Abstract
On exposure to mildly acidic conditions, apomyoglobin forms a partially folded intermediate, I. The A, B, G, and H helices are significantly structured in this equilibrium intermediate, whereas the remainder of the protein is largely unfolded. We report here the effects of mutations at helix pairing sites on the stability of I in three classes of mutants that: (i) truncate hydrophobic side chains in native helix packing sites, (ii) truncate hydrophobic side chains not involved in interhelical contacts, and (iii) extend hydrophobic side chains at residues not involved in interhelical contacts. Class I mutants significantly decrease the stability and cooperativity of folding of the intermediate. Class II and III mutants show smaller effects on stability and have little effect on cooperativity. Qualitatively similar results to those found in I were obtained for all three classes of mutants in native myoglobin (N), demonstrating that hydrophobic burial is fairly specific to native helix packing sites in I as well as in N. These results suggest that hydrophobic burial along native-like interhelical contacts is important for the formation of the cooperatively folded intermediate.
Apomyoglobin undergoes three-state acid-induced unfolding with the formation of an intermediate (I) at pH 4 (1). This intermediate has been the subject of intense scrutiny, and many of its properties are now known. Of the eight helices in myoglobin (labeled A through H), the A, G, and H helices are protected from hydrogen exchange (2, 3), and the B helix is partially formed (4). Mutagenesis of hydrophobic residues in the interhelical interfaces between the A, G, and H helices have shown that these contacts contribute significantly to the stabilization of I (5). These experiments establish the importance of hydrophobic burial as a source of stability in I, but do not address whether this burial is restricted to native-like helix packing sites or is nonspecific, as previously proposed in a “rolling hydrophobic” model (6).
In this study, we further characterize the nature of hydrophobic burial in I by making hydrophobic mutations at both native-like helix packing sites and at potential nonnative (surface-exposed) sites. Three classes of mutants were constructed: (i) hydrophobic truncation mutants of residues buried in the A–H and G–H helix interfaces in native myoglobin, (ii) hydrophobic truncation mutants of residues that are surface exposed in I, and (iii) hydrophobic extension mutants (Ala → Leu) at surface-exposed positions in I. These mutants address the question of whether the strong destabilizing effects seen in previous mutants are specific to native helix pairing sites or simply a result of the loss of nonspecific hydrophobic burial.
Analysis of Hydrophobic Burial in the Intermediate.
The pattern of hydrophobic burial in the globin family is the subject of several excellent studies (7–9). In this work, we examine all of the hydrophobic residues in the A, G, and H helices and calculate their solvent-accessible surface areas (10) at several different potential stages of folding (Table 1). The folding of apomyoglobin is divided into three stages for the purposes of this study: (i) formation of isolated helices, (ii) the collapse of the A, B, G, and H helices into the compact structure of the intermediate (the ABGH subdomain), and (iii) the formation of the fully folded native protein (including the binding of heme). The structure at each stage of folding is treated as a subset of the structure seen in the myoglobin crystal structure (11), with the remainder of the protein assumed to be completely unfolded. The helices are defined according to Lesk and Chothia (9): helix A, residues 3–18; helix B, residues 20–35, helix G, residues 100–118; and helix H, residues 124–149. Phe-123, a residue in the GH turn (which packs into the G–H helix interface) also is considered, but it does not belong to a helix and lacks an entry for local helix accessible surface areas. The ABGH subdomain of the intermediate is modeled as the sum of all residues in the four helices, plus residues in the AB and GH loops. The assumption of fully formed helices in the intermediate is an oversimplification, as recent NMR data has shown that all of the helices in I suffer from significant fraying (4). Given the lack of high-resolution structural data on I, the fully formed ABGH subdomain is used as the best available approximation to the structure of I and is an upper limit for the extent of structure present.
Table 1.
