Abstract
Motivation: Mutual information (MI) theory is often applied to predict positional correlations in a multiple sequence alignment (MSA) to make possible the analysis of those positions structurally or functionally important in a given fold or protein family. Accurate identification of coevolving positions in protein sequences is difficult due to the high background signal imposed by phylogeny and noise. Several methods have been proposed using MI to identify coevolving amino acids in protein families.
Results: After evaluating two current methods, we demonstrate how the use of sequence-weighting techniques to reduce sequence redundancy and low-count corrections to account for small number of observations in limited size sequence families, can significantly improve the predictability of MI. The evaluation is made on large sets of both in silico-generated alignments as well as on biological sequence data. The methods included in the analysis are the APC (average product correction) and RCW (row–column weighting) methods. The best performing method was APC including sequence-weighting and low-count corrections. The use of sequence-permutations to calculate a MI rescaling is shown to significantly improve the prediction accuracy and allows for direct comparison of information values across protein families. Finally, we demonstrate how a lower bound of 400 sequences <62% identical is needed in an MSA in order to achieve meaningful predictive performances. With our contribution, we achieve a noteworthy improvement on the current procedures to determine coevolution and residue contacts, and we believe that this will have potential impacts on the understanding of protein structure, function and folding.
Contact: cmb@qb.ffyb.uba.ar; mniel@cbs.dtu.dk
1 INTRODUCTION
Multiple sequence alignments (MSAs) of homologous proteins carry at least two levels of information. One is given by the amino acid conservation at each position in the protein sequence and the second is given by the inter-relationship between two or more positions. While the first type of information is relatively straightforward to calculate and interpret, the second type is more complex. Mutations at essential residues in a protein sequence may occur only if a compensatory mutation elsewhere in the protein takes place to preserve or restore the activity (Martin et al., 2005). As stated by DePristo et al. (2005) (and references therein), the frequency of compensatory mutations is very high, and involves not only functional but also biophysical properties like stability and tendency to aggregation. The extent of the mutual coevolutionary relationship between two positions in a protein family can be estimated using mutual information (MI) from information theory (Cover and Thomas, 1991; Gloor et al., 2005; Martin et al., 2005; Tillier and Lui, 2003). Even though in principle the calculation of MI is simple, its interpretation has been demonstrated to be very difficult and different approaches have been tested to benefit from that information (Chiu and Kolodziejczak, 1991; Cover and Thomas, 1991; Dunn et al., 2008; Gouveia-Oliveira and Pedersen, 2007; Korber et al., 1993; Shackelford and Karplus, 2007; Wollenberg and Atchley, 2000).
The problem faced when correlating MI values obtained from biological data to the extent of coevolution lies in the fact that protein sequences are not independent, but have an inherent signal defined by their evolutionary relationship. This has nicely been demonstrated in the work by Gouveia-Oliveira and Pedersen (2007) where they show how sequences that are related through a tree-formed history can result in a covariance signal that resembles coevolution. Likewise, it has been shown that the degree of sequence conservation of two positions correlates strongly with their estimated amount of MI (Fodor and Aldrich, 2004; Martin et al., 2005). To deal with these difficulties, a number of different approaches have been proposed to lower this high background signal imposed by phylogeny and noise, enabling more accurate identification of coevolving positions in protein sequences. A recent paper by Dunn et al. (2008) gives a detailed introduction to the field describing these different methods. However, no paper has compared the accuracy of these methods. Here, we perform such a benchmark calculation. We compare the predictive accuracy of the APC (average product correction) method proposed by Dunn et al. (2008), and the RCW (Row–column weighting) method proposed by Gouveia-Oliveira and Pedersen (2007) on a large set of both artificial and biological data. We demonstrate that both methods as well as the raw MI calculation can be significantly improved by including (i) sequence-clustering techniques to reduce sequence redundancy, and (ii) low count corrections to account for small number of observations in limited size sequence families. Further, we show how the use of sequence permutations can be applied to perform a MI rescaling to allow for a direct comparison of information values across protein families.
The software is written in C, it is fast and suitable to analyze a large number of sequences.
2 METHODS
2.1 Data
2.1.1 Artificial data
We use two datasets (where clustering and one-by-one refer to the model of evolution) constructed by Gouveia-Oliveira and Pedersen (2007). Each dataset consists of a collection of 100 MSA of length 300 amino acids. Twenty amino acid pairs are simulated to coevolve, and the remaining 44 830 pairs evolve independently. The size of the MSA includes 32, 64 and 256 sequences and the rates of evolution are set to 1 and 3, where 1 is the fastest. Details on the dataset are given in the work by Gouveia-Oliveira and Pedersen (2007). The datasets are available online at: http://www.cbs.dtu.dk/services/InterMap3D/Coevolution_Benchmarking_datasets.zip.
