Abstract
Although genomewide association studies have successfully identified associations of many common single-nucleotide polymorphisms (SNPs) with common diseases, the SNPs implicated so far account for only a small proportion of the genetic variability of tested diseases. It has been suggested that common diseases may often be caused by rare alleles missed by genomewide association studies. To identify these rare alleles we need high-throughput, high-accuracy resequencing technologies. Although array-based genotyping has allowed genomewide association studies of common SNPs in tens of thousands of samples, array-based resequencing has been limited for 2 main reasons: the lack of a fully multiplexed pipeline for high-throughput sample processing, and failure to achieve sufficient performance. We have recently solved both of these problems and created a fully multiplexed high-throughput pipeline that results in high-quality data. The pipeline consists of target amplification from genomic DNA, followed by allele enrichment to generate pools of purified variant (or nonvariant) DNA and ends with interrogation of purified DNA on resequencing arrays. We have used this pipeline to resequence ≈5 Mb of DNA (on 3 arrays) corresponding to the exons of 1,500 genes in >473 samples; in total >2,350 Mb were sequenced. In the context of this large-scale study we obtained a false positive rate of ≈1 in 500,000 bp and a false negative rate of ≈10%.
Keywords: multiplex, mismatch repair detection, single nucleotide polymorphism (SNP), mutation
Genetic variability is clearly a major contributor to many common diseases (1), although the relative importance of common and rare alleles has been hotly debated (2–4). Recent advances in array technologies have allowed large-scale association studies (with thousands to tens of thousands of samples) of common alleles in common diseases and confirmed that common alleles play a role (5–7). Generally these common alleles have low to moderate genetic effects. Although technology has been lacking to perform large-scale association studies of rare allele, there are several reasons to believe that they, too, play a fundamental role in common disease. First, studies of several candidate genes have shown that rare alleles can contribute to common diseases (8–11). Second, for many common diseases the failure of linkage studies to generate robust linkage signals argues against the presence of common alleles with large relative risk; the data are instead consistent with rare alleles of large or small relative risks or common alleles with small relative risks (12). Finally, although exhaustive common allele studies have been performed for several diseases, only a small fraction of the genetic contribution is explained (13). The remaining genetic contribution is likely caused by both common alleles with extremely small effects (which will require even larger association studies) and rare alleles (which may have substantial genetic effects) (14, 15).
Array-based platforms have allowed the study of tens of thousands of samples in whole-genome studies. Unfortunately, even though resequencing arrays have long been available (16) their use has been much more limited for 2 reasons: the lack of a fully multiplexed pipeline that allows high-throughput processing and the inability of the performance to reach the low false positive rate (<10−5) necessary for large-scale resequencing-based association studies (see SI Text and Fig. S1).
We have developed solutions to both of these limitations and propose arrays as a reasonable method of high-throughput resequencing. We have developed a pipeline that uses target amplification by capture and ligation (TACL) to amplify the specific loci of interest. TACL is then followed by an allele enrichment step with mismatch repair detection (MRD) in which amplicons (average size 160 bp) carrying variant and nonvariant alleles are separated into almost pure homozygous states. Enriched alleles are then sequenced on the array with an automated algorithm making sequencing calls. This pipeline is fully multiplexed with 10,000 amplicons processed in a single reaction, and sample handling (except the array handling steps) is done in 96-well plates.
With this pipeline, we have recently resequenced exons from 1,500 genes (≈5 Mb) in 473 samples; in total we sequenced 2,351 Mb. This technology enables the generation of high-quality sequencing data over many megabases in thousands of samples; these type of data are necessary for resequencing-based association studies.
Results
We have developed and implemented a resequencing pipeline that consists of target capture, allele enrichment, array detection, and automated variant calling. The target capture method, TACL, is capable of jointly amplifying many thousands of amplicons. The allele enrichment method, MRD, efficiently separates variant and nonvariant alleles from thousands of amplicons simultaneously. The resequencing array then enables the identification of the variant through an automatic base-calling algorithm.
Target Capture by TACL.