Calculated nonpolar solvent-accessible surface area at several potential stages of folding in myoglobin in Å2
| Residue | Helix position | Local helix | ABGH subdomain | Δ(Local helix-ABGH) | Native myoglobin |
|---|---|---|---|---|---|
| TRP 7 | A5 | 135 | 94.8 | 40.5 | 6.48 |
| LEU 9 | A7 | 121 | 54.9 | 66.4 | 54.9 |
| VAL 10 | A8 | 88.4 | 14.1 | 74.2 | 0.09 |
| LEU 11 | A9 | 103 | 103 | 0 | 41.9 |
| VAL 13 | A11 | 91.9 | 1.13 | 90.8 | 1.13 |
| TRP 14 | A12 | 128 | 107 | 20.8 | 11.0 |
| ALA 15 | A13 | 50.7 | 50.7 | 0 | 50.7 |
| VAL 17 | A15 | 70.7 | 9.29 | 61.4 | 2.25 |
| VAL 21 | B2 | 127 | 83.9 | 43.2 | 33.7 |
| ALA 22 | B3 | 73.5 | 73.5 | 0 | 23.8 |
| ILE 28 | B9 | 112 | 21.4 | 90.9 | 4.09 |
| LEU 29 | B10 | 94.3 | 83.1 | 11.3 | 6.33 |
| ILE 30 | B11 | 89.1 | 89.1 | 0 | 14.0 |
| LEU 32 | B13 | 115 | 37.2 | 77.7 | 12.8 |
| PHE 33 | B14 | 132 | 132 | 0 | 2.64 |
| ILE 101 | G2 | 146 | 59.6 | 86.7 | 40.1 |
| TYR 103 | G4 | 78.8 | 75.5 | 3.31 | 17.6 |
| LEU 104 | G5 | 120 | 38.7 | 81.1 | 17.0 |
| PHE 106 | G7 | 107 | 89.7 | 17.8 | 28.5 |
| ILE 107 | G8 | 111 | 50.1 | 60.5 | 33.6 |
| ALA 110 | G11 | 52.7 | 2.84 | 49.8 | 2.84 |
| ILE 111 | G12 | 90.4 | 34.2 | 56.2 | 9.69 |
| ILE 112 | G13 | 110 | 18.0 | 92.3 | 18.0 |
| VAL 114 | G15 | 74.7 | 11.1 | 63.5 | 11.1 |
| LEU 115 | G16 | 110 | 2.83 | 108 | 2.83 |
| PHE 123 | GH5 | 0 | 0 | 0 | 0 |
| ALA 125 | H2 | 72.7 | 72.7 | 0 | 72.7 |
| ALA 127 | H4 | 59 | 6.39 | 52.6 | 6.39 |
| ALA 130 | H7 | 54.9 | 1.13 | 53.7 | 0.49 |
| MET 131 | H8 | 115 | 6.54 | 108 | 2.07 |
| ALA 134 | H11 | 48.2 | 12.4 | 35.8 | 0.20 |
| LEU 135 | H12 | 84.6 | 26.5 | 58.1 | 2.65 |
| LEU 137 | H14 | 109 | 85.3 | 23.5 | 23.6 |
| PHE 138 | H15 | 129 | 105 | 24.5 | 20.8 |
| ILE 142 | H19 | 74.7 | 49.1 | 25.6 | 0.96 |
| ALA 143 | H20 | 51.6 | 30.5 | 21.2 | 15.7 |
| ALA 144 | H21 | 57.4 | 57.4 | 0 | 57.2 |
| TYR 146 | H23 | 111 | 78.6 | 32.5 | 6.34 |
| LEU 149 | H26 | 103 | 103 | 0 | 40.1 |
Local helix refers to the isolated helix in which the residue is located. The ABGH subdomain refers to these helices plus the AB and GH loops in the myoglobin structure. Native myoglobin refers to the fully folded protein (as described by the complete crystal structure). Only hydrophobic residues in the A, B, G, and H helices are listed.
