Abstract
The ability to inhibit an enzyme in a specific tissue with high spatial resolution combined with a readily available antidote should find many biomedical applications. We have accomplished this by taking advantage of the cis–trans photoisomerization of azobenzene molecules. Specifically, we positioned azobenzene moieties within the DNA sequence complementary to a 15-base-long thrombin aptamer and then linked the azobenzene-modified cDNA to the aptamer by a polyethylene glycol (PEG) linker to make a unimolecular conjugate. During the photoisomerization of azobenzene by visible light, the inhibition of thrombin is disabled because the probe hybridizes with the cDNA in the trans-azobenzene conformation so that the aptamer cannot bind its target thrombin. However, when UV light is applied, melting of the hairpin structure (duplex) is induced via trans-to-cis conversion, thereby changing conformation of the aptamer and making the aptamer free to bind to and inhibit its target thrombin. By using standard clotting assays, we measured the IC200 of various probe designs in both states and concluded the feasibility of using photon energy to temporally and spatially regulate these enzymatic reactions. Thus, we can report the development of DNA probes in the form of photon-controllable (thrombin) inhibitors, termed PCIs, and we expect that this approach will be highly beneficial in future biomedical and pharmaceutical applications.
Keywords: anticoagulation, aptamer, enzyme inhibitor, photo controllable, thrombine
Over the last decade, extensive research has been devoted to the study of phototransformable molecules based on photochromism chemistry. This science has been applied to photooptical technology and such photomodulated devices as eyeglasses called Transitions (1–4). A photochromic compound has 2 molecular states: stable and relatively unstable. The 2 states are interchangeable by the effect of irradiation using different wavelengths and differ from one another in terms of both physical and chemical properties. The photoconversion mechanism can largely be divided into 3 transformation types: photochromic tautomerism, cis–trans isomerization, and photocyclization. Briefly, photoisomerization is a process in which molecular structural change between isomers is caused by photoexcitation. Therefore, because isomerization causes a conformational change that can change the overall structure of a molecule, cis–trans isomerization is an intriguing mechanism that can be used to regulate mechanical devices and biological reactions (5–8).
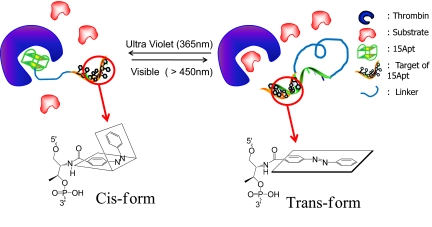
As one of the most popular phototransformable molecules in use today, azobenzene and its derivatives belong to the cis–trans isomerization category and are composed of 2 phenyl rings linked by a N N double bond (Fig. 1) (9). The 2 isomers can be switched with particular wavelengths of light: UV light at 365 nm, corresponding to the trans-to-cis conversion, and visible light at 465 nm, corresponding to the cis-to-trans isomerization. There are reports that demonstrate the possible applications of such a feature in the development of sensors (10), nanomotors (11–13), and even peptide engineering (14–16). These reports involved the use of enzymes that naturally act on DNA. However, we are interested in regulating enzymes that do not naturally act on DNA, and, at the same time, we want to take advantage of the special reactivity of azobenzene to photon energy. Therefore, we will focus our molecular design on using azobenzene to regulate the binding of DNA aptamers that have enzyme inhibitory capabilities.
N double bond (Fig. 1) (9). The 2 isomers can be switched with particular wavelengths of light: UV light at 365 nm, corresponding to the trans-to-cis conversion, and visible light at 465 nm, corresponding to the cis-to-trans isomerization. There are reports that demonstrate the possible applications of such a feature in the development of sensors (10), nanomotors (11–13), and even peptide engineering (14–16). These reports involved the use of enzymes that naturally act on DNA. However, we are interested in regulating enzymes that do not naturally act on DNA, and, at the same time, we want to take advantage of the special reactivity of azobenzene to photon energy. Therefore, we will focus our molecular design on using azobenzene to regulate the binding of DNA aptamers that have enzyme inhibitory capabilities.
Fig. 1.
Xcomp/Yazo probes. The working principle is that dissociation and association of the 2 domains report high and quenched fluorescence signal, respectively. We assign test probes the following nomenclature. Xcomp equals the number of complementary sequences, and Yazo equals the number of incorporated azobenzene molecules. The trans- and cis- conformation is reversibly regulated by different input of electromagnetic radiation of the energy. Thus, when probes are treated with visible light, the regulatory domain is hybridized, and the probe is released from its target, resulting in activation of the enzyme (thrombin). When treated with UV light, probes form the open conformation, and the inhibitory domain can bind to the target, causing low enzymatic activity. The use of Xcomp/Yazo creates combinations that afford the opportunity to optimize probe design in accordance with thermal stability, thrombin affinity, and the results of clotting assays.