2.1.2 Biological data
Three sets of biological data were used. One set consisted of 83 MSA constructed by Dunn et al. (2008). Each family in this dataset is very small, containing on average 222 protein sequences per family. Another set consisted of 85 Pfam families. The 85 Pfam families were selected from Pfam release November 2008 with > 2000 sequences per family, at least one PDB entry each, and an alignment length > 150 amino acids. Families were included only if the number of columns in the MSA with <50% gaps were 50 or more. The last set consisted of five protein families, randomly chosen with no previous knowledge of the sequence coverage. The only condition was to have a sample of each of the four major SCOP protein classes (all α, all β, α + β and α/β) and that the family should have at least one member with known 3D structure. The alignment of the chosen families was obtained after a Blast search with the proteins: 2TRX, 1O1N, 1URE, 1BMC and 1JWP used as seeds. For each sequence seed, an MSA with its homologs was prepared by scanning the non redundant (nr) protein sequence database at NCBI (August 2008) with the program PSI-BLAST, version 2.2.18 (Altschul et al., 1997) The scanning was performed without filtering compositionally biased segments, running three Blast iterations and E-value threshold equal to 0.001. Default values were used for all the other parameters. This last small dataset would illustrate the applicability of the method for the non-expert user that does not have the skill to prepare high accuracy MSA using advanced bioinformatics tools like hidden mark models etc.
In all cases MSAs were gap trimmed to remove positions with gaps in the seed sequence. In addition, all positions with >50% gaps, as well as sequences covering <50% of the seed sequence length were removed. Unlike other methods, with this procedure we allow gaps to occur.
2.2 The algorithm
The MI between two positions in an MSA is given by the relationship:
| (1) |
where P(ai, bj) is the frequency of amino acid a occurring at position i and amino acid b occurring at position j in the same sequence, P(ai) is the frequency of amino acid a at position i and P(bj) is the frequency of amino acid b at position j. We introduced a very simple correction for low number of sequences. The amino acid frequencies, P(a, b), are normalized from N(a,b), the number of times an amino acid pair (a, b) is observed at positions i and j in the MSA. From N(ai, bi), P(ai, bj) is calculated as (λ + N(ai, bj)))/N, where:
It is clear that for MSAs of limited size, a large fraction of the P(ai, bj) values will be estimated from a very low number of observations, and their contribution to MI could be highly noisy. To deal with such low counts, a parameter λ is introduced. The initial value for the variable N(ai, bj) = λ is set for all amino acid pairs. Only for MSAs with a small number of sequences, where a large fraction of amino acid pairs remain unobserved, will λ influence the amino acids occupancy calculation. For large MSAs, most amino acid pairs will be observed at least once, and the influence of λ will be minor. We investigated how the performance depended on the values used for λ on a small independent dataset. We tested a range of values 0–0.2 in steps of 0.01. The maximal performance was achieved for a value of λ equal to 0.05, but similar results are obtained in the range 0.025–0.075. This value was consistently found to be optimal for all datasets independently of size, evolutionary model, or rate of evolution (data not shown).When dealing with biological data, MSAs will often suffer from a high degree of unnatural sequence redundancy. It is hence expected that the sequence clustering would improve the accuracy of the MI calculation. We employed a Hobohm 1 algorithm (Hobohm et al., 1992) to define sequence clusters, and assign each sequence within a given cluster a weight corresponding to one divided by the number of sequences in the cluster. Clusters were defined at a sequence identity threshold of 62% (Shackelford and Karplus, 2007). Earlier work by us has demonstrated that this threshold is also optimal when using a Gibbs sampler approach to identify the motif for MHC class II binding (Nielsen et al., 2004). The performance remains stable for threshold values in the range 40–75%. When accumulating the amino acid concurrencies, the sequence weight, rather that the value one, was used.
Several methods have been suggested to correct for an inherent property of MI, namely, that the MI value between two residues depends strongly on the conservation or entropy of the two residues. In this work, we apply two recent methods suggested to deal with this problem. One such method, named RCW, was proposed by Gouveia-Oliveira and Pedersen (2007). This method divides all MI values by the average MI value of the two residues:
where MIi. is the average of the MI value of residue i to all other residues in the MSA. The other method, proposed by Dunn et al. (2008), defines an APC, and subtracts this value from MI:
where MIi−as before—is the average MI value of residue i to all other residues in the MSA, and MI.. is the average MI value over all pairs of residues in the MSA.
