Abstract
Summary: Jalview Version 2 is a system for interactive WYSIWYG editing, analysis and annotation of multiple sequence alignments. Core features include keyboard and mouse-based editing, multiple views and alignment overviews, and linked structure display with Jmol. Jalview 2 is available in two forms: a lightweight Java applet for use in web applications, and a powerful desktop application that employs web services for sequence alignment, secondary structure prediction and the retrieval of alignments, sequences, annotation and structures from public databases and any DAS 1.53 compliant sequence or annotation server.
Availability: The Jalview 2 Desktop application and JalviewLite applet are made freely available under the GPL, and can be downloaded from www.jalview.org
Contact: g.j.barton@dundee.ac.uk
1 INTRODUCTION
Sequences of DNA, RNA and proteins are the fundamental currency of modern biological research that links the different levels of the biological hierarchy, from gene to 3D structure. Multiple sequence alignments (MSAs) permit the identification of common features between species or identify functionally important residues. MSAs provide the foundation for a range of computational methods including the prediction of protein secondary structure and solvent accessibility, functional sites and interaction sites. MSAs are also the essential first step in studying molecular phylogeny and the identification of genomic rearrangements. In journal publications, MSAs provide a convenient framework for displaying common features and complex annotations relating to sequences and their functions. It is therefore important to obtain the best alignment possible. Many multiple alignment techniques exist (Notredame, 2007), but no single method is perfect for all situations (Blackshields et al., 2006; Raghava et al., 2003). As a consequence, all alignments require inspection and interpretation, and often adjustment by hand, in order to produce an alignment that best represents the biological context of the sequences. Editing tools are essential for this task, not least because they provide visual feedback on an alignment's quality in the light of all known and computationally predicted annotation.
Jalview Version 1.0 (Clamp et al., 2004) was an alignment editor first developed in 1996 as an advance over static alignment visualization tools such as ALSCRIPT (Barton, 1993). As well as alignment editing, colouring and generation of figures as postscript or HTML, it included methods for alignment conservation analysis, phylogenetic tree construction and a simple linked view of 3D structure that could colour residues in the same way as the alignment. Many alignment formats were supported, and feature annotation extracted from SwissProt (Boeckmann et al., 2003) flat-files could be plotted on the alignment to highlight important regions of a sequence. Jalview V1.0 gained a strong following, and was best known in its lightweight web applet form which was adopted as an alignment viewing/editing tool by many web sites worldwide including major databases such as PFAM (Finn et al., 2008) and SRS (Etzold et al., 1996). It has also been embedded in stand-alone analysis tools such as ModView (Ilyin et al., 2003). However, the original program had many limitations—only one multiple alignment could be edited at a time, and an alignment's colouring and tree-based conservation analysis could only be exported as a figure, not stored and returned to later. Introduction of new functionality to the program was also difficult. Jalview 1's software architecture was developed for optimum performance within the constraints of the Java runtime environment; and the addition of extensions could become complex and lead to unmaintainable code. In summary, Jalview V.1.0's capabilities are now insufficient for the larger, longer and more detailed analysis tasks that a researcher may now routinely perform. Stability, usability and extensibility are now also of prime importance for software used in research, and to this end, we re-engineered the original Jalview code to develop Jalview Version 2 (JV2).
2 IMPLEMENTATION
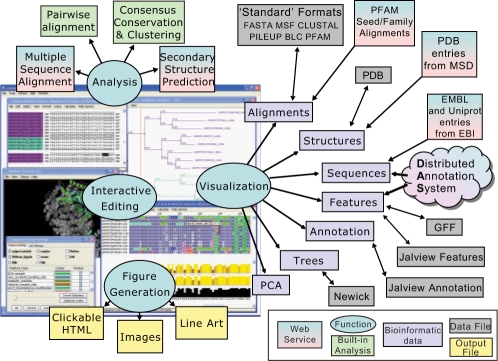
The new JV2 software architecture and alignment-rendering model provides the foundation for two JV2 program flavours: JalviewLite (JVL) and Jalview Desktop (JVD). JVL is a web optimized, Java 1.1 compliant applet that replaces Jalview V1.0 where it is used on a web page. In contrast, JVD is a fully-fledged desktop application that can be installed easily on the user's machine and launched in batch or interactive mode from the command line, or started via Java WebStart (Java 1.4 or later). The capabilities of the JVD that are summarized in Figure 1 and described below include the ability to generate high-quality alignment figures for publication, and to exploit web services for data retrieval and analysis. JVL and JVD both utilize Jmol, an open source molecular graphics viewer, to present linked views of PDB files associated with an aligned sequence.
Fig. 1.