The rational design of nucleic acid (DNA) aptamers for this purpose is, therefore, a key part of our developmental strategy. As a previously uncharacterized class of binding elements, nucleic acid aptamers selected by the SELEX (Systematic Evolution of Ligands by Exponential Enrichment) method have shown some outstanding features (17–20). For instance, DNA-aptamers possess a relatively small molecular weight coupled with high binding affinity. Structurally, DNA-aptamers are easily modifiable with robust structural restoration under ambient conditions (20). For these reasons, many nucleic acid sequences have been selected for various types of targets, and some of them have shown considerable clinical potential. Among them, thrombin-binding aptamers (TBAs) are some of the most popular DNA sequences (21, 22). Of the 2 known TBAs, 1 is 15 bases long (15Apt) and binds to exosite 1 of thrombin. Because it occupies the fibrinogen-binding pocket, only 15Apt can inhibit thrombin-mediated coagulation (23). Thrombin is important in the regulation of homeostasis and is associated with many types of cancer. Therefore, engineering probes that target thrombin and, moreover, whose function can be precisely regulated by photonic energy, can advance anticoagulation therapy for many diseases (24, 25).
Photoregulation of azobenzene conformation and DNA duplex structures using azobenzene is a well-known and studied phenomenon. We also believe that it is a key component to the functionalization of probes engineered to target thrombin. This conclusion is based on the work that has been done on synthesizing azobenzene phosphoramidite monomers and the fact that the NMR structures of both cis- and trans- states have been described (26–27). Most recently, a full thermodynamic description based on nearest-neighbor models of DNA duplex formation has been presented and successfully used to predict melting temperatures of azobenzene-incorporated DNA duplexes (29). Finally, because researchers have shown its ability to regulate enzymes (30, 31), we chose azobenzene as the fundamental component of our thrombin-binding aptamer, which we term photocontrollable inhibitor, or PCI.
Our PCI design has 3 functional domains: inhibitory, regulatory, and linking (Fig. 1). The inhibitory domain is a 15Apt sequence. As noted above, 15Apt binds to exosite 1 of thrombin. Because it occupies the fibrinogen-binding pocket, only 15Apt can inhibit thrombin-mediated coagulation (23). Next, the regulatory domain is composed of the complementary sequence to 15Apt, but with azobenzene molecules inserted into the phosphate backbone of the DNA sequence using an automated DNA synthesizer (31). Finally, the linking domain ties the inhibitory and regulatory domains together. Importantly, because the regulatory domain is composed of photochromic molecules, the affinity of the inhibitory domain can be altered by different light treatments, as discussed above. Briefly, UV light induces trans-to-cis isomerization, resulting in a low binding affinity of the regulatory domain to 15Apt. This alteration frees 15Apt for binding to exosite 1 of thrombin. On the other hand, visible light reverses the conformation of the regulatory domain, allowing it to hybridize 15Apt. This results in the low affinity of 15Apt for thrombin, thus enabling thrombin to hydrolyze fibrinogen for coagulation. Or, stated another way, the inhibition of thrombin is disabled because the probe hybridizes with the cDNA in the trans-azobenzene conformation so that the aptamer cannot bind its target thrombin.
To optimize the PCI, we designed and characterized a series of candidate molecules with different lengths of cDNA and different amounts of azobenzene in the inhibitory domain. The inhibitory potency of these probes in both the ground and excited states was measured by using prothrombin time (PT) measurement in human plasma. The results demonstrate the real-time ability of the cis-form of these probes to switch on thrombin's activity under visible illumination, or, conversely, switch off thrombin's activity under UV illumination. Moreover, by using a microscopic fluorescence-based PT assay, a second key part of our investigation sought to confirm the feasibility of sequential site-specific activation of coagulation by spatial control of thrombin inhibition.
Results and Discussion
Optimization of Probe Designs.
To design effective photon-controllable inhibitors, the complementary sequences to 15Apt (regulatory domain) were modified with azobenzenes, and the inhibitory and regulatory domains were linked by using a polyethylene glycol (PEG) linker composed of 5 units of hexa-ethylene glycols conjugated by using phosphoramidite chemistry. Several factors influenced our decision to incorporate azobenzene into the complementary sequences of 15Apt rather than the 15Apt sequence itself. First, the complementary sequence of 15Apt can function as an excellent antidote for 15Apt, as a result of its high binding affinity via Watson–Crick base pairing (32–33). In other words, had we decided to incorporate azobenzene moieties to 15Apt, we would have lost the complementary antidote effect. Second, even minor modifications to 15Apt can result in significant disruption of interacting configurations (34), which are generally unpredictable without computational simulation or experimental investigation. Thus, modifying the complementary 15Apt sequence with azobenzene is a more feasible approach. Five units of hexa-ethylene glycol (≈10 nm) were used to ensure close proximity of the inhibitory and regulatory domains (35). This complex then resulted in a unimolecular conjugate. In fact, it is the unimolecular nature of our design that allows the molecule to switch between active and inactive inhibitory states without the addition of another strand of DNA. In this way, reaction was allowed to occur between the regulatory and inhibitory domains in a manner that maximized the efficiency of dissociation and association between them according to the number and position of azobenzene insertions, which is explained later in this article. This was expected because small alterations are enough to disrupt intramolecular interactions, whereas more significant alterations are required to break intermolecular interactions.