2.3 Z-scores
In a Z-score transformation, all prediction scores are compared with a distribution of prediction scores obtained from a large set of randomized MSAs. The Z-score is then calculated as the number of standard deviations that the observed MI value falls above the mean value obtained from the randomized MSAs. Two procedures for permutation were tested, one column-based and one sequence-based. The first approach tests the hypothesis that the sequences are homologous and correctly aligned, but that the columns are not correlated, whereas the latter tests the hypothesis that the sequences are not homologous. It was hence a priori expected that the first approach would be most suited for the analysis of MI between columns of the MSA. The permutations were made with gaps fixed in their original positions and boundaries. The background mean and SD values were estimated from 100 such randomizations. It should be noted that sequence-based Z-score does not test for the appropriated null hypothesis (the columns not being correlated), and that sequence-based Z-score hence should be interpreted only as an additional prediction score.
3 RESULTS
3.1 Artificial data
We first tested our method on the artificial data constructed by Gouveia-Oliveira and Pedersen (2007). The predictive performance of the different methods was evaluated in terms of the area under the receiver operating characteristic (ROC) curve (AUC) (Swets, 1988). An AUC value of one indicates a perfect prediction and a value of 0.5 a random prediction. The result of the experiment is shown in Table 1.
Table 1.
Predicted performance values in terms the AUC of the different tested methods and the applied clustering and low count corrections with the artificial dataset
| Method | 32Kme | 32Ind | 64Kme | 64Ind | 256Kme | 256Ind |
|---|---|---|---|---|---|---|
| Benchmark using the dataset with rate of evolution 1 | ||||||
| MI | 0.597 | 0.647 | 0.603 | 0.663 | 0.661 | 0.717 |
| MI-C | 0.598 | 0.647 | 0.604 | 0.664 | 0.661 | 0.716 |
| MI-Lc | 0.757 | 0.816 | 0.753 | 0.817 | 0.740 | 0.808 |
| MI-C-Lc | 0.792 | 0.865 | 0.797 | 0.863 | 0.788 | 0.850 |
| RCW | 0.653 | 0.719 | 0.660 | 0.743 | 0.707 | 0.798 |
| RCW-C | 0.653 | 0.719 | 0.661 | 0.744 | 0.707 | 0.796 |
| RCW-Lc | 0.774 | 0.841 | 0.788 | 0.861 | 0.802 | 0.885 |
| RCW-C-Lc | 0.787 | 0.862 | 0.811 | 0.885 | 0.834 | 0.912 |
| APC | 0.710 | 0.843 | 0.768 | 0.885 | 0.850 | 0.943 |
| APC-C | 0.709 | 0.842 | 0.767 | 0.885 | 0.851 | 0.943 |
| APC-Lc | 0.783 | 0.863 | 0.821 | 0.908 | 0.897 | 0.967 |
| APC-C-Lc | 0.779 | 0.857 | 0.816 | 0.901 | 0.895 | 0.965 |
| Benchmark using the dataset with rate of evolution 3 | ||||||
| MI | 0.635 | 0.784 | 0.664 | 0.833 | 0.765 | 0.949 |
| MI-C | 0.635 | 0.784 | 0.664 | 0.833 | 0.764 | 0.949 |
| MI-Lc | 0.808 | 0.958 | 0.814 | 0.957 | 0.842 | 0.986 |
| MI-C-Lc | 0.851 | 0.998 | 0.863 | 0.994 | 0.880 | 0.997 |
| RCW | 0.712 | 0.928 | 0.747 | 0.965 | 0.840 | 0.998 |
| RCW-C | 0.712 | 0.928 | 0.747 | 0.965 | 0.839 | 0.998 |
| RCW-Lc | 0.830 | 0.989 | 0.857 | 0.996 | 0.906 | 1.000 |
| RCW-C-Lc | 0.846 | 0.999 | 0.878 | 1.000 | 0.927 | 1.000 |
| APC | 0.799 | 0.967 | 0.869 | 0.990 | 0.952 | 1.000 |
| APC-C | 0.797 | 0.967 | 0.868 | 0.990 | 0.952 | 1.000 |
| APC-Lc | 0.842 | 0.997 | 0.889 | 1.000 | 0.965 | 1.000 |
| APC-C-Lc | 0.837 | 0.998 | 0.882 | 1.000 | 0.961 | 1.000 |
The upper panel gives the AUC values for the fast evolving data, and the lower panel, the results for the slower evolving data. Kme refers to the cluster model of evolution, and Ind refers to the one-by-one model of evolution. The numbers before the model of evolution give the number of sequences in each alignment: 32, 64 and 256, respectively. The methods included in the benchmark are: MI, C is clustering, Lc is low count correction with λ = 0.05, RCW and APC. The best performing method for each benchmark dataset is highlighted in bold.
