Abstract
KcsA is a proton-activated, voltage-modulated K+ channel that has served as the archetype pore domain in the Kv channel superfamily. Here, we have used synthetic antigen-binding fragments (Fabs) as crystallographic chaperones to determine the structure of full-length KcsA at 3.8 Å, as well as that of its isolated C-terminal domain at 2.6 Å. The structure of the full-length KcsA–Fab complex reveals a well-defined, 4-helix bundle that projects ≈70 Å toward the cytoplasm. This bundle promotes a ≈15° bending in the inner bundle gate, tightening its diameter and shifting the narrowest point 2 turns of helix below. Functional analysis of the full-length KcsA–Fab complex suggests that the C-terminal bundle remains whole during gating. We suggest that this structure likely represents the physiologically relevant closed conformation of KcsA.
Potassium channels are ubiquitous integral membrane proteins found in all kingdoms of life. They play a critical role in setting electrical excitation in nerve and muscles and are involved in a wide range of important physiological processes, including epilepsy, diabetes, and cardiac dysfunction (1).
KcsA, a potassium channel from Streptomyces lividans, has attracted extensive attention because of the sequence similarity of its pore-forming region to eukaryotic channels. The channel is a tetramer consisting of 2 domains: a transmembrane (TM) portion of 120 residues and a cytoplasmic domain of about 40 residues. Crystal structures of a truncated form of KcsA lacking the C-terminal domain (residues 125–160) have been critical in establishing models for K+ selectivity and permeation (2, 3) and provided insights in analyzing the mechanisms of activation (4–6) and inactivation (7) gating.
Almost all known ion channels, including KcsA, contain cytoplasmic domains that affect channel function in different ways. For instance, Kir6.2 gating in pancreatic beta cells is modulated by ATP binding to its cytoplasmic domain (8). Ca2+ activates several different K+ channels through binding intracellular domains regulating K+ conductance (9). The cytoplasmic domain of Kir2.1 is involved in modulating the inward rectification (10). In KcsA, removal of the C-terminal cytoplasmic domain affects channel thermal stability, promotes higher basal activity (11, 12), and reduces the efficiency of channel folding and assembly (13). This suggests that the cytoplasmic portion of the channel contributes to the stability of the closed state, but the mechanistic nature of these functional effects has remained elusive in the absence of a high-resolution structure of full-length (FL) KcsA.
Previous EPR studies have shown that in a native-like environment, the C-terminal domain is formed mainly by a bundle of α-helices extending linearly toward the cytoplasm (11), and this model has been confirmed in detergent by X-ray, Neutron scattering data (14), and solution NMR (15, 16). However, efforts to crystallize FL KcsA have not been successful. This is presumably because of inherent flexibility in the C-terminal cytoplasmic domain structure (11). Thus, the high-resolution details of this structure, the nature of their intersubunit interactions, and how this C-terminal domain contributes to (or influences) the activation gating conformational changes are still to be determined.
Based on the low-resolution model (11), the extended nature of the C-terminal domain KcsA (CDKcsA) and its intrinsic flexibility make crystallization of the FL molecule improbable. To overcome these obstacles, we introduce a set of new technologies based on the use of synthetically derived antigen-binding fragments (Fabs) that can be used as crystallization chaperones. These bind tightly and specifically to a target molecule to promote its crystallization and provide phasing information. The technical breakthrough that forms the foundation of our method is an innovative synthetic antibody library design that employs a “reduced genetic code” to produce synthetic Fabs to an extraordinarily broad range of molecular entities (17, 18).
Here, we report the crystal structure of FL KcsA in complex with an antibody fragment at 3.8 Å, which reveals that the CDKcsA is a well-ordered, canonical, 4-helix bundle structure that extends 70 Å away from the membrane. By using electrophysiology, we show that in the presence of the Fab chaperone, KcsA is able to gate, and ion permeation is not affected. We have also solved the structure of the C-terminal domain alone (residues 129–158) in complex with another synthetic Fab at 2.6-Å resolution and showed that it can be directly superimposed on the FL domain with virtually no changes. This provides direct structural evidence that the Fabs do not induce structural changes in the C-terminal domain of the molecule. We propose that the present FL structure likely represents a physiologically relevant model of the closed state of the KcsA K+ channel.
Results
Fab Library Design and Selection.