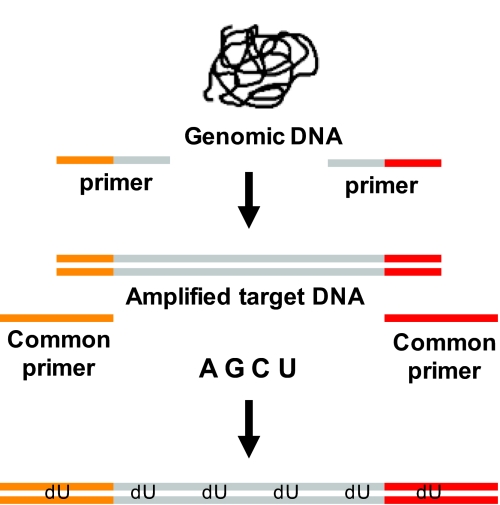
TACL uses probes to capture specific sequences of interest. Probes were made once and used with every sample studied. These dU probes are dsDNA containing “target” sequences unique to each dU probe flanked by 2 sequences common to all dU probes (Fig. 1 shows probe manufacture). The unique sequences range in size between 70 and 350 bp, with an average size of ≈160 bp, and match the target sequence to be captured from the genome except that all of the deoxythymidines (T) in the dU probe sequences are replaced by deoxyuridine (U).
Fig. 1.
Schematic of dU probe manufacturing process. Individual PCRs are performed for each locus. Genomic DNA is amplified with primers that contain sequences specific to the region of interest (gray) and 13-bp sequences common to all primers (orange and red) appended the 5′ end. After amplification of target DNA, all of the PCR products are pooled. A small aliquot is then used for a secondary amplification with common primers (orange and red). The common primers are 30 nt long and are able to amplify all of the amplicons because they have 13 nt in common with all of the amplicons. In this PCR dUTP is used instead of dTTP. The results in the generation of the dU probes that are double-stranded sequences with all of the Ts replaced by Us. Each dU probe has a unique sequence flanked by 30 nt common to all of the dU probes.
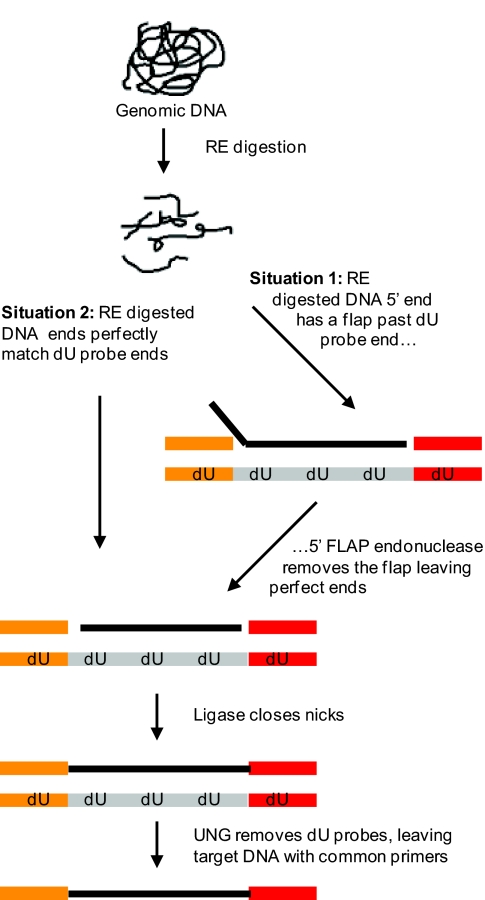
The dU probes were then pooled together and used to capture sequences of interest from genomic DNA; dU probes were hybridized with restriction enzyme (RE)-digested genomic DNA and common oligonucleotides and then incubated with 5′ flap endonuclease and ligase. The hybridization generates a structure in which one strand comes from the dU probe and the other comes from the common primers and genomic DNA (Fig. 2). This process creates a duplex in which one strand is the dU probe and the other is genomic DNA flanked by common primers. The dU probe is then removed with UNG.
Fig. 2.
TACL. Genomic DNA (black) is digested with RE and hybridized with the pool of dU probes and common primers. If the dU probe was designed with RE sites at each end (Situation 1), the digested DNA will hybridize with dU probe and common primers to create a double-stranded structure with 2 nicks. To increase genomic coverage, some dU probes were designed so that the 3′ end of digested DNA has a PM, but the 5′ does not, generating a structure with a flap up to 1,000 bp long (Situation 2). The use of 5′flap endonuclease makes the 5′ end a substrate for a thermostable ligase that is able to close the nicks. UDG and heat treatment destroy the dU probes, leaving only genomic DNA liagated to common primers that can be later amplified by using common primers.