From Table 1 it is clear that clustering and low count correction improve the accuracy of all three prediction methods. In particular, the performance for the raw MI method does improve dramatically from being close to random to produce highly significant predictions. It is apparent from the results that the clustering does not have any strong influence on the predictive performance for these datasets. This is to be expected, since the artificial MSAs contain very little sequence redundancy. For the small datasets (Kme32, and Ind32), all three methods achieve similar predictive performances when including clustering and correction for low count. On the other hand, for the large datasets with 256 sequences in the MSA, the APC method combined with low count correction (and sequence clustering) significantly outperforms both other methods (P < 0.001, binomial test).
3.2 What is the function of low count corrections?
One might ask how the highly simple approach correcting for low counts introduced in this work can have so strong an influence on the predictability of coevolving residue pairs. From Equation (1)—that defines the MI between two sites in an MSA—it is apparent that diversity is essential to achieve high MI values. Only if all amino acids are present in equal frequencies between two perfectly coevolving pairs will the MI achieve its maximum value. This leads to the observation that fast evolving sites tend to have high values of MI albeit being non-coevolving (Gouveia-Oliveira and Pedersen, 2007). Likewise, slowly evolving sites will only occupy a small fraction of the amino acid space, and hence tend to have low MI values. The extreme case is perfectly conserved amino acids that will always have a MI value of zero. By introducing a correction for low count this behavior is altered. This can be illustrated taking an example from the artificial dataset 32Kme1. This dataset is of limited size, and the use of low count corrections was shown to greatly improve the predictability of coevolving residues. In the dataset 4 of the 32Kme1 alignment set, columns 45 and 263 have the highest MI value (2.18) of all residue pairs in the alignment. These sites are non-coevolving. Calculating the information content, I =log(20)+∑apa · log(pa), for each of the two residues clearly demonstrates that the two sites are fast evolving (I45 = 0.17 and I263 = 0.26). Introducing the low count correction, the MI value is lowered to 0.91. The low count correction only gives minor changes to the MI between slowly evolving residue pairs. An example of this is the coevolving residue pair 287 and 288 in the multiple alignment. These two residues have information content of 2.18 and 1.82, respectively, manifesting their slow rate of evolution. The MI is 0.47. By introducing the low count correction, this value is maintained at 0.50. In summary, for limited size MSAs, the use of low count correction lowers the MI between fast evolving sites, and maintains the MI between slowly evolving sites.
3.3 Biological data
Next, we investigated the performance of the different methods on actual biological data. In this case, the knowledge of which residue pairs are coevolving is not available to us. As an approximation, we assumed that all residue pairs in contact (i.e. with a Cβ distance <8 Å) are coevolving. This is naturally a wrong assumption since many sites that are in contact are non-coevolving and many sites that are coevolving are not necessarily in contact in the final folded structure of the protein. However, the vast majority of coevolving pairs can be assumed to be in contact, so this approach seems reasonable when carrying out a comparable study of the performance of different prediction methods. One should note that the actual predictive performance of the different methods would most likely be underestimated in such a benchmark.
We tested the methods in three datasets, one constructed by Dunn et al. (2008), one built from Pfam and a third with a narrower set of five selected protein families to illustrate the procedure a user will perform using a Blast search (for details on the dataset, see Section 2).
For each protein family, we calculated the MI between each residue pair in the target sequence that is present in >50% of the sequences in the MSA. Next, for each prediction method, we used the 8 Å contact classification to calculate an AUC value for each of the proteins. The result of this calculation is shown in Table 2 for the Dunn and Pfam datasets. AUC values should be interpreted with caution when the dataset is highly unbalanced, due to the large number of negative values. In those cases, one should focus on the region of the ROC curve with low false positive rate, which is often of prime interest. For clarity, we only include the APC method combined with sequence clustering and/or low count correction.
Table 2.