Crystallization of FL KcsA was accomplished by using chaperone-assisted crystallization (19), aided by the use of a combinatorial phage display library that employs a “reduced genetic code” to produce synthetic Fabs. The library was designed to introduce limited sequence diversity into 4 of the 6 complementarity-determining regions (CDRs)—L3, H1, H2, and H3—in the Herceptin Fab scaffold that had been engineered for improved display and stability (see SI Methods). High-affinity Fabs to the FL KcsA were obtained by using a solution capture strategy (17). After 3 rounds of library sorting, we identified 4 unique Fabs binding to the FL KcsA. To specifically select Fabs that bound to the C-terminal domain, we compared their binding to the FL and the truncated KcsA lacking the C-terminal domain (residues 125–160). Interestingly, of the 4 clones identified, only 1 showed significant binding to the truncated KcsA, suggesting that the epitopes for the other 3 Fabs included the CDKcsA. We further characterized Fab2, Fab3, and Fab4 (Fig. 1A), which showed single nanomolar affinity to the FL KcsA.
Fig. 1.
Crystal structure of FL KcsA in complex with Fab2. (A) CDR sequences of the 3 Fabs selected against FL KcsA from a “reduced genetic code” phage display library. Numbering is according to the Kabat definition (36). Gly is green; Tyr, yellow, Ser, red; and nondiversified positions, gray. (B) Crystal packing of the KcsA–Fab2 complex at 3.8 Å. KcsA is in orange, and the light and heavy chains of the Fab are in cyan (light chain) and magenta (heavy chain). (C) Simulated annealing composite-omit 2Fo-Fc map (contoured at 1σ) of FL KcsA. The red trace shows the fitted model as Cα tracing. (D) The final model of the KcsA–Fab complex. Three regions distinguished by the level of symmetry are highlighted: the fourfold TM segments (blue color; residues 22–117), the twofold bulge helix (red color; residues 118–135), and the fourfold distal C-terminal bundle (gray color; residues 136–158). (E and F) Experimental (E) EPR mobility and (F) NiEdda accessibility parameters (11) from membrane-reconstituted FL KcsA, mapped on the crystal model of FL KcsA. The scales represent a linear increase in local dynamics (E) and accessibility to the aqueous media (F).
The FL KcsA Structure.
FL KcsA-Fab2 crystals grew in the orthorhombic space group I222 and diffracted to 3.8-Å resolution. The structure was solved by molecular replacement using both the Fab molecule (PDB ID code 1FVD) and the tetrameric KcsA TM domain (1K4C) as search models. In the crystal, FL KcsA packs head to head with 2 Fab2 molecules bound to KcsA tetramer and a symmetry mate in a similar manner to that observed previously in the C2 crystals of the truncated form without a bound Fab (1BL8) (Fig. 1B) (2). The rest of the lattice contacts are through an extensive set of interactions mediated through the Fab2 molecules and mainly involving the hinge regions connecting the variable and the constant domains. The initial maps indicated that the 4 copies of the C-terminal domain in KcsA tetramer were organized in an extensive 4-helix bundle. To guide our model building of the C-terminal domain, we introduced 3 SeMet residues in KcsA (Met-4, Met-96, and Met-154) (20) and collected single, anomalous, isomorphous data at Se-edge. By using the unambiguously defined position of M154 as a marker, we built the C-terminal domain model (Fig. 1C; see Fig. S1 for stereo). Additionally, we made single Cys mutations at positions 119 and 126, respectively, and derivatized them with Hg++. The strong Hg++ anomalous signals confirmed the register of the C-terminal helix in our model. The FL KcsA model contains residues 22–160 and is refined to Rwork and Rfree values of 0.27 and 0.33, respectively, at 3.8-Å resolution. Full refinement statistics are provided in (Table S1).
The overall architecture of FL KcsA molecule can be divided into 3 sections based on the degree of noncrystallographic symmetry among the subunits. The first section (22–117) is the pseudo-fourfold TM domain that closely matches the conformation of its counterpart in the 1K4C structure (Fig. 1D and Fig. S2) (3). We term the second section (residues 118–135) the “bulge helix,” and unlike the rest of the molecule, it has twofold symmetry (Fig. 1D). From residue 136 to the C terminus of the chain at position 160, the 4 copies of the C-terminal domains are assembled into a canonical 4-helix bundle structure with pseudo-fourfold symmetry (Fig. 1D). The N-terminal 22 aa could not be built because of intrinsic disorder, given the location of this region at the membrane–water interface (11), similar to the structures 1BL8 and 1K4C.
The TM domain of the FL channel narrows down at Val-115, and the helices bend significantly, adopting a conformation parallel to the axis of symmetry that extends to the end of the C-terminal domain (Fig. 1D). The stretch of helix connecting the TM domain with the 4-helix bundle section (118–135) is clearly bulged out and displays a higher degree of flexibility than the other parts of the channel, as judged by EPR data (Fig. 1E). This flexibility may play an important role during the opening of the channel by providing the necessary mobility for the conformational changes that occur on channel activation (see Discussion).