Optimization of probe design also depends on maximizing the efficacy of photoisomerization. This involves adjusting both the number and position of azobenzene molecules and base pairs in the regulatory domain. Previous reports have shown that the insertion of azobenzene molecules (trans conformation) to the DNA chain can destabilize or stabilize duplexes of DNAs depending on their positions. Thus, the most common method of regulating DNA duplex conformations is to alternate every 2 bases with a single azobenzene phosphoramidite. Although this strategy works well at high temperatures, a maximum of 7 azobenzene molecule insertions did not result in a kinetically favorable duplex transition within the 15-bp stem under the reaction conditions necessary to perform the PT assay (37 °C and physiological salt). Therefore, we investigated the feasibility of alternating azobezene moieties between every other nucleotide. Using this protocol, we could potentially have a probe with 15 or 16 azobenzene incorporations within the regulatory domain.
These conditions combined with the potential of azobenzenes to destabilize our probe design required us to test a series of molecular probes having different numbers of azobenzene and base pairings [supporting information (SI) Table S1]. Each probe contained a FRET pair (fluorescein and dabcyl) as a signaling element to monitor the hybridization and dehybridization between the regulatory and inhibitory domains (36). The working principle is that dissociation and association of the 2 domains report high and quenched fluorescence signal, respectively. Our protocol can best be understood if we assign probes the following nomenclature. Let Xcomp equal the number of complementary sequences, and let Yazo equal the number of incorporated azobenzene molecules. A series of combinations, such as 8comp-4azo or 10comp-4azo, then arise that enable us to measure, compare, and characterize these probe combinations by (i) melting temperature, (ii) binding affinity to thrombin, and (iii) clotting assay. Depending on the number of azobenzene incorporations, these molecular probes had significantly different properties, as summarized in Table S2. Specifically, in this experiment, we investigated what prototype probe design would work best at room temperature (RT) and whether different ranges of photoalteration could be achieved by adjusting the number of base pairing.
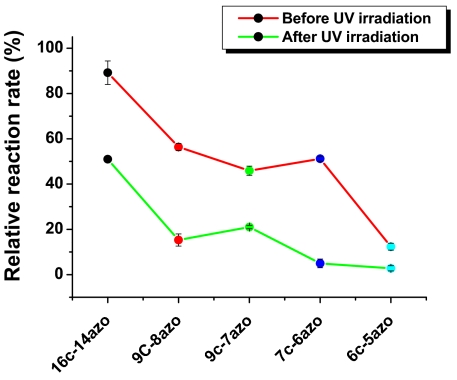
First, we will consider the property of thermal stability. We found that probes with ≤40% azobenzenes per nucleotide, e.g., 10c-4azo, 8c-4azo, 7c-3azo, and 6c-3azo, showed higher melting temperatures than non-azobenzene-incorporated probes. It is a well-known phenomenon that the insertion of azobenzenes between at least 2 nucleotides can enhance duplex stability by enhanced π–π stacking interaction. On the other hand, the continued insertion of azobenzenes causes significantly reduced stability. The reduced thermal stability is clearly shown by comparing the Tms of probes with the same number of base pairs, but different percentages of azobenzene incorporations, e.g., 7c-6azo < 7c-4azo < 7c-3azo. In this example, a negative correlation is established because thermal stability decreases with the increase of azobenzene molecules, even though the number of base pairs stays the same. Next, we ran a fluorescent assay to determine the maximum number of azobenzene incorporations consistent with duplex stability under either UV or visible (Vis) light. Results showed that molecular probes containing a low percentage of azobenzenes did not respond differentially to either UV or Vis light, indicating that the duplex had not dissociated. On the other hand, probes possessing the maximum number of azobenzenes per regulatory domains (16c-15azo, 14c-13azo, 12c-11azo, 10c-9azo, 9c-8azo, 7c-6azo, and 6c-5azo) showed dramatic decreases in duplex stability after illumination with UV light (Fig. 2). Taking these findings together, we concluded that probe optimization depended on incorporation of at least 1 azobenzene between every 2 nucleic acids in the regulatory domain and that the prototype probes demonstrate different ranges of photoalteration based on the number of base pairings.
Fig. 2.
Photo-regulating inhibitory function of probes.
Probes Can Act as a Photoswitching Anticoagulant.
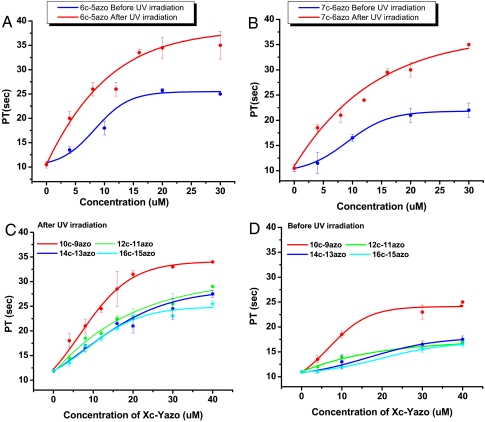
Although the above results showed that the duplex structure of our probe design thus far can be regulated with UV and Vis light under physiological conditions (i.e., RT), complete optimization still required an additional step. Specifically, to demonstrate that these probes are also capable of regulating human blood clotting reaction, the PT of each probe was measured in the cis- or trans-conformations (37). PT measures how long it takes for blood to clot. As such, PT testing is used frequently as a standard and accurate method to assess bleeding problems and the efficiency of anticoagulants. In our experiment, the results obtained from the PT tests were plotted against probe concentration and fitted to a sigmoid curve. Then, the concentration most effective in doubling the plasma clotting time, the IC200, was calculated for each probe. By the theory underlying our probe engineering, it was expected that probes in the trans state would show shorter PTs and higher IC200, whereas probes in the cis state would have relatively longer PTs with lower IC200 under identical sample preparation and experimental conditions. Typical results are shown in Fig. 3 A and B. In accordance with the reasoning indicated above, the PTs of trans probe samples are much shorter than those of cis probe samples. Also, a general trend of increased clotting time correlated with increased number of base pairs as can be seen by comparing probes 6c-5-azo with 10c-9azo (Fig. 3 A and D). However, probes 12c-11azo through 16c-15azo showed only negligible difference in PT (Fig. 3 C and D).