Several interesting patterns emerge from the data in Table 1. First, as has been observed previously, there is not strict partitioning of hydrophobic residues into the core of the protein (8). Although polar and charged residues are almost invariably paired or solvent-exposed, about one-third of the hydrophobic residues in myoglobin have significant (>20 Å2) solvent-exposed surface area. The only hydrophobic groups that are nearly completely solvent-exposed (>70%) are three Ala residues (15, 125, and 144). This incomplete partitioning allows us to examine hydrophobic residues in both the core and at the surface of myoglobin. Several bulky hydrophobic groups become highly solvent exposed in I (L11, V21, A22, L29, I30, F33, I101, F106, L137, F138, and L149) as the central helices (C–F) are not formed, and burial against these helices is lost.
For the purposes of this study, it also is important to distinguish the burial of hydrophobic residues occurring in the interhelical interfaces of the ABGH subdomain (A–H, G–H, and B–G) from burial that occurs locally on helix formation. Local burial in a helix can stabilize the helix but is unlikely to contribute to the specification of tertiary structure of I. In contrast, burial occurring between helices, whether native or nonnative, is predicted to be important for determining the tertiary fold of I.
Based on the analysis above, three classes of mutants can be constructed to test the hypothesis that native-like hydrophobic burial contributes significantly to the stability of I and to the formation of its specific tertiary fold. In class I mutants, the side chains are deeply buried in the core of the ABGH subdomain by interhelical contacts. Only one member of this class has been studied previously (M131A). Members of this class are characterized by a large decrease in solvent-accessible surface area in going from the isolated helix to the complete ABGH subdomain. Although these residues bury a large portion of their surface area in tertiary contacts, they also bury significant area within their own helices. In general, the hydrophobic accessible surface area buried during helix formation is of similar magnitude to the burial during the tertiary assembly of helices.
The side chains of class II mutants are largely surface-exposed, bulky hydrophobic groups that do not contact the interhelical interfaces within the ABGH subdomain. These sites generally become more buried on completion of folding to the myoglobin native state. The exposure of a large number of hydrophobic groups in the ABGH subdomain probably contributes to its limited solubility. These sites are mutated to alanine to study the effects of hydrophobic truncations at the protein surface. Class III side chains are alanines that are surface-exposed, both in I and N. These residues are mutated to leucine to study the effects of adding hydrophobic side chains at surface-exposed sites, distant from helix packing.
Several criteria were applied in picking representative mutants from within each class. First, the B helix was avoided in this study because of its less well defined structure in I. In particular, although NMR chemical shift analysis reveals significant helix formation (4), hydrogen exchange data indicate that this helix must be of marginal stability (3, 12). This loosened structure could muffle the effects of changes in hydrophobic burial. Mutants were also chosen, as much as possible, in the center of helices to avoid possible fraying effects present at the ends of all helices in I, as seen by recent NMR chemical shift analysis (4). Finally, as much as possible, mutations that would result in large helix propensity changes were avoided. Because of the limited numbers of members in each class, not all of these criteria could be accommodated for all mutants.
MATERIALS AND METHODS
Preparation of Proteins.
Mutants were constructed by using the MORPH mutagenesis system (5 Prime → 3 Prime). Sequences were confirmed by using standard dideoxy methods at the Stanford University Protein and Nucleic Acid Facility. The myoglobin synthetic gene was originally a gift from S. G. Sligar (University of Illinois) (13). Proteins were >95% pure as judged by using SDS/PAGE. Apomyoglobin preparation and concentration measurements were performed as described (12).
CD, Fluorescence, and Soret Measurements.
CD data were collected on an Aviv 62DS circular dichroism spectropolarimeter at 4°C at 222 nm. All CD measurements were taken in a 1 cm × 1 cm quartz cuvette with approximately 2 μM apomyoglobin (apoMb). CD data were averaged over 300 s of acquisition time, after >10 min equilibration on ice. Fluorescence data was acquired on a SLM-Aminco Bowman Series 2 luminescence spectrometer in a 1 × 0.5 cm cell with 1 μM apoMb. All measurements were taken at 4°C. All experiments for both CD and fluorescence were performed in 4 mM citrate, pH 4.2.