The 4-helix bundle structure of the CDKcsA (residues 136–160) allows for the extensive set of ionizable side chains to decorate the surface of the bundle, and it is in agreement with the aqueous accessibility data from EPR NiEdda collision frequency measurements in membrane-reconstituted KcsA (Fig. 1F), suggesting that Fab binding to the CDKcsA had a minimal effect on its conformation.
Structural Changes Caused by C-Terminal Domain Truncation.
Despite its limited resolution, the FL KcsA structure allows for a comparison with the truncated version of the channel to establish overall changes between the 2 forms of the channel at neutral pH. It was clear from the early stages of refinement that the orientation of last ≈14 residues of the 1K4C structure (residues 110–124) was not compatible with that in the FL structure. To build this segment and to minimize bias, we used a series of omit maps to trace the helix from residue 110 to 124. These residues in the truncated structure are splayed out compared with their FL conformations. Superposition of the main chain Cα between residues 60 and 100 in the 1K4C and FL KcsA models show that in the truncated channel, TM2 bends ≈15° starting at about residue L110 (Fig. 2A). As a consequence, the inner bundle gate is wider in the truncated channel as it exits the membrane on the cytoplasmic side, shifting the position of its narrowest region to residue T107, immediately before entering into the channel inner aqueous cavity (Fig. 2B). Consequently, in FL KcsA, the narrowest point in the TM region is 2 helical turns down (Fig. 2B), where the 4 Val-115 side chains are close to van der Waals contact. Together with residues at the preceding 2 turns of the helix (Ala-108 and Ala-111), this conformation creates an obvious hydrophobic barrier to ion permeation. This is consistent with other channels where hydrophobic residues form the constriction point of the pore (10, 21, 22).
Fig. 2.
Influence of the C-terminal truncation on the conformation of the inner helix bundle gate. (A) Cα superposition of the high-resolution truncated KcsA structure (1K4C; red ribbons) with FL KcsA (blue ribbons). Inset highlights the splaying out of the inner helix bundle gate between residues 110 and 115, resulting in a 15° outward tilting. (B) Radius profile (calculated with the program HOLE; ref. 29) of truncated (red) and FL KcsA (blue).
Structure of the KcsA C-Terminal Domain at 2.6-Å Resolution.
Crystal trials using the FL KcsA and another synthetic Fab (Fab4) produced crystals in space group I4 and diffracted to 2.6 Å. Unexpectedly, these crystals were formed from a fragment of KcsA containing only the last 30 residues of the CDKcsA (residues 129–158), with 1 Fab and 1 C-terminal fragment in the asymmetric unit (Fig. 3A). Molecular replacement using the Fab framework as the search model produced interpretable electron density maps that enabled the 30 residues to be built unambiguously (Table S1). This higher-resolution structure provided a detailed view of how the C-terminal 4-helix bundle is constructed. The core of the bundle contains sets of tightly packed, hydrophobic side chains starting at Tyr-137 (Fig. 3B) and including Leu-144, Phe-148, Leu-151, and Leu-155, all pointing toward the core of the bundle with fourfold symmetry and a gradual opening of the bundle toward its N terminus (Fig. 3B).
Fig. 3.
Structure of the C-terminal bundle (residues 129–158) in complex with Fab4. (A) Top view of the CDKcsA structure as a tetramer with 4 Fab molecules bound. Each monomer of the CDKcsA is represented by different shades of orange. The heavy and light chains of Fab are shown in magenta and cyan, respectively. (B) Side chain packing of the CDKcsA core. Hydrophobic residues that constitute the core of the bundle are shown as green stick models. The distances between Cα–Cα are shown for A133 and L151. There is a 2-Å (15.4-11.4)/2 increase in the distance of Cα–Cα from the axis of symmetry at the N terminus of the CDKcsA relative to the C terminus. (C) Intersubunit electrostatic H-bonding interactions observed in the C-terminal 4-helix bundle. (Left) A top view of the network. (Right) A close-up, side view of the interactions between 2 subunits. A water molecule bridging several residues through H bonds is shown as a red sphere.
The bundle contains a set of H bonds formed by surface-exposed side chains that knit the individual helices together, as shown in Fig. 3C. Arg-147 plays a pivotal role in the interaction. It forms salt bridges with Asp-149 and Glu-152 and an H bond with His-145 from an adjacent helix (Fig. 3C). A well-defined water molecule also mediates the interactions between Arg-147 and Glu-152 between adjacent helices. The presence of a His-145 opens the possibility that this arrangement could be perturbed at acidic pH, where these groups would have altered protonation states. This could help lower the energy costs of allowing some helix bending upon proton-dependent gating. However, present evidence suggests that the bundle does not change dramatically during gating (see below).