Fig. 3.
PT measurement using each probe -cis and -trans (A–D). Dose–response of 6c-5azo or 7c-6azo -cis and -trans was plotted in each graph (A) or (B), respectively. Dose–responses of 10c-9azo, 12c-11azo, 14c-13azo, and 16c-14azo -cis or -trans are compared (C) or (D), respectively. We then obtained the dose–response curve, and IC200s of each -cis and -trans are summarized in Table 1.
The difference in probes is more obvious when we compare the concentration most effective in doubling the plasma clotting time, or the IC200 value (Table 1). That is, to determine the optimal probe design, we looked for probes showing the highest differential IC200, thereby showing the largest difference in anticoagulation response upon treatment with UV or Vis light. We expected the IC200 values of both states of the 6c-5azo probe samples to be the lowest. We also expected that IC200 values for the cis forms would increase with an increasing number of base pairs. We found that probes with >10 bp were too stable to have a detectable difference in IC200 between the trans and cis states. The difference between IC200-cis and IC200-trans was maximized with the 10c-9azo and 9c-8azo probe (Table 1). On the other hand, probes with >10 bp showed abrupt increase of IC200-cis (from 9.5 to 16 μM), which is highly unfavorable, even though these probes formed highly stable duplexes when Vis light was treated. This tendency causes nondetectable difference in IC200 between the trans and cis states of those probes, but it does not refer to the huge cis-to-trans or trans-to-cis transition. Thus, we concluded that the 10c-9azo and 9c-8azo probes were the optimal probe designs because they showed the highest differential IC200, thereby showing the largest difference in anticoagulation response upon treatment with UV or Vis light. In subsequent experiments on real-time and spatial (submillimeter) control of clotting activity, we chose probe 9c-8azo over 10c-9azo because probe 9c-8azo had a lower melting temperature and faster overall kinetics than the 10c-9azo probe (51 °C and 60 °C, respectively).
Table 1.
IC200s of each -cis and -trans
| IC200 | 16c-15azo, μM | 14c-13azo, μM | 12c-11azo, μM | 10c-9azo, μM | 9c-8azo, μM | 7c-6azo, μM | 6c-5azo, μM |
|---|---|---|---|---|---|---|---|
| IC200-cis | 20 | 19 | 16 | 9.5 | 8.3 | 7.6 | 6 |
| IC200-trans | >40 | >40 | >40 | 22 | 21 | 18 | 12 |
| Difference | >20 | >20 | >20 | 12.5 | 12.7 | 11.6 | 6 |
Dynamic Photoconversion to Restore Coagulation Process.
In most cases, it is difficult to reverse an enzyme's activity once a drug molecule has bound, but this conversion can be useful in many respects, especially in clinical applications. For instance, immediate deactivation of drugs can limit side effects in medical treatment and potentially save lives. In some cases, there are antidotes that disrupt inhibitor–enzyme interactions, but there is still no general way of finding molecules that can perform this function. In this sense, aptamers can be advantageous compared with other molecule- or antibody-based enzyme inhibitors because an aptamer's cDNA sequences naturally function as effective antidotes. This can occur because the cDNA sequence of the aptamer can replace the protein to form a DNA duplex, thus disabling the aptamer's inhibition function with target proteins. However, this example also illustrates a key potential drawback of this approach in that a large quantity of antidote sequences must be administered to overcome the slow reaction time of diffusion and hybridization in the case of in vivo applications. Another attempt used caged thrombin aptamers to deactivate thrombin. However, because this mechanism is the opposite of restoring the protein's function, it does not fit the concept of an antidote (38). With this in mind, the covalent linkage of the inhibitor to its antidote, which corresponds to the inhibitory domain (15Apt) and regulatory domain (cDNA) in our design, is highly beneficial in maximizing the hybridization efficiency between an aptamer and its cDNA sequence. In brief, this is precisely the capability that would be necessary to realize clinical inhibitor–antidote applications. Therefore, we tested whether the inhibitory nature of our probes could be controlled in real time.