Preparation and study of cyano-Mb was as follows: briefly, cyano-Mb was reconstituted from apoMb by the addition of 1 equivalent of cyano-heme (pH 10), followed by ultracentrifugation (90,000 g for 10 min at 4°C) to remove precipitates. Protein concentration was determined by absorbance at 280 (35 cm−1⋅mM−1) and 423 nm (109.7 cm−1⋅mM−1). Only protein stocks with good agreement in absorbance between Soret and UV measurements (>90% holo-Mb) were used in unfolding experiments. CD data were taken at 222 nm, and Soret absorbance data were taken at 423 nm. Native cyano-Mb was studied at pH 5 with 10 mM sodium acetate, 0.5 mM sodium cyanide at 1 μM protein concentration. All cyano-Mb samples were allowed to equilibrate in urea for 1 hour before CD or Soret measurements.
Data Analysis.
All curve fitting was performed by using kaleidagraph software for the Macintosh (Synergy Software, Reading, PA). Urea unfolding curves were fitted by using the procedure of Santoro and Bolen (14), where the two-state transition was defined by using data both inside and outside the transition zone. Linear folded and unfolded baselines were used. Urea unfolding data for I is normalized by dividing all points by the CD value at 0 M urea, giving a normalized scale of 0 to 1. This procedure does not remove baseline contributions, allowing the observation of altered baselines. CD values for I at 0 M urea ranged from −12,500 to −14,000 deg cm2 dmol−1 at 222 nm.
Urea unfolding data for native cyano-Mb is reported as fraction folded, where the data are fitted to a two-state model and the baseline contributions are removed. This treatment is appropriate for data that does not appear to deviate from being fully cooperative. The fitted CD or Soret value of N is set to 1.0 in Fig. 4. CD values for cyano-Mb at 0 M urea ranged from −23,000 to −27,000 deg cm2 dmol−1 at 222 nm. Urea unfolding results are reported as urea midpoint of unfolding (Cm). This value is reported instead of ΔG to avoid the large error associated with the uncertainty in m-value in a two-state fit (15, 16). A constant m-value of 1,500 cal M−1 and 1,600 cal M−1 was used to fit all intermediate and native cyano-Mb data, respectively.
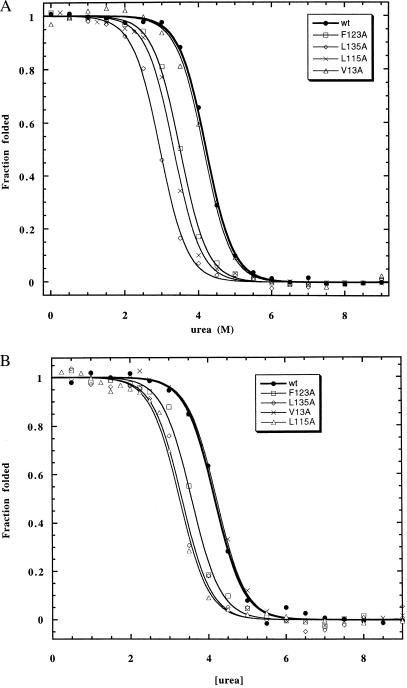
Figure 4.
(A) Hydrophobic truncation mutants at native interhelical contact sites (class I) in cyano-Mb as measured by using CD. (B) Hydrophobic truncation mutants at native interhelical contact sites (class I) in cyano-Mb as measured by using Soret absorbance.
Solvent accessibility calculations used the program naccess (17) on a Silicon Graphics workstation.
RESULTS
Stability of I in Mutants as Measured by CD and Fluorescence.