Although the same Fab surface is (in principle) available to bind and generate crystal contacts, Fab2 and Fab4 have similar but not identical epitopes. Fab4 binds to 2 adjacent C-terminal helices with no common amino acids, allowing for next-neighbor Fab–KcsA interaction. Fab2 epitope sterically overlaps with the C-terminal helix in the counterclockwise neighbor, with several overlapping residues in the biding interface, thus disrupting Fab binding to 2 of the 4 faces of the C-terminal bundle. The reason why the Fab4–KcsA complex only showed density for the C-terminal helical bundle is unknown. We suspect that perhaps the TM and bulge-helix regions were proteolyzed by unknown contaminants. Superimposing the Cα positions of the independently refined structures of the C-terminal domain at 2.6 Å and 3.8 Å resulted in an rmsd value of <1.0 Å throughout most of the length of the C-terminal bundle. This argues convincingly that Fab binding does not structurally perturb the C-terminal domain.
The KcsA–Fab Interaction Interfaces.
The structures of the complexes of FL KcsA with Fab2 and Fab4 show that Fabs make extensive interactions with the C-terminal domain of the channel. As expected from the amino acid bias in the phage display library, a major feature of the binding interface is interactions involving Tyr residues in Fab CDRs (Fig. 1A). For Fab2, 53% of the buried surface area involves Tyr; for Fab4, it is 35% (Fig. 4 A and B). In CDR-H3, Trp is extensively used, appearing in 12% of the contacts in Fab2 and 20% in Fab4, even though it was represented in the library at only 3% (Fig. 4 A and B).
Fig. 4.
Side-chain interactions at the KcsA–Fab4 interface. (A) Pie chart representation of the amino acid composition of the binding interface of the Fab4–CDKcsA complex. The buried surface area on Fab or KcsA upon complex formation for each amino acid type is shown. Atomic structure of Fab–KcsA interface. (B) The helical cartoons with the transparent molecular surface representation in orange show CDKcsA. CDRs with residues within 4.5 Å of KcsA are shown as sticks in magenta for the heavy and in cyan for the light chain. (C) Fab4 is shown as molecular surfaces. Residues from CDKcsA contacting (closer than 4.5 Å) their respective antibody are shown as sticks for basic (blue) and for acidic (red) residues. The Cα traces of 2 of the KcsA helices are shown.
A striking feature of KcsA interface (i.e., epitope) is that it is highly charged and characterized by a clustering of Arg residues at the binding interface with both Fab2 (51%) and Fab4 (52%). Asp is another major component, contributing 21% of the contact surface with Fab2 and 19% in the case of Fab4 (Fig. 4A and Fig. S3A). The binding C-terminal-binding epitopes to Fab2 and Fab4 overlap and involve many of the same side chain residues (Fig. S3B).
Because of resolution issues, only the Fab4 interface can be analyzed in detail. Even though the Fab4 interface is rich in Tyr and Trp, many of the contacts involve extensive H-bonding interactions. In all, the interface contains a network of more than 25 H bonds, with many charged residues interacting with multiple H-bonding partners. Six water molecules are found in the interface and presumably perform a bridging function tying together hydrophilic groups from both molecules. Significant ring stacking is observed, as well as ring–Arg stacking. Fab4–CDKcsA interface is composed of a highly organized set of electrostatic and van der Waals interactions that demonstrate how specific reduced genetic code-derived interfaces can be.
Fab Binding and KcsA Function.
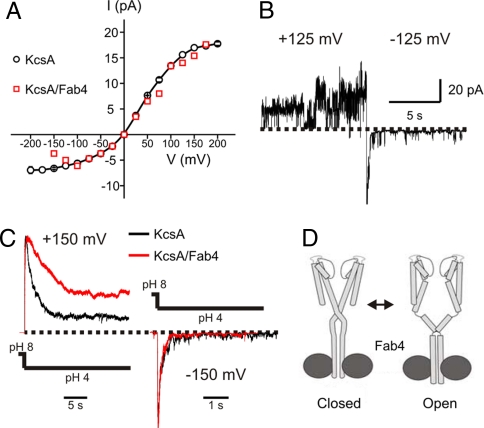
To establish whether the structures of KcsA–Fab complexes represent a functionally relevant state of the channel, we determined the functional influence of Fab binding. Unfortunately, KcsA–Fab2 complex turned out to be pH-labile, and it dissociates at pH 5.5 (from gel-filtration chromatography). This is not surprising, given the number of ionizable groups in Fab2–KcsA interface. In contrast, KcsA–Fab4 complex stayed as a monodisperse complex at pH 4, allowing for a series of electrophysiological measurements. In patch–clamp experiments with reconstituted KcsA–Fab4 complex, neither the chord conductance (≈105 pS; ref. 23) (Fig. 5A), nor the voltage-dependent modulation of the pore (Fig. 5B) (24) were significantly affected by Fab binding, in comparison with KcsA.