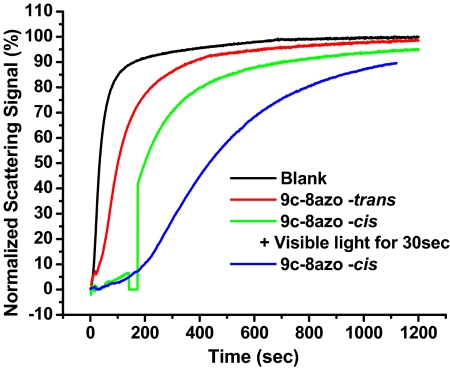
To show the dynamic transition of the aptamer's function from active to inactive, the following experiment was designed. First, the 9c-8azo-cis probe was incubated with thrombin for 5 min under 365-nm excitation in the UV spectrometer. To initiate the clotting reaction, fibrinogen substrate was added, and the reaction was monitored by measuring the decrease in light intensity due to scattering (Fig. 4). After 100 s, visible light (553 nm) was directed onto the sample in the UV spectrometer for 30 s by using a regular laser pointer. Then, the clotting reaction was continuously monitored until a plateau was reached. The samples that did not contain probes, 9c-8azo-trans and 9c-8azo-cis without light treatment, were used as references. As shown in Fig. 4, the blank sample showed fast reaction kinetics. The sample containing cis-form probes showed greatly inhibited reaction kinetics and took a much longer time to reach the plateau compared with the sample containing trans-form probes. Especially, during the first 100 s, the reaction rate was very slow. Thus, this time point was chosen to show the dramatic alteration of a reaction rate. As shown in Fig. 4, the 30-s treatment with Vis light considerably accelerates the clotting reaction. The reaction after light treatment had the same slope as the trans probe; thus, we can see that the cis-state probe and thrombin inhibition can be efficiently controlled in real time by photons. This experiment clearly demonstrated that our optimized probe designs can quickly respond to light exposure such that the deactivation of the coagulation process can be restored in a short amount of time, thus confirming the probes' capability to realize clinical inhibitor–antidote applications.
Fig. 4.
Dynamic alteration of thrombin's activity by switching 9c-8azo-cis to -trans.
Spatially Controllable Activation of Coagulation Reaction in Microfluidic Channel.
A second key part of our investigation sought to confirm the feasibility of sequential site-specific activation of coagulation by spatial control of thrombin inhibition. The regulation of reactions using photons in the UV/Vis region, instead of other mechanisms, such as heat, is advantageous because light from these wavelengths allows high spatial resolution, measuring the ability to activate a molecule's function in a spatially confined area (39). This can be therapeutically beneficial as side effects, and unnecessary drug activation in undesired areas can be eliminated. For example, the activity of a specific enzyme that causes cell death could be up-regulated within a tumor but not changed within healthy tissue. In the case of thrombin, when the cis-probes are introduced into the body, local Vis light treatment can activate the clotting reaction, resulting in selective clotting of blood vessels, whereas nontargeted sites (no Vis light) are not affected. In general, the spatial resolution of this method is limited to the resolution of the light source (the wavelength of light and optics used to deliver the Vis light). Theoretically, nanometer spatial resolution can be achieved using near-field optics (40). Using a confocal microscope, we should be able to achieve nanometer spatial resolution.
To investigate whether our PCI probes were capable of spatially controlled clotting, we made use of a microfluidic system to model 2-dimensional clot formation. First, we demonstrated that the clotting reaction could be visualized with fluorescent fibrinogen (Fig. S1A) (41). Fluorescent fibrinogen and the 10c-9azo-cis probe were mixed with the standard activated partial thromboplastin time (aPTT) (37) assay solutions and injected into a microfluidic channel. After the serine protease hydrolyzed soluble fibrinogen, the insoluble strands of fibrin could be visualized as a fluorescent protein network (Fig. S1A). The time required to form this network is an indication of the clotting reaction. Fig. S1 depicts fluorescent time-lapse images of clotting assays, which clearly show the difference in clotting times from solutions with no probe, 10c-9azo-trans, and 10c-9azo-cis probes. Fig. S1B shows the results using a blank sample without probes. In the beginning, there was homogeneous fluorescence because fibrinogen is a highly water-soluble protein. Thirty seconds after the reaction was initiated, the fluorescence signal became heterogeneous as a result of fibrin aggregation. This result is consistent with the data obtained by using UV absorption measurement. Fig. S1C shows the same time-lapse by using the 10c-9azo-trans sample. The clotting reaction was started at approximately 105 s. Fig. S1D shows the results for the 10c-9azo-cis sample. As shown, the heterogeneous fluorescence took place at ≈135 seconds. Interestingly, we noticed that there was an observable asymmetry in the time-lapse corresponding to the 10c-9azo-cis PCI probes. Fig. S2 shows the real-time fluorescence signal change of the left (a blue box in Fig. S1D) and right (a red box in Fig. S1D) sides of the images, with a longer aPTT on the right side of approximately 40 s more than the left side. We speculated that this difference arose from a problem intrinsic to the device and measurement method. Specifically, the residual fluid flow allows a small volume of solution to pass from left to right during the time-lapse recording. Thus, once cis-probes enter the field of view from the left side, they undergo conformation switch from opened to closed hairpin structure during the time-lapse, causing an asymmetric clotting pattern as the inactive form of the probe is moving from left to right.