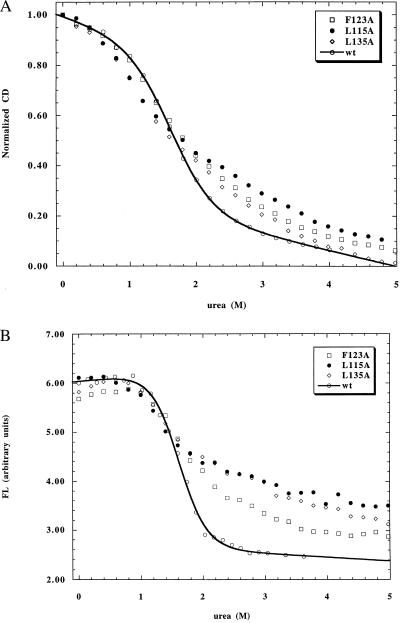
Table 2 summarizes the CD and fluorescence-monitored urea unfolding data for all of the mutants studied. Probes of both secondary (CD) and tertiary (fluorescence) structure are used to determine whether folding is fully cooperative (i.e., two-state). Three of the four class I mutants (L115A, F123A, and L135A) exhibited broad transition curves when measured by CD with steeply sloping unfolded baselines, indicative of a deviation from two-state folding (Fig. 1A). These unfolding curves are difficult to analyze quantitatively because of their broad transitions. One can estimate, however, the Cm values of these mutants by fitting the data with a steep unfolded baseline. These measurements are not particularly meaningful except to show that the Cm values of these mutants are decreased significantly from that of wild type, and the values obtained by fitting CD and fluorescence show poor agreement. Further evidence of loss of cooperativity is seen in the fluorescence-monitored unfolding curves, which are also broad, have small amplitudes, and do not coincide with the CD-monitored transitions (Fig. 1B). Two other class I mutants, V13A and M131A (measured previously in ref. 5), exhibit normal two-state transitions by both CD and fluorescence, and the transitions are superimposible obtained by the two probes. V13A is only slightly destabilized compared with wild type (0.1 kcal/mol), whereas M131A was previously reported to be strongly destabilized (0.9 kcal/mol).
Table 2.
Cm values (in M urea) for hydrophobic mutants in I and N
| Mutant | Intermediate
|
Native (cyano-Mb)
|
||||
|---|---|---|---|---|---|---|
| CD Cm, M | Fluorescence Cm, M | ΔΔG kcal/mol | CD Cm, M | Soret Cm, M | ΔΔG, kcal/mol | |
| wildtype | 1.60 | 1.56 | 0 | 4.21 | 4.17 | 0 |
| Class I | ||||||
| V13A | 1.58 | 1.49 | −0.1 | 4.15 | 4.22 | 0 |
| L115A | 1.06 | 0.74 | −1.0 | 3.33 | 3.26 | −1.4 |
| F123A | 1.46 | 1.27 | −0.3 | 3.49 | 3.57 | −1.1 |
| L135A | 1.28 | 0.81 | −0.8 | 2.96 | 3.32 | −1.7 |
| Class II | ||||||
| L11A | 1.20 | 1.31 | −0.5 | 3.93 | 3.68 | −0.6 |
| F106A | 1.35 | 1.51 | −0.2 | 3.71 | 3.75 | −0.7 |
| L137A | 1.55 | 1.48 | −0.1 | 4.04 | 3.98 | −0.3 |
| Class III | ||||||
| A15L | 1.72 | 1.66 | +0.2 | 4.26 | 4.20 | +0.1 |
| A125L | 1.81 | 1.84 | +0.4 | 4.56 | 4.57 | +0.6 |
| A144L | 1.74 | 1.60 | +0.1 | 4.44 | 4.45 | +0.4 |
Cm values were measured at pH 4, 4 mM citrate, 4°C (intermediate) and at pH 5, 10 mM acetate, 0.5 mM sodium cyanide, 25°C (native cyano-Mb) and are given in M. ΔΔG is the difference between the average Cm given by CD and fluorescence and the Cm of wildtype, multiplied by the average m-value for all mutants, 1,500 cal mol−1⋅M−1 for I and 1,600 cal mol−1⋅M−1 for N.
Figure 1.