Given that a RE site is required at the 3′ end of the target fragment, this method is not completely flexible, and it is not possible to capture some specific sequences (those on very large or very small restriction fragments). With 1 RE, however, ≈84% of the exonic bases are in restriction fragments of the correct size (between 70 and 1,350 bp), and with 3 enzymes, 99% of exonic bases are in such fragments. To study the exons of 1,500 genes, we constructed 3 panels each using a different RE (AluI, DdeI, and HpyCH4V). In each of the panels there were slightly >10,000 amplicons covering ≈1.65 Mb; in total the panels covered ≈5 Mb.
For each of 3 REs, we performed TACL to amplify simultaneously 10,000 amplicons encompassing ≈1.65 Mb of DNA. The capture was very specific. When DNA such as herring sperm DNA was used instead of human, no amplification product was obtained, which is indicative of very low intrinsic background, because protocols that involve amplification often amplify some material even in the absence of proper targets. Because of the high specificity small amounts of input DNA could be successfully amplified. We typically used 150 ng per reaction, but we have successfully used as little as 3 ng of DNA. Capture success did not have a substantial dependence on amplicon or flap size (Fig. S2), within the range of sizes we allowed (probes 70–350 bp; flap <1,000 bp).
Allele Enrichment by MRD.
The allele enrichment technology using MRD has been described (17–19); it relies on the exquisitely-precise ability of bacteria to detect and repair mismatches. Bacterial repair of a mismatch (which only occurs when there is a variant) triggers the corepair of a marker gene, cre, which in turn removes genes for antibiotic resistance and sensitivity. After transformation 2 pools are generated through bacterial selection, the variant pool (VP) and the nonvariant pool (NVP). These pools are hybridized on separate resequencing arrays. The utilization of MRD leads to dramatic improvement of the data quality from arrays for 2 main reasons: the information on whether a fragment was present in the VP or NVP, and the enrichment of the variant allele to near homozygousity in the VP. All steps including transformation are performed in a 96-well format.
Standards.
The allele enrichment by MRD relies on hybridization of the test sequences with cloned references (standards). The same pools of individual PCR products that were used in making the dU probes were also used to make standards (details in SI Text). Making the standards used the same labor-intensive step of individual PCR as for making the dU probes, but once constructed, they could be used with every sample that was studied, making the investment in the generation of these reagents reasonable when many hundreds or thousands of samples are to be studied. Many steps (PCR with U for the dU probes and ligation and 2 transformations of the standards) were performed en masse at >1,000 plex. A highly efficient alternative method for standard construction that eliminates the labor intensive step of individual PCR is discussed in Fig. S1.
Allele enrichment through bacterial selection.
All of the standards were hybridized to the TACL PCR products from test samples and vector sequences lacking inserts (17). For our study of the exons of 1,500 genes, >10,000 amplicons covering ≈1.65 Mb were hybridized in each of the 3 panels (one per RE used in TACL). The hybridization formed heteroduplex molecules with 2 nicks that were closed by a thermostable ligase. This heteroduplex mix was transformed en masse into a bacterial Escherichia coli strain engineered to respond in a specific way the presence of a mismatch. In the presence of a mismatch (or variant) the bacterium grew in 1 medium (VP) and in its absence it grew in another (NVP). The VP and NVP were prepared and the plasmid inserts were amplified through the use of common primers. The contents of these inserts were analyzed by hybridization on arrays.
Bacterial cloning is often criticized as inefficient. Inefficiency of this process is caused by the colony picking and processing steps and inefficiency of intermolecular ligation. We largely avoided both problematic steps. No individual colony was processed in the standard making or the MRD assay; instead we used cultures of bacteria carrying many inserts. Whereas making the standards required an intermolecular ligation reaction, in the MRD assay the hybridization event forms the appropriate molecule requiring only the more efficient intramolecular nick closure to generate the heteroduplex molecule. The ligation for making the standards can be less efficient as it is performed only once.
Differential detection of different types of mismatches.
E. coli detects single-base mismatches and small insertions/deletion (indels) 1–3 bp in length (there is partial detection of 4-bp indels). We evaluated the enrichment of the different classes of single-base variants. We demonstrated that we are able to robustly detect all of the different types of mismatches (SI Text).
Detection by Resequencing Array.