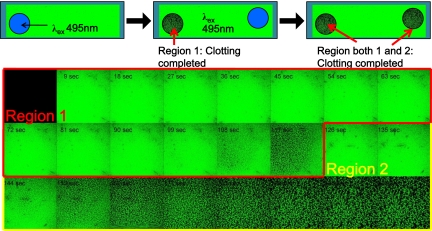
After observing the asymmetry noted above, we designed experiments to test whether we could initiate clotting in multiple regions of the channel in a temporally and spatially resolved manner. To accomplish this, the first region of the channel was imaged until fibrin fibers were observed. The stage was then moved manually to a different region of the channel to image a second zone that had not been illuminated by Vis light during the time-lapse of the first region. In the first region, the clot formed within 117 s, and the second region did not show signs of clotting (Fig. 5). After observing the second region for >30 s, a clot finally formed at 153 s. We repeated these experiments several times, and other experiments that used the confocal microscope software to illuminate a small region of the field of view before time-lapse also showed spatial resolution of clot formation. Overall, these lines of evidence lead us to conclude the feasibility and efficacy of regulating the clotting reaction in a site-specific manner with submillimeter resolution using Vis light.
Fig. 5.
Regulating the clotting reaction in a site-specific manner with submillimeter resolution using visible light. Site-specific activation of thrombin's activity by using a laser equipped to the microscope.
Conclusions
We have engineered photoswitchable nucleic acid probes modified with azobenzene for enzyme inhibition using photon energy control. Taking advantage of the principles of photoisomerization, we optimized the aptamer probes to inhibit and restore clotting reactions reversibly, depending on photon energy, in a self-regulating manner. In fact, the conversion of cis to trans was so noticeably rapid that the activity of thrombin could be instantly restored by Vis light. This quick restoration of activity of an enzyme allowed us to demonstrate that the clotting reaction within a microfluidic channel could be temporally and spatially regulated by photons in a controlled manner. In this report, we only focused on using azobenzenes to demonstrate the principle, and thus, there might be some concern that UV and Vis light may harm biological structures or have low tissue-penetration efficacy. However, evolution of azobenzene design to achieve near-infrared-responsive property can solve these problems, as demonstrated recently (42). Thus, we anticipate that such high resolutions will potentially allow fabricating small-scale architectures and devices conveniently.
Furthermore, from a therapeutic point of view, manipulating the clotting reaction with light can provide sophisticated tissue-specific clotting activation and inhibition. In detail, when cis-probes/thrombin complexes are introduced to the body, local Vis light treatment can activate the clotting reaction, resulting in selectively blocking the blood vessels, whereas nontargeted sites (no Vis light) remain unaffected. We expect that this approach will be highly beneficial in future biomedical and pharmaceutical applications. For example, as a class of cancer treatment regulating angiogenesis has been highlighted as a means of minimizing nutrition delivery to the tumor site. Based on the working principle of our PCI probes, this nutritional supply line to the tumor site could be cut off in a similar manner.
Materials and Methods
Chemicals and Reagents.
DNA synthesis reagents were purchased from Glen Research. A physiological buffer contained 25 mM Tris·HCl (pH 7.4), 150 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 5% (vol/vol) glycerol for the buffer experiment. Human α-thrombin was obtained from Haematologic Technologies. Fibrinogen was obtained from Sigma–Aldrich. Universal coagulation reference plasma (UCRP) and thromboplastin-DL for human sample testing were purchased from Pacific Hemostasis.
Preparation of Probes.
We designed, prepared, and characterized a series of molecular probes, which are shown in Table S1. The probes were synthesized by using an ABI 3400 DNA/RNA synthesizer (Applied Biosystems) at 1-μmol scale with the standard synthesis protocol. Details of the synthesis procedures are given in SI Text.
Real-Time Monitoring of Clotting Reaction.
To monitor the clotting time alteration in response to different light treatments, we designed a simple buffer experiment that contained only thrombin, one of the nucleic acid inhibitors, and fibrinogen substrate in physiological buffer. The underlying principle of the experiment is based on the mixture of sample that becomes nonfluidic and tends to scatter light by fibrin aggregation, the product of thrombin's catalytic reaction. As a result, the clotting process can be measured by the decrease of light penetration through the sample cell. To compare the efficacy of inhibition, each probe was pretreated with UV (365 nm) or Vis (white light). The mixture containing 1 μL of 10 μM thrombin and 1 μL of 100 μM concentrations of each inhibitor in 200 μL of physiological buffer was then incubated for 10 min under either UV (365 nm) or Vis (500 nm) light in the spectrometer. In sequence, 4 μL of 20 mg/mL fibrinogen was quickly added and mixed while the UV spectrometer was monitoring either the UV or the Vis wavelength as a function of time. Reaction mixtures containing only thrombin and fibrinogen with or without 15Apt were always tested together with other samples as internal standards. All clotting times were normalized based on the internal standard and compared with it.
Human Plasma Tests.
To evaluate the feasibility of the inhibitor as a potential anticoagulant reagent, we determined PT for each ligand using human plasma samples. Details are given in SI Text.