(A) Hydrophobic truncation mutants at native interhelical contact sites (class I) in I as measured by using CD. (B) Hydrophobic truncation mutants at native interhelical contact sites (class I) in I as measured by using fluorescence.
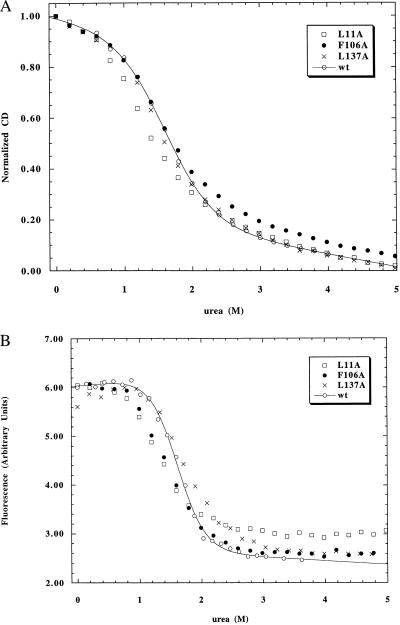
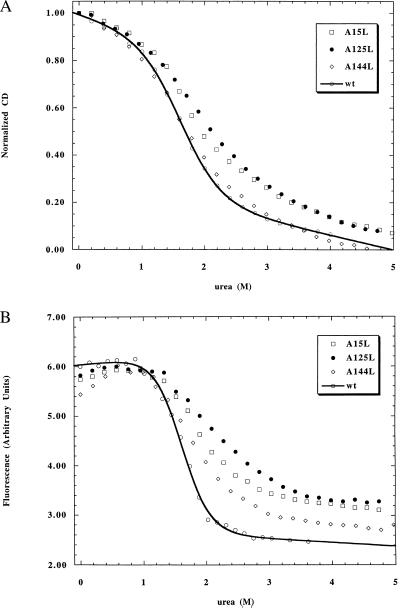
The class II and III mutants all display apparent two-state transitions with reasonable agreement between CD and fluorescence monitored urea unfolding (Figs. 2 and 3). Two of the class II mutants are mildly destabilized (0.1 to 0.15 M decrease in Cm), but one mutant (L11A) is moderately destabilized (0.3 M decrease in Cm). Class III mutants display moderate stabilization (0.1–0.3 M increase in Cm). Some of these mutants also exhibit slightly broadened transitions (Fig. 3) with steeper unfolded baselines, indicative of some deviation from pure two-state behavior.
Figure 2.
(A) Hydrophobic truncation mutants at surface-exposed sites (class II) in I as measured by using CD. (B) Hydrophobic truncation mutants at surface-exposed sites (class II) in I as measured by using fluorescence.
Figure 3.
(A) Hydrophobic extension mutants at surface-exposed sites (class III) in I as measured by using CD. (B) Hydrophobic extension mutants at surface-exposed sites (class III) in I as measured by using fluorescence.
Stability of Native Cyano-Mb in Mutants Measured by Using CD and Soret Absorbance.
All of the mutants in this study also were examined for their effects on the native state (cyano-Mb). In the previous study of hydrophobic packing, the apomyoglobin native state (pH 7.8) was chosen to compare the effects of mutants in N and I. Measurements of stability changes in apoMb N require a three-state analysis because of the three-state unfolding of N (N → I → U). Furthermore, the N → I and I → U transitions overlap in urea unfolding (as opposed to acid-induced unfolding, where they are well separated). This form of the native state (cyano-Mb) was chosen because of problems with irreversibility in the unfolding of holo-Mb (18). These problems have, however, recently been overcome (unpublished results) by using cyano-Mb. The presence of cyanide solubilizes heme and allows reversible heme binding during unfolding. An additional benefit of this method is that the unfolding of cyano-Mb at pH 5 was shown to be two-state, greatly facilitating quantitative analysis of stability changes in mutants.