Capabilities of the Jalview 2 desktop application. Ovals depict major capabilities: visualization, interactive editing, analysis and WYSIWYG figure generation. Arrows connect bioinformatics data handled by JV2 with flat-file or web-service data sources. Analysis includes built-in alignment conservation and tree building algorithms, and web services for MSA and secondary structure prediction methods. The screenshot of the application shows two sets of sequences for proteins in the lactate dehydrogenase family. One contains an alignment of protein sequences retrieved from the Uniprot database, the other their coding sequences retrieved from the EMBL database. Interactive highlighting shows the region corresponding to the amino acid or codon position near the mouse pointer in both the alignment windows and the Jmol structure display of a PDB record associated with one protein.
The core JV2 functionality present in JVL and JVD provides significantly enhanced editing and viewing capabilities when compared to JV1. Interactive editing, colouring and annotation can be performed via the mouse or in a keyboard-editing mode. Alignment edits can be undone, and any number of independent views may be created in tabs or as separate windows opened on the same alignment. Navigation in a view is facilitated by an overview window, and each view also has its own layout and display settings. Specific sequences or columns can be hidden from a view, and arbitrary regions may be selected for analysis either by the built-in algorithms or remote web services, cut or copied to another alignment, or defined as named groups and coloured with one of 11 built-in or user-defined alignment colour schemes, or shaded by conservation or quantitative alignment annotation. Annotation rows may be interactively created and displayed below the columns of the alignment. They may contain labels, secondary structure symbols, coloured histograms or line graphs. Sequence features may also be overlaid onto an alignment. Non-positional information, such as sequence database accession numbers and literature references, are viewed as a tooltip displayed when the mouse hovers over a sequence's ID. Positional features such as metal ion binding sites are rendered as transparent or opaque shading over the visible regions of the alignment. Features may also be edited interactively or created from the results of a regular expression search over the alignment. New JV2 file parsers have been developed to generate annotated alignments from Stockholm alignment files, to import features from GFF, and to read and write Newick formatted phylogenetic trees. Three new formats have also been developed. Sequence regions and groups, colouring and alignment annotation are recorded in Jalview annotation files, whereas Jalview feature files are used to exchange sequence feature annotation. The JVD also supports an additional XML document format, the Jalview Project Archive, which enables all alignments, trees, structures, views and DNA/protein/structure mappings to be recorded and returned to at a later date.
2.1 Embedding JV2 in web applications
The JVL applet provides JV2's core MSA visualization, annotation, analysis and editing facilities as a lightweight web application component. Input data and initial display settings are specified using the comprehensive set of start-up parameters. Furthermore, a Javascript API (described on the web site) allows access to user selections, alignment and annotation data, and control of group and feature display settings in a particular alignment view. JVL has been successfully deployed on many servers, including MyHits (Pagni et al., 2007), and the structural genomics target optimization pipeline, TarO (Overton et al., 2008). Interaction with web application developers has been an important influence on development. For example, the sequence feature settings interface arose to support the needs of MACSIMS (Thompson et al., 2006).
2.2 Web services access from the Jalview desktop
Figure 1 provides an overview of the capabilities of the JVD. Its primary role is to support alignment creation, editing and in-depth analysis, and enables the visual integration of local and distributed sources of sequence annotation and structural data. Command-line parameters passed via Java Webstart provides a route for the JVD to be launched from the JVL or directly by a bioinformatics web application, but it can also access public sequence, structure and alignment databases with WSDbFetch (Pillai et al., 2005) to retrieve or transfer database accessions and annotation from their records. Menus on the JVD interface enable the researcher to gather sequence and annotation data from external databases, and utilize Jalview's own dedicated SOAP web services for sequence alignment with ClustalW (Thompson et al., 1994), Muscle (Edgar, 2004) and MAFFT (Katoh et al., 2005) and secondary structure prediction with Jpred3 (Cole et al., 2008).
JVD also interacts with Distributed Annotation System (DAS) servers conforming to the DAS 1.53 specification (Dowell et al., 2001; Prlic et al., 2007). Sequence and annotation servers can be manually added or discovered from the public DAS server registry. Sequences can be retrieved from or matched against any registered sequence source, and features retrieved from annotation sources mapped onto the alignment sequence's local coordinate frame.
3 DISCUSSION
The first version of JV2 appeared in May 2005, and after the release of Jalview 2.4 in September 2008, a search for ‘Jalview’ in Google returns over 450 000 hits. Furthermore, estimates derived from HTTP logs suggest that the Jalview Desktop is launched between 1500 and 2500 times per week. Naturally, Jalview is not the only program to have editing/analysis and display features, though it is perhaps surprising that relatively few of the 25 or so interactive programs distributed since 1985 appear as widely used. Many, such as HOMED (Stockwell and Petersen, 1987) and MALIGNED (Clark, 1992), seem not to be actively supported or undergoing further development. Programs that are maintained include DCSE RNA alignment editor (De Rijk and De Wachter, 1993), which is now a component of RnaViz (De Rijk et al., 2003), and CINEMA (Parry-Smith et al., 1998), which is now distributed as part of Utopia (Pettifer et al., 2004). SeaView (Galtier et al., 1996) is a specialized cross-platform alignment editor developed for molecular phylogeny studies. ClustalX (Thompson et al., 1997) provides a graphical interface to the clustal multiple alignment algorithm, but does not allow manual manipulation of the alignment. A more recent introduction is the PFAAT Java alignment editor (Johnson et al., 2003), which has novel residue-level annotation tools and uses Jmol for protein structure display. Like Jalview it also provides tree viewing options, and the PFAAT authors kindly acknowledge Jalview as a source of their inspiration.