Fig. 5.
Functional effect of Fab4 binding on KcsA function and inner gate conformation. (A) I–V relationship in symmetric conditions (pH 4) of FL KcsA (black) and FL KcsA–Fab4 complex (red), as determined from patch-clamp experiments. (B) Change in FL KcsA–Fab4 single-channel activity in symmetric solutions (pH 4) upon a 150-mV jump. (C) The pH jump experiment at +150 mV (Left) and −150 mV (Right). (D) A model for FL KcsA activation gating. The channel (light gray) is shown in 2 conformations, both bound to the Fab4 (dark gray). The model assumes that opening of the inner bundle gate proceeds without dissociation of the distal C-terminal bundle, but implies a rearrangement of the “bulge” helix region to allow for a large TM2 movement.
The effect of pH on activation and inactivation kinetics was evaluated by using pH jumps from pH 8 to pH 4 at ±150 mV. Fig. 5C shows that the complex has an activation kinetics slightly slower than WT KcsA (7, 25), but displays a 2-fold reduction in the rate of entry into the inactivated state (τ = 4.8 s) at positive potentials (there is no major effect at negative voltages). Moreover, we find a large increase in the steady-state activity of KcsA–Fab4 (Fig. 5C). Although Fab binding to the C-terminal helix bundle does not appear to impinge on the channel's ability to open its gate, it somehow can exert a remote influence on the selectivity filter, enhancing the return from the inactivate state and thus enhancing steady-state open probability. The Fab–bundle interface is composed of helices from different channel subunits, so that a bound Fab should preclude large conformational changes in the C-terminal bundle. Indeed, binding of the Fab to the C-terminal bundle clearly stabilizes the fully closed conformation when compared with truncated KcsA. Yet, the bound Fab still allows the gate to open, strongly suggesting that Fab4 binding does not preclude the helical movements in TM2 necessary for activation gating. Therefore, we conclude that the C-terminal segment retains its 4-helix bundle structure throughout the gating cycle (Fig. 5D).
Discussion
In KcsA, evidence for a C-terminal helical bundle protruding straight toward the cytoplasm first came from computational modeling of EPR accessibility and dipolar broadening data (11). This low-resolution model was later supported by neutron and X-ray solution scattering (14) and NMR approaches (15, 16). Here, we have successfully determined the crystal structure of FL KcsA by the application of chaperone-assisted crystallography. The use of a novel combinatorial phage display library created by using a single antibody scaffold with a “restricted amino acid diversity” approach (17) proved to be critical to the present implementation of this methodology. This library has been used recently to successfully crystallize soluble proteins and structured RNA (17, 18), showing not only the power and convenience of this approach, but also its versatility. In this project, in vitro phage display library sorting allowed us to select Fabs specifically targeted to the C-terminal domain of KcsA, presumably reducing its flexibility (11). This induced rigidity, and additional crystal contacts afforded by the Fab chaperones were key determinants in improving the resolution of FL KcsA crystals, which ultimately led to solving the structure.
But could binding of the Fab perturb the structure of FL KcsA to produce a nonnative conformation at the site of binding? This issue has often been raised concerning the potential effects of lattice contact points and their influence on the conformation of the contacting residues (26, 27). The debate is particularly contentious in the case of membrane proteins, which are known to exhibit a high degree of flexibility and where the crystalline lattice is typically generated in the absence of a membrane bilayer. Examples like the voltage-dependent K+ channel KvAP have generated fierce controversies regarding the role of Fab chaperones in stabilizing distorted, high-energy conformations (28). However, in the vast majority of cases (see http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html) there is now solid evidence to suggest that neither the crystallization chaperones nor the lattice itself exerts systematic distorting forces on target molecules. Indeed, thermodynamics argue against these processes introducing high-energy, nonnative conformations. What is typically captured is one of several low-energy states, perhaps not always the lowest-energy state, but a major one that exists among a family of conformations that describe the dynamics of the molecule at steady state. Any deviation from the native conformational ensemble and the lattice contact or the protein–protein interface is likely to pay the price of a lower affinity that matches the energy difference. In this work, we were able to estimate the extent to which the Fabs influence the C-terminal segment by comparing 2 crystallographically distinct structures generated with 2 independent Fab chaperones (Fab2 and Fab4; Figs. 1 and 3). The 2 C-terminal helical bundles are nearly identical (between residues 131 and 158), strongly suggesting that the helical bundle in the FL KcsA structure represents a native, low-energy state. The limited effects of Fab4 on KcsA function (Fig. 5) further strengthen this interpretation.