Average AUC values for the different methods evaluated on the families in the Dunn and Pfam datasets
| Dunn |
Pfam |
|||
|---|---|---|---|---|
| AUC | AUC0.1 | AUC | AUC0.1 | |
| APC | 0.700 | 0.218 | 0.723 | 0.259 |
| APC-C | 0.699 | 0.217 | 0.748 | 0.292 |
| APC-Lc | 0.712 | 0.224 | 0.730 | 0.261 |
| APC-C-Lc | 0.711 | 0.219 | 0.763 | 0.307 |
| APC-C-Lc-Z seq | 0.710 | 0.218 | 0.781 | 0.341 |
| APC-C-Lc-Z col | 0.698 | 0.206 | 0.768 | 0.315 |
Dunn refers to the 83 MSAs constructed by Dunn et al. (2008), and Pfam refers to the 85 Pfam families described in Section 2. AUC and AUC0.1 refer to the AUC corresponding to the full dataset or those data falling in the 10% lowest false positive rates. The methods included in the benchmark are: APC, C is clustering, Lc is low count correction with λ = 0.05, and Z seq and Z col are the sequence- and column-based Z-score sequence permutation described in the text. The best performing method for each benchmark dataset is highlighted in bold.
For the Dunn set, it is clear that the sequence clustering has very limited effect on the predictive performance. This is to be expected since this dataset has been preprocessed to remove sequence redundancy. The Pfam dataset is in that respect unbiased, and here it is apparent that both clustering and low count correction has strong impact on the performance values. For the Pfam dataset, the sequence-based Z-score transformation led to a significant improvement in the overall prediction accuracy (P < 0.0001, binomial test). Moreover, did the sequence-based permutation significantly outperform the column-based (P < 0.0001, binomial test). This is a surprising result since the a priori expectation as described earlier was that the column-based permutation test would be most appropriate for the detection of MI. Due to the low performance of the column-based permutation, we will in the remaining part of the article use sequenced-based permutations only to estimate Z-scores. It should be stressed that since the sequence-based permutations do not test for the appropriate hypotheses, the corresponding Z-scores should be interpreted with caution.
For the 75 Pfam families with >400 clusters, we further found that the average sequence-based Z-score threshold defining a sensitivity of 0.4 and a specificity of 0.95 was 6.5 ± 2.5.
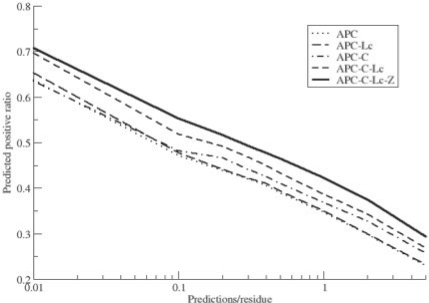
For datasets with a high ratio of negative values, an illustrative manner to rank the predictive performance of different methods is to plot the predicted positive ratio as a function of predictions per residue on a log scale as the x-axis (Shackelford and Karplus, 2007). Figure 1 shows the average of such curves for the 85 families in the Pfam dataset for the five methods included in the benchmark. Again, here we find the exact same ranking of the five methods, as found using the AUC analysis.
Fig. 1.
Average of predicted positive ratio as a function of predictions per residue in a semi-log plot for the 85 MSA's in the Pfam dataset. C, clustering and Z, Z-score sequence-based permutation.
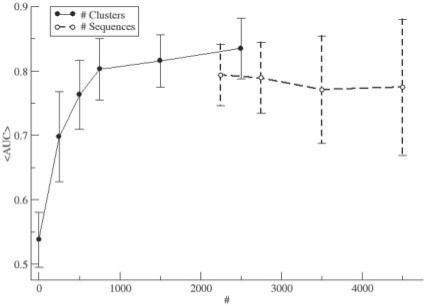
The Pfam dataset is relatively large and covers a broad range of family sizes (2000–10 000 proteins per family). This allowed us to investigate to what extent the performance—in terms of the ability to predict amino acid contacts—depends on the number of sequences in the MSA and how this dependency is altered by the data redundancy. The result of such analysis is shown in Figure 2, where the predictive performance is displayed as a function of the number of sequences and clusters, respectively, in the MSA. It is clear that the predictive performance is strongly related to the number of clusters in the MSA, and that MSAs with <400 clusters tend to show very low predictive performance values (AUC < 0.75).
Fig. 2.
Average AUC values (<AUC>) as a function of the number (#) of clusters or sequences in the MSA from the Pfam dataset. For clusters, the # refers to the definition by the Hobohm 1 algorithms using 62% sequence identity. For sequences, the # refers to individual sequences from protein families defined in the Pfam database. Performance values are calculated using sequence-based Z-score permutations.
