in Computer Science. Vol 4905. Berlin: Springer; 2008. Abstract interpretation of cellular signalling networks; pp. 83–97. [Google Scholar]
- 20.Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. A network model of early events in epidermal growth factor receptor signaling that accounts for combinatorial complexity. BioSystems. 2006;83:136–151. doi: 10.1016/j.biosystems.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Cousot P, Cousot R. Abstract Interpretation: A Unified Lattice Model for Static Analysis of Programs by Construction or Approximation of Fixpoints. New York: ACM Press; 1977. pp. 238–252. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.