In this article, we have described the new capabilities and features available in Jalview 2, which enable both the expert bioinformatician and novice alike to perform sequence analysis investigations of ever-increasing size and complexity. Exploiting distributed access to computation and data resources is integral to modern bioinformatics, and to our knowledge, JVD was the first program capable of retrieving and visualizing DAS annotation on MSA. The new JV2 architecture also provides a solid foundation to extend the program further, to provide specialized support for next generation sequencing, improved support for the rendering of quantitative and symbolic annotation, and exchange data with other molecular visualization and analysis applications.
ACKNOWLEDGEMENTS
The authors would like to thank everyone who has contributed to Jalview 2 and previous versions of Jalview, including James Cuff and Steve Searle. Andreas Prlic contributed the DAS client library ‘dasobert’ as part of BioJava. Benjamin Schuster-Bockler developed the first version of Jalview's Stockholm parser. We also gratefully acknowledge all JV2's users for their encouragement, bug reports, helpful comments and suggestions.
Funding: Biotechnology and Biological Sciences Research Council (grant number BBSB16542).
Conflict of Interest: none declared.
REFERENCES
- Barton GJ. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- Blackshields G, et al. Analysis and comparison of benchmarks for multiple sequence alignment. In Silico Biol. 2006;6:321–339. [PubMed] [Google Scholar]
- Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, et al. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Clark SP. MALIGNED: a multiple sequence alignment editor. Comput. Appl. Biosci. 1992;8:535–538. doi: 10.1093/bioinformatics/8.6.535. [DOI] [PubMed] [Google Scholar]
- Cole C, et al. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijk P, De Wachter R. DCSE, an interactive tool for sequence alignment and secondary structure research. Comput. Appl. Biosci. 1993;9:735–740. doi: 10.1093/bioinformatics/9.6.735. [DOI] [PubMed] [Google Scholar]
- De Rijk P, et al. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics. 2003;19:299–300. doi: 10.1093/bioinformatics/19.2.299. [DOI] [PubMed] [Google Scholar]
- Dowell RD, et al. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzold T, et al. SRS: information retrieval system for molecular biology data banks. Methods Enzymol. 1996;266:114–128. doi: 10.1016/s0076-6879(96)66010-8. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, et al. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Ilyin VA, et al. ModView, visualization of multiple protein sequences and structures. Bioinformatics. 2003;19:165–166. doi: 10.1093/bioinformatics/19.1.165. [DOI] [PubMed] [Google Scholar]
- Johnson JM, et al. Protein family annotation in a multiple alignment viewer. Bioinformatics. 2003;19:544–545. doi: 10.1093/bioinformatics/btg021. [DOI] [PubMed] [Google Scholar]
- Katoh K, et al. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C. Recent evolutions of multiple sequence alignment algorithms. PLoS Comput. Biol. 2007;3:e123. doi: 10.1371/journal.pcbi.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton IM, et al. TarO: a target optimisation system for structural biology. Nucleic Acids Res. 2008;36:W190–W196. doi: 10.1093/nar/gkn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagni M, et al. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 2007;35:W433–W437. doi: 10.1093/nar/gkm352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry-Smith DJ, et al. CINEMA—a novel colour interactive editor for multiple alignments. Gene. 1998;221:GC57–GC63. doi: 10.1016/s0378-1119(97)00650-1. [DOI] [PubMed] [Google Scholar]
- Pettifer SR, et al. UTOPIA-user-friendly tools for operating informatics applications. Comp. Funct. Genomics. 2004;5:56–60. doi: 10.1002/cfg.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S, et al. SOAP-based services provided by the European Bioinformatics Institute. Nucleic Acids Res. 2005;33:W25–W28. doi: 10.1093/nar/gki491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlic A, et al. Integrating sequence and structural biology with DAS. BMC Bioinformatics. 2007;8:333. doi: 10.1186/1471-2105-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghava GP, et al. OXBench: a benchmark for evaluation of protein multiple sequence alignment accuracy. BMC Bioinformatics. 2003;4:47. doi: 10.1186/1471-2105-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell PA, Petersen GB. HOMED: a homologous sequence editor. Comput. Appl. Biosci. 1987;3:37–43. doi: 10.1093/bioinformatics/3.1.37. [DOI] [PubMed] [Google Scholar]
- Thompson JD, et al. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, et al. MACSIMS: multiple alignment of complete sequences information management system. BMC Bioinformatics. 2006;7:318. doi: 10.1186/1471-2105-7-318. [DOI] [PMC free article] [PubMed] [Google Scholar]