The class I mutants show similar destabilizing effects in N (cyano-Mb) as in I (Fig. 4, Table 2). In contrast to the unfolding data in I, these mutants exhibit cooperative unfolding in N. The three mutants (L115A, F123A, and L135A) that show broadened folding transitions in I are strongly destabilized in N, with losses of stability in the 1–2 kcal/mol range. Two of the these three mutants (L115 and F123A) show good agreement between CD- and Soret-monitored unfolding, whereas the Cm values of L135A differ by about 10% between the two probes, indicating a small deviation from two-state unfolding. In general, the effects of all mutants are larger in N than in I.
The class II mutants show similar qualitative changes in stability in N and I (Table 2), with mild to moderate destabilization (0.1–0.7 kcal/mol). The residues mutated in the class II mutants, although largely solvent-exposed in the ABGH subdomain, become more buried on formation of the full native structure (Table 1). Class III mutants also show similar behavior in I and N (Table 2), with mild stabilization observed for all three mutants (0.2–0.6 kcal/mol). The residues mutated in all class III mutants remain largely surface exposed even after the folding of the C–F helices.
DISCUSSION
In the present work, we study the specificity of hydrophobic burial in the apomyoglobin intermediate and cyano-Mb. Three classes of mutants are examined to determine the relative importance of hydrophobic burial in native interhelical contacts vs. sites predicted to be solvent-exposed in the ABGH subdomain. The effects of these mutants are qualitatively similar in both I and N (cyano-Mb), demonstrating that the intermediate possesses a loosened but native-like set of interhelical hydrophobic contacts. This work confirms and extends previous work (5) showing that these hydrophobic helix contact sites are native-like and are major contributors to stability and cooperativity in I.
Effects of the Three Mutant Classes on Stability.
In agreement with previous work, almost all mutants targeting these native contact sites are found to be significantly destabilized in both N and I. The one exception in this study, V13A, is probably not much destabilized because of the low helix propensity of Val and replacement by the strong helix former Ala (19). The resulting stabilization of the A helix may offset the loss of hydrophobic burial in the interhelical interface. The previously studied M131A (a class I mutant) is strongly destabilized but maintains its cooperativity. A possible explanation for this difference from other class I mutants is that only M131 has significant burial in both of the two major interhelical interfaces (A–H and G–H) in I. By destabilizing both interfaces equally, M131A may maintain cooperativity by propagating its destabilizing effect more evenly throughout the protein.
The three destabilized class I mutants all show severe loss of stability in N (cyano-Mb) (1.1–1.7 kcal/mol). This result confirms the expectation from the crystal structure that, because these residues are completely buried in the native state, they should contribute greatly to stability. The advent of a system for studying the reversible unfolding of holo-Mb in a two-state manner allows testing of the importance of hydrophobic burial in the native state. It is important to use these results obtained in the native state to provide a reference for the properties of mutants in I and to determine to what extent interactions present in the intermediate are native-like. As found in a previous study comparing native apomyoglobin to I (5), hydrophobic interactions are found to be stronger in N, implying that these interactions are only partly formed in I.
Quantitation of the destabilization of class I mutants in I is difficult because of the breakdown of cooperative two-state unfolding. The urea unfolding curves measured by only one probe can generally be fitted to a two-state equation, but the unfolded baseline is much steeper than observed for wild type, decreasing the amplitude of the transition both in CD- and fluorescence-monitored unfolding. The poor agreement between CD- and fluorescence-monitored unfolding shows that the unfolding of secondary and tertiary structure is no longer simultaneous, indicating a loss of cooperativity.
The loss of cooperativity seen here in class I mutants strongly resembles a previous study (15). In the earlier study, helix-destabilizing mutations (Gly and Pro) were made in the middle of the A and G helices. These mutations were severely destabilizing, resulting in broadened transitions with loss of superposition between CD- and fluorescence-monitored unfolding. Interestingly, these mutants also exhibited a markedly decreased amplitude of fluorescence change on unfolding. The fluorescence of the helix-destabilizing mutants was reduced by ≈75% at 0 M urea. In contrast, class I mutants have decreased amplitude of the fluorescence transition caused by an increase in the steepness of the unfolded baseline. In the helix-destabilizing mutants, this loss of amplitude appears to reflect a decreased population of folded I at 0 M urea, whereas for class I mutants it appears to reflect the presence of other intermediates or nonnative states at moderate urea concentrations.