In FL KcsA, the transition between the TM and the cytoplasmic helices is accompanied by changes in the level of internal symmetry of the bundle. The helical stretch (residues 118–135) connecting the 2 fourfold symmetric helical bundles, TM2 and C-terminal domain, is not only twofold symmetric, but conformationally different from the C-terminal bundle. This is likely an intrinsic property of the channel, but it could also be a consequence of the different stoichiometry in Fabs binding. Immediately below the inner bundle gate at position 115, the helices bend significantly and adopt a conformation parallel to the axis of symmetry (Fig. 1D). This region clearly “bulges” out while displaying a higher degree of flexibility than the other regions of the channel (11) (Fig. 1E). We believe that this helical segment may play an important role during the opening of the channel by providing the necessary flexibility for the conformational changes that occur during channel activation (see below).
Truncating the channel at position 124 effectively reduces this strain by allowing the helices to splay outward by about 15° (Fig. 2A). This helical rearrangement has a direct impact on the conformation of the closed gate. In the C-terminal-truncated KcsA, the narrowest segment of the permeation pathway, (about ≈4 Å in diameter; ref. 29) is located around position Thr-107 (Fig. 2B). In contrast, the radius plot of FL KcsA indicates that the narrowest point along the pore is not only tighter (≈2.5 Å in diameter), but is also shifted 2 turns of the helix to Val-115 (Fig. 2B). At this position, the configuration of the side chains optimizes intersubunit van der Waals contacts, sealing off the channel. In the truncated channel, these interactions would weaken, changing the conformation of the gate and destabilizing the closed conformation. This suggestion is supported by 2 independent lines of evidence. First, truncation of the cytoplasmic domain of KcsA produces channels with reduced thermal stability (11, 12), and it has also been associated with enhancing tetramerization efficiency during folding and assembly (13). Second, in bulk flux assays, the steady-state 86Rb+ flux at rest (pH 7) increases in C-terminal-truncated KcsA when compared with the FL channel (11). Based on the present set of results, we propose that the structure of the inner bundle gate in FL KcsA represents a physiologically relevant closed conformation of the channel, an important consideration in future mechanistic and computational analyses of the closed state. Existing truncated structures (1BL8, 1K4C, etc.), although functionally nonconductive, could instead correspond to the very first steps in the transitions toward the open conformations. Indeed, a possible role for the C-terminal helix would be to exert a steric bias on the gate and the bulged helix region, tightening the inner bundle gate and therefore stabilizing the closed conformation.
Still, FL KcsA and the high-resolution CDKcsA crystal structures have allowed us to derive a detailed map of interhelix C-terminal interactions in the closed state of KcsA. The cytoplasmic domain (residues 137–158) has a canonical 4-helix bundle that buries hydrophobic amino acids while exposing the charged residues to solvent. H bonding between polar residues with salt bridges formed by charged residues from adjacent helices may be important for the thermal stability of the 4-helix bundle. These interactions would be prime targets for disruption by ionic detergents. Of course, the large difference in resolution between the truncated (2.6-Å) and FL (3.8-Å) structures could potentially decrease the structural significance of the comparisons described above. Nevertheless, these comparisons are helped by the fact that they involve long and relatively straight helices.
In addition to aiding crystallization, the synthetic Fabs were a powerful tool to probe the mechanistic role of the C-terminal domain during activation gating. The stability of FL KcsA–Fab4 complex at pH 4 was critical in demonstrating that Fab4 did not significantly affect activation kinetics. Because Fab4 epitope encompasses helices from adjacent subunits, these results are a strong indication that the cytoplasmic domain remains essentially intact during channel opening. This observation has several implications. It suggests that the cytoplasmic domain does not “dissociate” during activation, contrary to earlier proposals based on solution determinations (14, 16). This in turn implies that the close-to-open transition in TM2 must occur in concert with a rearrangement around the membrane-proximal, bulged helix region of the C-terminal domain, although not in the distal helix bundle segment (Fig. 5D). If this is indeed the case, K+ should permeate through putative side windows created between residues 115 and 137, as has been suggested before in a variety of ion channels, including Kv channels (30, 31) and the acetylcholine receptor (21, 32).
Interestingly, although Fab had a relatively minor effect on activation kinetics, inactivation kinetics decreased 2-fold, whereas the steady-state current (a reflection of the rate of inactivation recovery; ref. 25) increased 2- to 3-fold (Fig. 5C). Given that in KcsA, inactivation occurs at the selectivity filter (7), at 50–60 Å, the remote Fab influence on the kinetics of inactivation hints at a subtler effect that might affect the degree of gate opening, and thus the coupling between activation and inactivation gates (33).