Monitoring Site-Specific Activation of Enzymatic Reaction in Microfluidic Channel.
To demonstrate site-specific activation of thrombin's activity, we used a microfluidic channel to create a 2-dimensional model of the clotting process using fluorescent fibrinogen to visualize fibrin networks. Fibrinogen was labeled with Alexa Fluor 488. Fibrinogen (10 mg) was dissolved in 1 mL of 0.1 M sodium bicarbonate buffer. Then, 0.5 mg of Alexa Fluor 488 in dimethyl sulfoxide (DMSO) was added to the protein,followed by stirring for 1 h. The conjugated protein was separated from unreacted Alexa Fluor 488 by using a Nap-25 column (GE Healthcare Bio-Sciences). The purified protein had approximately a single fluorophore on each protein.
Microfluidic channels were prepared by the following procedure. Microscope slides and 18-mm-square no. 1 coverglasses (Fisher) were rinsed with deionized H2O and dried under nitrogen. Strips of double-sided tape (3M) were placed 4 mm apart on a microscope slide, and the coverglass was placed on top. Devices were filled by capillary action. Solution exchange was performed by simultaneously pipetting solution at one end and withdrawing fluid from the other end with P8 filter paper (Fisher). An Olympus FV500-IX81 confocal microscope was used to both illuminate and image specific regions of the channel using a 10× objective and the 494-nm laser line. The length scale of the 10× field of view was calibrated by using a micrometer, and the image was 1.286 × 1.286 mm. A human plasma sample (10 μL) was prepared for aPTT measurement containing 10 μM concentrations of cis-probes and Alexa 488-labeled fibrinogen, and the mixture was loaded into the channel and incubated at RT. The aPTT is a performance indicator for measuring the efficacy of both the “intrinsic” pathway (now referred to as the contact activation pathway) and the common coagulation pathway. The fluorescent time-lapse images were obtained every 15 s until the reaction was completed.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants GM066137 and CA122648, the National Science Foundation, and the Center for Bio-Nano Sensors, a Florida state-supported Center of Excellence.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812402106/DCSupplemental.
References
- 1.Sherman JL. New Research on Optical Materials. Hauppauge, NY: Nova Science Publishers; 2007. pp. 119–156. [Google Scholar]
- 2.Heinz D, Henri B-L. Photochromism: Molecules and Systems. Amsterdam, The Netherlands: Elsevier Science and Technology; 2003. [Google Scholar]
- 3.Pala RA, Shimizu KT, Melosh NA, Brongersma ML. A nonvolatile plasmonic switch employing photochromic molecules. Nano Lett. 2008;8:1506–1510. doi: 10.1021/nl0808839. [DOI] [PubMed] [Google Scholar]
- 4.Raymo FM, Tomasulo M. Optical processing with photochromic switches. Chem Eur J. 2006;12:3186–3193. doi: 10.1002/chem.200501178. [DOI] [PubMed] [Google Scholar]
- 5.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 6.Habuchi S, et al. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc Natl Acad Sci USA. 2005;102:9511–9516. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katsonis N, Lubomska M, Pollard MM, Feringa BL, Rudolf P. Synthetic light-activated molecular switches and motors on surfaces. Prog Surf Sci. 2007;82:407–434. [Google Scholar]
- 8.Gorostiza P, Isacoff EY. Optical switches for remote and noninvasive control of cell signaling. Science. 2008;322:395–399. doi: 10.1126/science.1166022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yager KG, Barrett CJ. Novel photo-switching using azobenzene functional materials. J Photochem Photobiol A. 2006;182:250–261. [Google Scholar]
- 10.Mohr GJ. A tricyanovinyl azobenzene dye used for the optical detection of amines via a chemical reaction in polymer layers. Dyes Pigm. 2004;62:7–81. [Google Scholar]
- 11.Browne WR, Feringa BL. Making molecular machines work. Nat Nano. 2006;1:25–35. doi: 10.1038/nnano.2006.45. [DOI] [PubMed] [Google Scholar]
- 12.Liang XG, Nishioka H, Takenaka N, Asanuma H. A DNA nanomachine powered by light irradiation. ChemBioChem. 2008;9:702–705. doi: 10.1002/cbic.200700649. [DOI] [PubMed] [Google Scholar]
- 13.Yamada M, et al. Photomobile polymer materials: Towards light-driven plastic motors. Angew Chem Int Ed. 2008;47:4986–4988. doi: 10.1002/anie.200800760. [DOI] [PubMed] [Google Scholar]
- 14.Christophe D. Cis–trans Isomerization in Biochemistry. New York: Wiley-VCH; 2006. [Google Scholar]
- 15.Bose M, Groff D, Xie J, Brustad E, Schultz PG. The incorporation of a photoisomerizable amino acid into proteins in E. coli. J Am Chem Soc. 2006;128:388–389. doi: 10.1021/ja055467u. [DOI] [PubMed] [Google Scholar]
- 16.Woolley GA. Photocontrolling peptide alpha helices. Acc Chem Res. 2005;38:486–493. doi: 10.1021/ar040091v. [DOI] [PubMed] [Google Scholar]