Array design is shown in Fig. S3. On each chip, there were ≈1.65 M perfect match (PM) probes that matched the reference sequence of the human genome. Each reference base in the genome had a PM probe in which it lay in the middle position (13th as probes were 25 bp long). Each PM probe had 3 matching mismatch (MM) probes in which the middle base had been replaced with each of the 3 other bases (e.g., if the PM probe had C in the middle position the MM probes were identical except they had A, G, or T in the middle). (Each chip has ≈4.95 M MM probes.) Because probes were tiled along the genome, each position in the genome was represented in 25 different PM probes (at the first position on one probe, the second position in another probe, … the 25th position in yet another probe).
The probes were complementary to only one of the strands, and the strand switched at every adjacent position. We previously found that correlation of signals from probes in different strands is lower than for neighboring probes on the same strand, and therefore the switching provides more information than solely using 1 strand while requiring only half as many probes as standard resequencing arrays that tile both strands at each position.
Finally, to obtain more information in regions near known SNPs, we made extra tiling probes in these regions. For each PM probe, if there was a known SNP in the single nucleotide polymorphism database (dbSNP) within the 25-mer sequence, then there was 1 additional probe for each allele of the SNP. It was identical to the PM except that at the SNP position, the base matched the nonreference allele of the SNP allele. The use of these probes is discussed below.
Automated Variant Calling.
The variant calling pipeline had 3 layers of analysis. Each analysis was performed separately and generated a score. Results from each layer of analysis were then combined to generate a final combination score. The combination score was compared with a threshold number to determine whether the call was variant or not.
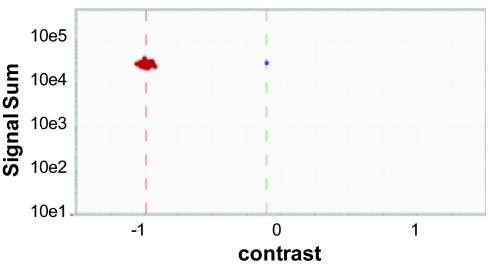
Ratio analysis was performed at the scale of the whole amplicon. If an amplicon contained a variation, it was enriched in the VP, and hence the “ratio” of the amplicon in the VP to that in the NVP was greatly increased relative to samples with no variation for that amplicon (example in Fig. 3). In this analysis, only the PM probes were considered. For each amplicon, the VP signal (Vs) and NVP signal (NVs) were computed as the median signal among all of the PM probes. For an amplicon of size X bp, there were X − 16 PM probes used in this analysis (probe at the ends were ignored). The minimum value of X was 70, and the average was 160. Hence ratio score was based on signal from at least 54 probes and was fairly robust. This analysis requires data from the VP and NVP from every sample. The other layers of analysis focus on the VP and do not require NVP data for each sample. The use of the NVP data in the ratio analysis helps discriminate heterozygous samples (present in both VP and NVP) from homozygous samples (present only in VP).
Fig. 3.
Ratio analysis. The x axis shows the contrast between the Vs and the NVs. The contrast is computed as (Vs − NVs)/(Vs + NVs). Therefore, if the fragment is nonvariant, variant, or heterozygous, the contrast is expected to be −1, +1, or 0, respectively. The y axis is the signal sum (Vs + NVs).
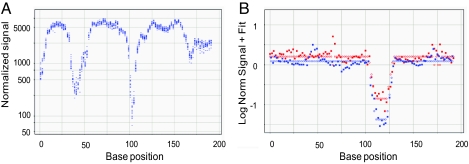
Dip analysis localized that variant to a region of the amplicon. Variants were enriched to near homozygosity in the VP. Therefore, variant sequences exhibit markedly reduced hybridization to the PM probes. This reduction in hybridization occurs for the PM probes when the variant base is within 10 bp of the center of the probe (i.e., for probes with the variant base at positions ≈3–23 in the probe); the most dramatic reduction is seen when the variant is up to 6 bases from the middle position. In most cases this “dip” in hybridization localizes the variant to ±3 bp, but for some variants the uncertainty is as much as ±8 bp. Like the VP/NVP ratio analysis, this dip analysis uses only the PM probes. However, unlike the ratio analysis, the dip is focused mainly on the VP. Only a small number (≈20) of NVP samples are analyzed and these serve as a reference to normalize the signal from different probes as shown in Fig. 4. Because the dip in signal is measured over multiple probes (15–20), it is reasonably robust.
Fig. 4.