Our results have important implications in the role of the cytoplasmic domain in activation gating. The structure described here will help establish the structural linkages between the TM and C-terminal domains in KcsA. The next critical piece of the puzzle will be to relate this FL channel structure with its open-state counterpart, setting the stage for a comprehensive understanding of how these channels open and close.
Experimental Procedures
Selection and Characterization of Antibodies Against KcsA.
Library design, construction, and manipulation have been described previously (17). KcsA was biotinylated by using EZ-Link Sulfo-NHS-SS-Biotin (Pierce Biotechnologies). Three rounds of selection were performed at room temperature. In the first round, 1 μM biotinylated FL KcsA was immobilized with streptavidin-coated magnetic beads and then mixed with ≈1013 cfu phages in 1 mL of buffer A (5 mM dodecyl maltopyranoside; 50 mM Tris, pH 8.0; and 150 mM KCl) supplemented with 0.1 mg/mL BSA. After 15-min incubation, the beads containing captured phages were recovered and washed 3 times with buffer A. In subsequent rounds, to eliminate clones that bind to streptavidin beads, amplified phages were first incubated with streptavidin beads for 15 min in buffer A, and the supernatant was then used in the subsequent selection with a Kingfisher magnetic particle processor (Thermo Electron Corporation) as described previously (17). The concentrations of KcsA in the second and third rounds of selection were 100 and 10 nM, respectively.
After selection, individual clones were grown as described to produce phages for phage ELISA to detect individual binding clones (17, 34). The amino acid sequences of clones that bound to FL KcsA but not to streptavidin were derived from DNA sequencing. Of the 12 clones selected, 4 were unique. Three of these Fabs were expressed and purified as described previously (17, 35).
Crystallization and Crystal Cryoprotection.
FL KcsA and Fab2, Fab3, and Fab4 (7 mg/mL) complexes were separately crystallized at 20 °C. Fab2–FL KcsA complex produced rod-shaped crystals (0.1 × 0.3 mm) by using a sitting-drop vapor diffusion method of mixing equal volumes of the Fab2–FL KcsA complex with reservoir solution [340 mM (NH4)2SO4, 11% PEG 4000 (Hampton Research), and 100 mM Na3C6H5O7, pH 5.6]. Crystals belonged to orthorhombic space group I222, with cell dimensions a = 115 Å, b = 177 Å, and c = 339 Å, and contained 4 subunits of the FL KcsA and 2 Fab molecules in the asymmetric unit. Crystallization screening at 20 °C for the FL KcsA–Fab4 complex was carried out by using a sitting-drop vapor diffusion method. Crystals (0.05 × 0.1 mm) appeared after 2 weeks of incubation over a well solution containing 20% PEG2000 (Hampton Research), and 20 mM Bis-Tris propane, pH 7. Crystals belonged to the tetragonal space group I4 with cell dimensions a = b = 115.492 Å, and c = 76.817 Å. Both I222 and I4 crystals were cryoprotected by passing through a series of modified well solutions with increasing amounts of glycerol. Crystals were directly flash frozen in liquid nitrogen. Details of data collection and structure determination are in SI Methods.
Supplementary Material
Acknowledgments.
We thank Dr. A. Koide for helpful discussions; V. Cancasci, X. Yang, and P. Rice for assistance and crystallographic suggestion; D. M. Cortes for Fab purification; and S. Chakrapani, L. G. Cuello, V. Jogini, J. Cordero-Morales, and the members of the E.P. laboratory for experimental advice and comments on the manuscript. We are thankful to the staff at the GM/CA 23ID beamline at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL). This work was supported in part by National Institutes of Health Grants R01-GM72688, R01-GM57846, and U54 GM74946, and by the University of Chicago Cancer Research Center (Chicago, IL).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structures described here have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3EFF and 3EFD for FL-KcsA-Fab2 and CTD-KcsA-Fab4, respectively).
This article contains supporting information online at www.pnas.org/cgi/content/full/0810663106/DCSupplemental.