Next, we turn to the small set of five selected families analyzed using the conventional Blast search program to build the MSAs. The result of this analysis is shown in Table 3.
Table 3.
Average AUC values for the different methods evaluated on the five selected families taken as example
| Method | 1BMC | 2TRX | 1JWP | 1O1N | 1URE | Ave |
|---|---|---|---|---|---|---|
| # Seq | 4171 | 5614 | 2522 | 1824 | 481 | |
| # Cluster | 2809 | 3227 | 980 | 429 | 121 | |
| APC | 0.807 | 0.776 | 0.765 | 0.715 | 0.686 | 0.750 |
| APC-Lc | 0.815 | 0.778 | 0.768 | 0.715 | 0.704 | 0.756 |
| APC-C | 0.812 | 0.769 | 0.807 | 0.787 | 0.686 | 0.772 |
| APC-C-Lc | 0.820 | 0.771 | 0.814 | 0.789 | 0.722 | 0.783 |
| APC-C-Lc-Z | 0.814 | 0.776 | 0.812 | 0.791 | 0.729 | 0.785 |
The columns refer to the PDB code of the five different protein families included in the benchmark. # Seq states the number of sequences in the MSA, # Cluster states the number of clusters in the MSAs, as defined by the Hobohm 1 reduction (Hobohm et al., 1992) at 62% sequence identity. Ave represents the average values across the different protein families. The methods included in the benchmark are the same as those in Table 2. The best performing method for each benchmark dataset is highlighted in bold.
Also, for this small dataset is it apparent that the clustering and correction for low count generally improve the predictive performance. However, for some protein families, there is hardly any gain obtained by applying correction for low counts and sequence clustering (1BMC, 2TRX). Both these families are populated with a very high number of diverse sequences (they both have more than 2800 sequence clusters), so it is expected that low count correction will not play a major role for these families. It is also apparent from the results in Table 3 that the accuracy of MI calculation depends on the number of sequence clusters available. The protein 1URE represents a family with a very low number of clusters (121), making accurate estimations of the 400 concurrent amino acid frequencies rather uncertain. For the remaining two protein families (1JWP and 1O1N), the number of clusters is sufficient to yield accurate predictions, provided the calculation includes sequence clustering and low count correction. One should bear in mind that despite this being just an illustrative (user-oriented) test exercise on limited size data, it confirms the results obtained using the Pfam MSAs.
For the four protein families with a number of clusters >400 (all families except 1URE), we find an average Z-score threshold of 6.1 ± 1.1 defining a sensitivity of 0.4 and a specificity of 0.95. The threshold value corresponds well with the value obtained earlier for the Pfam dataset.
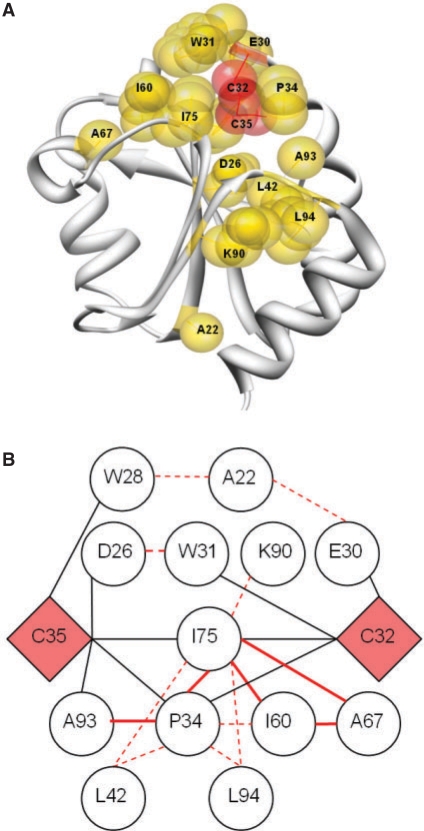
3.4 Mapping to a 3D structure: 2TRX as a case study
When mapping residue pairs for the 2TRX protein family onto the 3D structure, we found that 14 out of the 20 pairs with highest Z-score values and a sequential distance greater than four residues were interconnected forming a network around the conserved catalytic cystein residues 32 and 35 in the active site of thioredixin (Fig. 3). The pair of residues P34–A93, I60–A67, I60–I75, A67–I75 and P34–I75 are in direct sidechain contact (with a Cβ distance <8 Å), whereas the other pairs, although not in direct contact distance, all (with the exception of A22) are in contact with other residues of the network. Due to the proximity to the active site of the protein, the first large network might map residues of functional (rather than structural) importance. Experimental studies would be necessary to test this hypothesis.