Dip analysis. The x axes show position in the amplicon for a 200-bp amplicon. (A) The y axis shows the NVPs obtained at each position with data shown for 19 different samples. Even though the signal differs drastically among positions, the signal for each position among different samples is relatively tight. Hence data from some NVP samples can be used to build a model of the expected signals for each position and data from the VP can be compared to find regions of poor hybridization that may contain a variant. (B) The y axis shows the comparison of the VP pool with the model generated from the NVP samples; data from both strands are shown (solid red and blue circles). Open circles show processed data after dip fitting.
Base analysis identifies the exact sequence change within the dip using all probes (PM and MM) within 8 bp of the center of the dip. At each of 17 positions, there are 3 MM probes and for each we computed the contrast value (MM − PM)/PM + MM), generating 51 contrast values. Because MRD enriches variants to near homozygozity, in the VP there will be little DNA that matches the PM probe, resulting in low PM signal. If there is a variant, however, the matching MM probe should show much stronger signal that the PM probe. The MM probe with the largest contrast value was used to identify the variation.
Performance.
We have recently finished a project resequencing ≈5 Mb of DNA in each of 473 genomic samples. The 5 Mb covered exons of ≈1,500 genes that were chosen based on their potential role in cancer. All of the exons and 10 bp of the surrounding introns were targeted. The targets were covered in at least 1 of 3 pools, each with a specific RE at the TACL step. Each pool had slightly >10,000 amplicons.
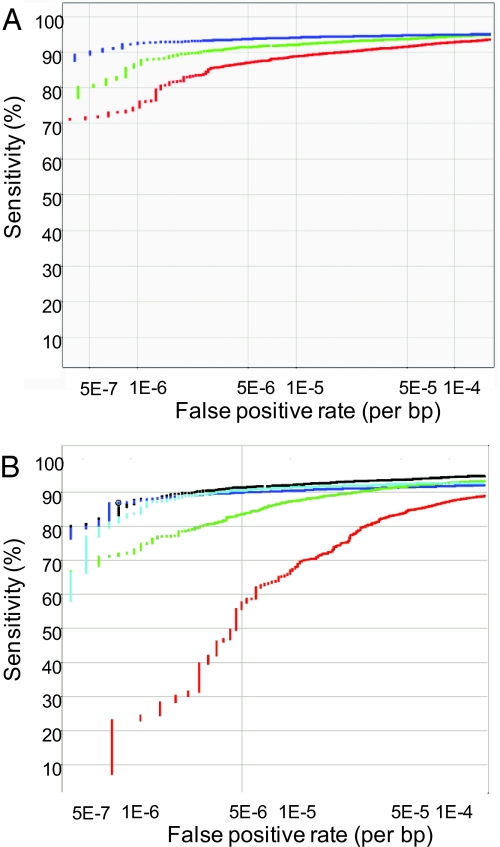
We assessed amplicon performance by measuring several metrics (e.g., average amplicon signal, percentage passed samples for amplicon) and then assessed data quality in the top 84% of amplicons. Twenty-four HapMap samples were used to identify false positives and false negatives. Fig. 5 shows the receiver operator curve (ROC) analysis representing the false positive/false negative tradeoff. As can be seen, at a false positive rate of ≈1 per 500,000 bp, ≈90% sensitivity can be obtained. Of called variants, for >98% we could identify the specific base change; for the remaining 1–2% we localized the variant to either an amplicon or a small dip. Dataset S1 provides variant calls for the 24 HapMap samples at the false positive rates shown in Table S1.
Fig. 5.
ROC analysis. (A) The ROC curve showing tradeoff between false positive and negative for the 3 different enzyme panels/arrays. The 3 panels/arrays have somewhat different performance with the best performance (blue) seen for the Dde panel that carries the MutS-overexpressing strain. The average performance among the 3 panels shows a false positive rate of 1/500,000 bp at a sensitivity of ≈90%. (B) This plot shows more specific performance data for the intermediate performance in A (Hpy). The combination score (dark blue) defines the performance of the technology (and is the same as the green line in A). Much of the power comes from the robust ratio analysis (light blue) that uses data from least 54 probes. Because they use data from fewer features, the dip (green) and the base (red) analyses have lower power. However, they add to the power of the ratio particularly at low false positive rates (comparison of light blue and dark blue lines) and localizing and identifying the specific change. The dark blue curve shows the combo calls that have a base call (i.e., it excludes the class where a combo call is present but the base could not be determined).