- 17.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded-DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 18.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment—RNA ligands to bacteriophage-T4 DNA-polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 19.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 20.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: An emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 21.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 22.Tasset DM, Kubik MF, Steiner W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J Mol Biol. 1997;272:688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 23.DeAnda A, et al. Pilot study of the efficacy of a thrombin inhibitor for use during cardiopulmonary bypass. Ann Thorac Surg. 1994;58:344–350. doi: 10.1016/0003-4975(94)92206-3. [DOI] [PubMed] [Google Scholar]
- 24.Griffin LC, Tidmarsh GF, Bock LC, Toole JJ, Leung LL. In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood. 1993;81:3271–3276. [PubMed] [Google Scholar]
- 25.Rickles FR, Patierno S. Fernandez PM. Tissue factor, thrombin, and cancer. Chest. 2003;124:58S–68S. doi: 10.1378/chest.124.3_suppl.58s. [DOI] [PubMed] [Google Scholar]
- 26.Asanuma H, et al. Enantioselective incorporation of azobenzenes into oligodeoxyribonucleotide for effective photoregulation of duplex formation. Angew Chem Int Ed. 2001;40:2671–2673. doi: 10.1002/1521-3773(20010716)40:14<2671::AID-ANIE2671>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 27.Asanuma H, Ito T, Yoshida T, Liang XG, Komiyama M. Photoregulation of the formation and dissociation of a DNA duplex by using the cis–trans isomerization of azobenzene. Angew Chem Int Ed. 1999;38:2393–2395. doi: 10.1002/(sici)1521-3773(19990816)38:16<2393::aid-anie2393>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 28.Liang XG, et al. NMR study on the photoresponsive DNA tethering an azobenzene. Assignment of the absolute configuration of two diastereomers and structure determination of their duplexes in the trans-form. J Am Chem Soc. 2003;125:16408–16415. doi: 10.1021/ja037248j. [DOI] [PubMed] [Google Scholar]
- 29.Asanuma H, Matsunaga D, Komiyama M. Clear-cut photo-regulation of the formation and dissociation of the DNA duplex by modified oligonucleotide involving multiple azobenzenes. Nucleic Acids Symp Ser. 2005;49:35–36. doi: 10.1093/nass/49.1.35. [DOI] [PubMed] [Google Scholar]
- 30.Liu MZ, Asanuma H, Komiyama M. Azobenzene-tethered T7 promoter for efficient photoregulation of transcription. J Am Chem Soc. 2006;128:1009–1015. doi: 10.1021/ja055983k. [DOI] [PubMed] [Google Scholar]
- 31.Asanuma H, et al. Synthesis of azobenzene-tethered DNA for reversible photo-regulation of DNA functions: hybridization and transcription. Nat Protocols. 2007;2:203–212. doi: 10.1038/nprot.2006.465. [DOI] [PubMed] [Google Scholar]
- 32.Rusconi CP, et al. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 33.Tuddenham E. Medicine: RNA as drug and antidote. Nature. 2002;419:23–24. doi: 10.1038/419023a. [DOI] [PubMed] [Google Scholar]
- 34.Ikebukuro K, Okumura Y, Sumikura K, Karube I. A novel method of screening thrombin-inhibiting DNA aptamers using an evolution-mimicking algorithm. Nucleic Acids Res. 2005;33:e108. doi: 10.1093/nar/gni108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y, Cao Z, Tan W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc Natl Acad Sci USA. 2008;105:5664–5669. doi: 10.1073/pnas.0711803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang XH, Sen A, Vicens M, Tan WH. Synthetic DNA aptamers to detect protein molecular variants in a high-throughput fluorescence quenching assay. ChemBioChem. 2003;4:829–834. doi: 10.1002/cbic.200300615. [DOI] [PubMed] [Google Scholar]
- 37.Proctor RR, Rapaport SI. Partial thromboplastin time with kaolin—A simple screening test for first stage plasma clotting factor deficiencies. Am J Clin Pathol. 1961;36:212–219. doi: 10.1093/ajcp/36.3.212. [DOI] [PubMed] [Google Scholar]
- 38.Heckel A, Mayer G. Light regulation of aptamer activity: An anti-thrombin aptamer with caged thymidine nucleobases. J Am Chem Soc. 2005;127:822–823. doi: 10.1021/ja043285e. [DOI] [PubMed] [Google Scholar]
- 39.Lutz C, et al. Holographic photolysis of caged neurotransmitters. Nat Methods. 2008;5:821–827. doi: 10.1038/nmeth.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan W, Shi ZY, Smith S, Birnbaum D, Kopelman R. Submicrometer intracellular chemical optical fiber sensors. Science. 1992;258:778–781. doi: 10.1126/science.1439785. [DOI] [PubMed] [Google Scholar]
- 41.Guthold M, et al. Visualization and mechanical manipulations of individual fibrin fibers suggest that fiber cross section has fractal dimension 1.3. Biophys J. 2004;87:4226–4236. doi: 10.1529/biophysj.104.042333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadovski O, Beharry AA, Zhang F, Woolley GA. Spectral tuning of azobenzene photoswitches for biological applications. Angew Chem Int Ed. 2009;48:1484–1486. doi: 10.1002/anie.200805013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.