Fig. 3.
(A) Ribbon representation of 2TRX. Represented as gold spheres, 14 of the 20 highest Z-score transformed MI scoring pairs of residues are shown. Orange spheres: catalytic C32 and C35. Molecular graphic images were generated using UCSF Chimera package (University of California; Meng et al., 2006). (B) Schematic representation of the 14 pairs of interactions scoring highest in Z-score transformed MI values. Red lines are high MI scoring residue pairs. Full lines denote physical contact (Cβ distance <8 Å).
Notably we have found that, in the five families of proteins chosen for illustration belonging to different classes, networks are formed. The extreme case is the 1JWP family of proteins, where 9 out of the 10 highest scoring pairs are interconnected forming a network and these nine pairs lie close in the 3D structure as well (they all map to the β-hairpin formed between residues 99 and 115 of the β-lactamase). This analysis demonstrates the power of the proposed MI approach to identify residue contacts and nets of interaction in biological sequences.
4 DISCUSSION AND CONCLUSIONS
Here, we have compared two recently published approaches to lessen the influence of phylogeny and signal noise into the calculation of MI or coevolution between residues. Furthermore, we have shown how including simple techniques of sequence clustering and low count correction can significantly enhance the estimation of MI between residue pairs. Large-scale benchmarking including both artificial (in silico generated) and biological data demonstrated that this improved method could be applied to achieve accurate prediction of coevolving sites and contacts. Our results demonstrate that raw MI was the worst predictor of coevolution. The RCW method of Gouveia-Oliveira and Pedersen (2007) outperformed MI. The APC background correction method by Dunn et al. (2008) achieved the highest performance. In this context, the inclusion of low count correction and clustering was shown to improve all three methods. The best performing method for both artificial and natural sequences was the combination of APC correction, clustering and low count correction. We demonstrated that Z-score transformation calculated from sequence-based permutations significantly improved the prediction accuracy of the method, and allows an interpretation of predictions across different protein families. Further, we demonstrate how the predictive performance of the method depends strongly on the number of sequence clusters rather than the number of sequences in the MSA, and those MSAs with <400 clusters tend to display very low predictive performance values.
A direct correlation of MI to the degree of coevolution has been shown to be extremely difficult. Tree-based evolution, phylogeny, conservation and noise are important factors that make difficult the identification of MI between residues. The usefulness of the information deposited in an MSA is remarkable in the protein-modeling field. During the last decade, many researchers have intended to take advantage of this information. A method to predict residues that coevolve may be a guide for assembling local structure prediction into full tertiary prediction (Shackelford and Karplus, 2007). Tertiary restraints derived from an MSA have been incorporated into software for ab initio folding of proteins to provide decoys closer to the native-like structure (Ortíz and Skolnick, 2000; Ortíz, et al., 1998). In addition, estimation of MI might emerge as an important tool for protein–protein interaction predictions (Dunn et al., 2008; Fares and Travers, 2006; Ramani and Marcotte, 2003). Furthermore, an MI analysis can prove very useful to further characterize protein structure or function and to guide the design of mutagenesis studies.
Pairs of residues that show high MI values are often postulated to coevolve and, in the vast majority of cases, they are assumed to be close in space. While the assumption of spatial proximity of coevolving residue pairs is appealing, it is apparent that residues that coevolve are not necessarily close in contact or, conversely, residues close in contact are not necessarily coevolving. One could speculate that residues that are far away in the folded protein structure might have been close in folding intermediates or might share a common outside neighboring molecule, like another protein or ligand. Lockless and Ranganathan (1999) have demonstrated both theoretically and experimentally that statistically coupled residues may be distantly positioned in the structure, and there are many examples of allosterically interacting residues (Shi et al., 2006).
We illustrated on one particular protein family (thioredoxins), a practical use of the proposed method to gain better understanding the role of coevolution. Here, the high-scoring MI pairs were found to order into a network or cluster of residues (Byung-Chul et al., 2008; Korber, et al., 1993). This finding might bear general significance for the understanding of protein structure and function.
The proposed method for MI calculation significantly filters the signal from the noise, not only on artificial datasets, but also when applied to biological data.
ACKNOWLEDGEMENTS
We acknowledge Anders Gorm Pedersen and Rodrigo Gouveia-Oliveira for fruitful discussion and valuable insights into the problem of relating MI to coevolution and for sharing the set of artificial sequence data with us. We thank S. D. Dunn and coworkers for sharing their dataset with us.