Our process and algorithms have no bias toward detecting high-frequency alleles as opposed to low-frequency alleles (except for the fact that a sample with the minor allele must be tested). For example, we did not perform clustering, a process that tends to perform better when there are numerous variant and nonvariant calls. Instead for ratio, dip, and base, we defined a nonvariant cluster and then compared each sample individually to that cluster to generate a score. To confirm the absence of bias, we looked at the performance for SNPs at different frequency bins and found essentially no difference in performance (Table S2).
Variants Identified.
This experiment was designed primarily to detect somatic mutations in tumors and hence does not represent the best sample set for the study of germ-line variants and population genetics. For instance, tumors often have large genomic regions with loss of heterozygosity, greatly decreasing the number of variants they carry. However, given the large size of the dataset we felt there was still value in looking at the pattern of variations identified. In total, there were 817,521 variant calls, among which 735,578 (90%) were in previously known SNP sites (Table S1). There were 3.5 novel (i.e., not present in dbSNP) variant calls per 100,000 bp. Given the rates of somatic mutations in tumors, we expected that most of these were germ line (20, 21). The 817,521 variant calls corresponded to 36,939 unique variants, 29,519 of which (80%) were novel (Table S3). As expected, the allele frequency distribution of the novel SNPs was strongly shifted toward the rarer alleles when compared with known SNPs (Fig. S4).
Discussion
We demonstrate here a high-throughput, high-quality array-based pipeline for resequencing; TACL allowed us to eliminate singleplex PCR, generating a scalable process; MRD dramatically improved array data quality by providing the ratio information on amplicons and enriching variant alleles to near homozygosity. The use of TACL and MRD ameliorated the 2 hurdles that have plagued the large-scale use of resequencing arrays. Genotyping arrays have enabled large association studies through genotyping tens of thousands of samples. By creating appropriate “upfront” processes for resequencing arrays we have created the potential to conduct similar large-scale resequencing-based association studies. In this work we studied ≈5 Mb of DNA representing exonic sequences of ≈1,500 genes in 473 samples with high accuracy, generating sequence for 2,351 Mb in total. Most of the samples studied were tumor samples in which we detected ≈2,000 nonsynonymous somatic mutations validated by independent methods. These confirmed mutations provide additional validation to the technology.
We previously used MRD with tag arrays to identify variant fragments; identifying the exact sequence change required follow-up sequencing. Our current study's use of resequencing arrays streamlines this process. In addition >1 variant can now be identified in a fragment.
Although our current process has allowed us to efficiently generate 2,351 Mb of sequence data, there are 5 ways in which we hope to improve the process in the future (details in SI Text and Fig. S5). Reducing the number of arrays by 1/3 through combining the NVP arrays should be straightforward, as should improving amplicon pass rate by optimizing hybridization conditions for AT-rich amplicons. Preliminary results for the other 3 methods (reducing dU probe and standard cost through array synthesis followed by cleavage; reducing array size/cost through enzymatic base discrimination rather than hybridization; and further reducing array size/cost through decreasing feature size) are promising.
Recently, several parallel sequencing techniques have been developed (22). So far, these techniques have not yet been applied for large-scale resequencing (thousands of genes in hundreds to thousands of samples). Even though the costs of these techniques are dropping, all require high levels of redundancy for highly-accurate detection of heterozygous variants, in part because different genomic fragments will be present at different concentration after amplifications. We note that the 3 modules of our technology (TACL, MRD, and array) are all independent of each other; hence TACL and/or MRD can be used in conjugation with the parallel sequencing technologies as these techniques become more cost effective.
Materials and Methods
Analysis.
After the ratio, dip, and base analyses are completed, calls are made through the use of a combo score based on all 3 scores.
Triple combo analysis.
Our calling algorithm combines the above 3 analyses. For each analysis, a score is given for each amplicon in every sample. A combined score is then generated from all 3 scores, and then a threshold is set above which data points are called variants and below which the data points are called nonvariants. In a small fraction (2%) of the calls with positive combo calls, the base could not be determined.
Double combo analysis.
When >1 variant is present in the same sample for the same amplicon, a modification of the algorithm is required. The simple modification is after identification of the first variant, one can look for another variant using the second and third analysis layers (dip and base) but not the ratio because the ratio pertains to the amplicon as a whole and is thus already affected by the first variant. In this case we can only combine the dip and base analyses into a single score for calling these variants. We have limited the search for >1 variant in the same sample and amplicon to those cases where one of the variants corresponds to a variation at a SNP already known in dbSNP.