References
- 1.Hille B. Ion Channels of Excitable Membranes. 3rd Ed. Sunderland, MA: Sinauer; 2003. [Google Scholar]
- 2.Doyle DA, et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 4.Kelly BL, Gross A. Potassium channel gating observed with site-directed mass tagging. Nat Struct Biol. 2003;10:280–284. doi: 10.1038/nsb908. [DOI] [PubMed] [Google Scholar]
- 5.Perozo E, Cortes DM, Cuello LG. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285:73–78. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu H, et al. Global twisting motion of single molecular KcsA potassium channel upon gating. Cell. 2008;132:67–78. doi: 10.1016/j.cell.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 7.Cordero-Morales JF, et al. Molecular determinants of gating at the potassium-channel selectivity filter. Nat Struct Mol Biol. 2006;13:311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 8.Tucker SJ, Gribble FM, Zhao C, Trapp S, Ashcroft FM. Truncation of Kir6.2 produces ATP-sensitive K+ channels in the absence of the sulphonylurea receptor. Nature. 1997;387:179–183. doi: 10.1038/387179a0. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Y, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 10.Pegan S, et al. Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat Neurosci. 2005;8(3):279–287. doi: 10.1038/nn1411. [DOI] [PubMed] [Google Scholar]
- 11.Cortes DM, Cuello LG, Perozo E. Molecular architecture of full-length KcsA: Role of cytoplasmic domains in ion permeation and activation gating. J Gen Physiol. 2001;117:165–180. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pau VP, Zhu Y, Yuchi Z, Hoang QQ, Yang DS. Characterization of the C-terminal domain of a potassium channel from Streptomyces lividans (KcsA) J Biol Chem. 2007;282:29163–29169. doi: 10.1074/jbc.M703277200. [DOI] [PubMed] [Google Scholar]
- 13.Molina ML, et al. Influence of C-terminal protein domains and protein-lipid interactions on tetramerization and stability of the potassium channel KcsA. Biochemistry. 2004;43:14924–14931. doi: 10.1021/bi048889+. [DOI] [PubMed] [Google Scholar]
- 14.Zimmer J, Doyle DA, Grossmann JG. Structural characterization and pH-induced conformational transition of full-length KcsA. Biophys J. 2006;90:1752–1766. doi: 10.1529/biophysj.105.071175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker KA, Tzitzilonis C, Kwiatkowski W, Choe S, Riek R. Conformational dynamics of the KcsA potassium channel governs gating properties. Nat Struct Mol Biol. 2007;14:1089–1095. doi: 10.1038/nsmb1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chill JH, Louis JM, Miller C, Bax A. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Protein Sci. 2006;15:684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fellouse FA, et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Ye JD, et al. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc Natl Acad Sci USA. 2008;105:82–87. doi: 10.1073/pnas.0709082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunte C, Michel H. Crystallisation of membrane proteins mediated by antibody fragments. Curr Opin Struct Biol. 2002;12:503–508. doi: 10.1016/s0959-440x(02)00354-8. [DOI] [PubMed] [Google Scholar]
- 20.Qiu Y, et al. The crystal structure of Aq_328 from the hyperthermophilic bacteria Aquifex aeolicus shows an ancestral histone fold. Proteins. 2006;62:8–16. doi: 10.1002/prot.20590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 22.Kuo A, et al. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 2003;300:1922–1926. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 23.LeMasurier M, Heginbotham L, Miller C. KcsA: It's a potassium channel. J Gen Physiol. 2001;118:303–314. doi: 10.1085/jgp.118.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordero-Morales JF, Cuello LG, Perozo E. Voltage-dependent gating at the KcsA selectivity filter. Nat Struct Mol Biol. 2006;13:319–322. doi: 10.1038/nsmb1070. [DOI] [PubMed] [Google Scholar]
- 25.Chakrapani S, Cordero-Morales JF, Perozo E. A quantitative description of KcsA gating I: Macroscopic currents. J Gen Physiol. 2007;130:465–478. doi: 10.1085/jgp.200709843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rapp CS, Pollack RM. Crystal packing effects on protein loops. Proteins. 2005;60:103–109. doi: 10.1002/prot.20492. [DOI] [PubMed] [Google Scholar]
- 27.Wagner G, Hyberts SG, Havel TF. NMR structure determination in solution: A critique and comparison with X-ray crystallography. Annu Rev Biophys Biomol Struct. 1992;21:167–198. doi: 10.1146/annurev.bb.21.060192.001123. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 29.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graphics. 1996;14:354–360. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 30.Kobertz WR, Williams C, Miller C. Hanging gondola structure of the T1 domain in a voltage-gated K(+) channel. Biochemistry. 2000;39:10347–10352. doi: 10.1021/bi001292j. [DOI] [PubMed] [Google Scholar]
- 31.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 32.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 33.Panyi G, Deutsch C. Cross talk between activation and slow inactivation gates of Shaker potassium channels. J Gen Physiol. 2006;128:547–559. doi: 10.1085/jgp.200609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidhu SS, Lowman HB, Cunningham BC, Wells JA. Phage display for selection of novel binding peptides. Methods Enzymol. 2000;328:333–363. doi: 10.1016/s0076-6879(00)28406-1. [DOI] [PubMed] [Google Scholar]
- 35.Lee CV, et al. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J Mol Biol. 2004;340:1073–1093. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 36.Kabat EA, Wu TT, Redi-Miller M, Perry HM, Gottesman KS. Sequences of Proteins of Immunological Interest. Bethesda, MD: National Institutes of Health; 1987. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.