Funding: UBACyT; CONICET; ANPCyT; National Institutes of Health (contract HHSN266200400025C to M.N.).
Conflict of Interest: C.M.B., J.S. and J.M.D. are career researchers of the National Research Council of Argentina (CONICET). The other author has declared none.
REFERENCES
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byung-Chul L, et al. Analysis of the residue-residue coevolution network and the functionally important residues in proteins. Protein Struct. Funct. Bioinform. 2008;72:863–872. doi: 10.1002/prot.21972. [DOI] [PubMed] [Google Scholar]
- Chiu DKY, Kolodziejczak T. Inferring consensus structure from nucleic acid sequences. Comput. Appl. Biosci. 1991;7:347–352. doi: 10.1093/bioinformatics/7.3.347. [DOI] [PubMed] [Google Scholar]
- Cover TM, Thomas JA. Elements of information theory. Wiley; 1991. [Google Scholar]
- DePristo MA, et al. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat. Rev. Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- Dunn SD, et al. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008;24:333–340. doi: 10.1093/bioinformatics/btm604. [DOI] [PubMed] [Google Scholar]
- Fares MA, Travers SAA. A novel method for detecting intramolecular coevolution: adding a further dimension to selective constraints analyses. Genetics. 2006;173:9–23. doi: 10.1534/genetics.105.053249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor AA, Aldrich RW. Influence of conservation on calculations of amino acid covariance in multiple sequence alignments. Proteins. 2004;56:211–221. doi: 10.1002/prot.20098. [DOI] [PubMed] [Google Scholar]
- Gloor GB, et al. Mutual information in protein multiple sequence alignments reveals two classes of coevolving positions. Biochemistry. 2005;44:7156–7165. doi: 10.1021/bi050293e. [DOI] [PubMed] [Google Scholar]
- Gouveia-Oliveira R, Pedersen A. Finding coevolving amino acid residues using row and column weighting of mutual information and multi-dimensional amino acid representation. Algorithms Mol. Biol. 2007;2:12. doi: 10.1186/1748-7188-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobohm U, et al. Selection of representative protein data sets. Protein Sci. 1992;1:409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber BT, et al. Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc. Natl Acad. Sci. USA. 1993;90:7176–7180. doi: 10.1073/pnas.90.15.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- Martin LC, et al. Using information theory to search for co-evolving residues in proteins. Bioinformatics. 2005;21:4116–4124. doi: 10.1093/bioinformatics/bti671. [DOI] [PubMed] [Google Scholar]
- Meng EC, et al. Tools for integrated sequence-structure analysis with UCSF chimera. BMC Bioinformatics. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, et al. Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics. 2004;20:1388–1397. doi: 10.1093/bioinformatics/bth100. [DOI] [PubMed] [Google Scholar]
- Ortíz AR, Skolnick J. Sequence evolution and the mechanism of protein folding. Biophys. J. 2000;79:1787–1799. doi: 10.1016/S0006-3495(00)76430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortíz AR, et al. Combined multiple sequence reduced protein model approach to predict the tertiary structure of small proteins. Pac. Symp. Biocomput. 1998:377–388. [PubMed] [Google Scholar]
- Ramani AK, Marcotte EM. Exploiting the co-evolution of interacting proteins to discover interaction specificity. J. Mol. Biol. 2003;327:273–284. doi: 10.1016/s0022-2836(03)00114-1. [DOI] [PubMed] [Google Scholar]
- Shackelford G, Karplus K. Contact prediction using mutual information and neural nets. Protein Struct. Funct. Bioinfom. 2007;69:159–164. doi: 10.1002/prot.21791. [DOI] [PubMed] [Google Scholar]
- Shi Z, et al. Networks for the allosteric control of protein kinases. Curr. Opin. Struct. Biol. 2006;16:686–692. doi: 10.1016/j.sbi.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Swets J. Measuring the accuracy of diagnostic systems. Science. 1988;3:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- Tillier ER, Lui TW. Using multiple interdependency to separate functional from phylogenetic correlations in protein alignments. Bioinformatics. 2003;19:750–755. doi: 10.1093/bioinformatics/btg072. [DOI] [PubMed] [Google Scholar]
- Wollenberg KR, Atchley WR. Separation of phylogenetic and functional associations in biological sequences by using the parametric bootstrap. Proc. Natl Acad. Sci. USA. 2000;97:3288–3291. doi: 10.1073/pnas.070154797. [DOI] [PMC free article] [PubMed] [Google Scholar]