SNP analysis.
In addition to the above levels of analysis, another analysis is done for known SNPs. In the array there are extra probes for all of the SNPs (and insertion/deletions) in dbSNP. Each of the 2 alleles of all SNPs is tiled in all 25 positions with respect to location of the SNP within the probe sequence. We use the 15 probes where the SNP is <8 bases from the probe center to do the SNP analysis. The status of the allele can be surmised through the median (from 15 probes) contrast between the 2 alleles in the VP of the specific sample compared with the contrast of all of the sample that are determined from the ratio to be nonvariant. This SNP analysis is done for all known SNPs. In those samples with identified variant alleles in the known SNPs, a search is done for any other variants at all other potential sites in the same amplicons.
Variant Detection.
The pipeline for detecting variants in samples of interest is as follows: (i) Do SNP analysis on all of the known SNPs. This will find variants that occur at known SNPs. (ii) Based on this result use 1 of 2 strategies to find variants that occur at unknown SNPs.
For triple combo analysis, combine the ratio, dip, and base analyses together for all cases where a sample has no variant alleles in the known SNPs (for the given amplicon). This represents ≈94% of all of the variants that occur at unknown SNPs, because in most cases there is no variation present at the known SNPs for each amplicon.
In double combo analysis, for those cases where a sample has 1 (or more) variations in the known SNPs of the given amplicon (based on the SNP analysis) do a double combo analysis (dip and base only) to search for additional variants at other sites than the known SNPs. This only represents ≈6% of the variants that occur at unknown SNPs, because in most cases there is no variation present at the known SNPs for each amplicon.
Calibration.
To calibrate the efficiency of the above variant detection pipeline we run 3 separate kinds of calibration analyses:
SNP calibration.
Use known genotypes of HapMap SNPs to determine the sensitivity and false positive rate of the SNP analysis.
Triple combo calibration.
We restrict this analysis to amplicons with at most 1 known (HapMap) SNP in the entire amplicon. Wherever HapMap indicates the sample is variant at the SNP we perform the triple combo analysis (ratio, dip, and base) and ignore the SNP analysis. This effectively determines the sensitivity for detecting a new variant that is not in the presence of a variant at a known SNP for the given amplicon and sample, i.e., as in triple combo analysis) of the variant detection pipeline above. The false positive rate is determined by using amplicons that have been Sanger-sequenced to find samples where there are no variants across the entire amplicon. These nonvariant cases are then subjected to the triple combo analysis (ratio, dip, and base). Because we know that no variants are expected from the Sanger sequencing, any detected variants are false positives.
Double combo calibration.
For finding new variants in the presence of variants at known SNPs (for the given amplicon and sample), i.e., as in double combo analysis) of the variant detection pipeline above, we must use a different approach to calibrate our efficiency of detecting the second (new) variant. In this case we use amplicons with exactly 2 HapMap SNPs and no other known SNPs. We consider cases where HapMap indicates the sample is variant at both SNPs. We then perform, at both SNPs, a double combo analysis (dip and base only) that ignores the SNP analysis and the ratio analysis. This allows us to calibrate the sensitivity for finding the second variant because we are using exactly the same information (dip and base) as in double combo analysis of the variant detection pipeline. The false positive rate is determined by using the same Sanger-sequenced nonvariant data as in triple combo calibration but instead applying the double combo analysis (dip and base only) to find any false variants.
At first glance it might seem troubling to use known (HapMap) SNPs to calibrate the sensitivity for detecting variants at unknown SNPs. However, in both the triple and double combo analyses, no information from the SNP analysis or any of the SNP specific probes are used. In addition, all of the metrics used in the ratio, dip, and base analyses are explicitly chosen so that they do not depend on the frequency of the variation within the sample population. This ensures that the sensitivity does not depend on allele frequency, which would otherwise create a bias because known SNPs are of generally higher allele frequency than unknown SNPs. Nonetheless, it is a fair criticism to suggest that HapMap SNPs have some level of selection bias in that they have been found to perform well on at least 1 genotyping platform. Given the similarity of different genotyping platforms, detection of these is potentially “easier” to detect than “average” SNPs. The effect is expected to be small, particularly because the allele enrichment is unlikely to be well correlated with the ability to genotype on other platforms.
Determining False Positive and Negative Rates.
To determine false positive and false negative rates we needed data representing true positives and true negatives. The true positives were easily available through the HapMap. Among the 473 studied samples, there were 8 HapMap trio samples (i.e., 24 in total). To avoid potential confounding when 2 variants were in the same amplicon, we limited analysis to amplicons with only 1 HapMap variant and no reported additional dbSNP variants. (When there are 2 variants in an amplicon, both contribute to the ratio score.) The 38,561 known variants from HapMap represent true positives allowing us to compute the false negative rate of this technology. (Although the array contained probes specific for known SNPs to allow us to identify variant near SNPs, for the false positive analysis we called SNPs using the same method used to detect other variants, a combination of ratio, dip, and base scores. Using the SNP-specific probes could artificially inflate our ability to detect known SNPs compared with other variants.)
The false negatives were harder to obtain because SNPs not assessed by the HapMap may be present in the studied samples. We have used Mendelian inheritance to identify true negatives. Fragments that are clearly not carrying a variant in the 2 parents are expected to have no variant in the child and hence are treated as true negatives. The data shown in Fig. 5 were obtained by using 10 Mb per enzyme per panel from this true negative data. We have also done traditional Sanger sequencing followed by extensive manual review to obtain a true negative data for ≈750,000 bp. Data from this smaller set of true negative data were consistent with what was obtained by using the Mendelian inheritance true negative data. As noted above, even though there were extra probes corresponding to the SNP sites, we did not use them in this analysis and hence the performance should be the same as for previously-unknown variants.
Supplementary Material
Acknowledgments.
We thank Sumathi Venkatapathy, Laura Miller, and Wipapat Kladwang for assistance with laboratory work and Francisco Useche for bioinformatics support.
Footnotes
Conflict of interest statement: This work was part of the research done at Affymetrix Laboratory, the research arm of Affymetrix. However, there are no current products or specific plans to make products of work described in this manuscript.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901902106/DCSupplemental.
References
- 1.Chakravarti A, Little P. Nature, nurture, and human disease. Nature. 2003;421:412–414. doi: 10.1038/nature01401. [DOI] [PubMed] [Google Scholar]
- 2.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 3.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: Common disease–common variant or not? Hum Mol Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 5.Plenge RM, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxena R, et al. Genomewide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 7.Zeggini E, et al. Replication of genomewide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 9.Vaisse C, et al. Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J Clin Invest. 2000;106:253–262. doi: 10.1172/JCI9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CHEK2 Breast Cancer Case-Control Consortium. CHEK2*1100delC and susceptibility to breast cancer: A collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74:1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 12.Jones HB, Faham M. Evidence and implications for multiplicative interactions among loci predisposing to human common disease. Hum Hered. 2005;59:176–184. doi: 10.1159/000086118. [DOI] [PubMed] [Google Scholar]
- 13.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyengar SK, Elston RC. The genetic basis of complex traits: Rare variants or “common gene, common disease?”. Methods Mol Biol. 2007;376:71–84. doi: 10.1007/978-1-59745-389-9_6. [DOI] [PubMed] [Google Scholar]
- 15.Polychronakos C. Common and rare alleles as causes of complex phenotypes. Curr Atheroscler Rep. 2008;10:194–200. doi: 10.1007/s11883-008-0031-1. [DOI] [PubMed] [Google Scholar]
- 16.Hacia JG. Resequencing and mutational analysis using oligonucleotide microarrays. Nat Genet. 1999;21(Suppl 1):42–47. doi: 10.1038/4469. [DOI] [PubMed] [Google Scholar]
- 17.Faham M, et al. Multiplexed variation scanning for 1,000 amplicons in hundreds of patients using mismatch repair detection (MRD) on tag arrays. Proc Natl Acad Sci USA. 2005;102:14717–14722. doi: 10.1073/pnas.0506677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters BA, et al. Highly efficient somatic-mutation identification using Escherichia coli mismatch-repair detection. Nat Methods. 2007;4:713–715. doi: 10.1038/nmeth1081. [DOI] [PubMed] [Google Scholar]
- 19.Bentivegna S, et al. Rapid identification of somatic mutations in colorectal and breast cancer tissues using mismatch repair detection (MRD) Hum Mutat. 2008;29:441–450. doi: 10.1002/humu.20672. [DOI] [PubMed] [Google Scholar]
- 20.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 21.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: Clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 22.Holt RA, Jones SJ. The new paradigm of flow cell sequencing. Genome Res. 2008;18:839–846. doi: 10.1101/gr.073262.107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.