The mutations at the surface-exposed sites of the protein (classes II and III) have smaller effects on the stability of I and N. These effects are, however, measurable, and indicate that hydrophobic burial does not occur exclusively at native packing sites. This result may be caused by the flexibility of I, which can allow portions of large hydrophobic groups to become buried in the interhelical interface or within their own helices. Interestingly, the effects of class II and III mutations in I and N are very similar, implying that both forms have hydrophobic groups on the surface and arguing against recruitment of surface-exposed hydrophobic groups to native-like helix contact sites in I. One class II mutant, L11A, shows an unexpectedly large loss of stability (0.5 kcal/mol) in I. This residue is expected to be largely exposed in I and significantly exposed in N. There is no obvious explanation for the large effect of this mutation on I. One possibility is that L11 forms an i, i+3 interaction with W14 and an i, i−4 interaction with W7, burying significant surface area. These two tryptophan residues do not, however, make direct contacts with L11 in the myoglobin crystal structure (i.e., this explanation invokes nonnative contacts).
Implications for Mb Folding.
These results support the idea that hydrophobic burial along native-like interhelical contact sites is important for the formation of the cooperatively folded intermediate. Disruption of these contacts dramatically destabilizes the intermediate, disrupts the cooperativity of folding, or both. Similar hydrophobic mutations away from these native interhelical contact sites have much less effect on both stability and cooperativity in I as well as N. These results discount the idea of a “rolling hydrophobic” interaction that can adapt easily to disruptive mutations. The importance of specific hydrophobic interactions has also been demonstrated recently for the ferricytochrome c (20) and α-lactalbumin molten globules (21, 22).
The precise nature of the hydrophobic burial examined in this study remains to be determined. In particular, it is unclear whether the hydrophobic interfaces simply exclude water or are well packed, as in the native protein. A good tool for making this distinction is overpacking mutants, where small hydrophobic groups in interhelical contacts are replaced by larger hydrophobic groups. These mutants are expected to be stabilizing if burial of hydrophobic surface area is of primary importance and if tight packing is important. One such mutant, A130L, has been studied (5). This mutant was slightly stabilizing (0.1 kcal/mol), indicating a compromise between increased burial and steric hindrance. In native apomyoglobin, this mutation was destabilizing (0.8 kcal/mol), showing the primary importance of tight van der Waals contacts in the native state. More studies of this type of mutant are needed to generalize these conclusions. Subtle hydrophobic mutations (e.g., L → I) in helix–helix packing interfaces will also be helpful for determining the degree of specificity in these interactions (23).
Acknowledgments
We thank Heinrich Roder for helpful comments on the manuscript. M.S.K. is a trainee of the National Institute of General Medical Sciences Medical Scientist Training Program. C.H.I.R. is a Pew Trust Latin American Fellow and thanks Fundaçao de Amparo à Pesquisa do Estado de Sao Paulo for a previous fellowship. This work was supported by a National Institutes of Health grant to R.L.B.
ABBREVIATION
- Mb
myoglobin
References
- 1.Griko Y V, Privalov P L, Venyaminov S Y, Kutyshenko V P. J Mol Biol. 1988;202:127–138. doi: 10.1016/0022-2836(88)90525-6. [DOI] [PubMed] [Google Scholar]
- 2.Jennings P A, Wright P E. Science. 1993;262:892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- 3.Hughson F M, Wright P E, Baldwin R L. Science. 1990;249:1544–1548. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- 4.Eliezer D, Yao J, Dyson H J, Wright P E. Nat Struct Biol. 1998;5:148–155. doi: 10.1038/nsb0298-148. [DOI] [PubMed] [Google Scholar]
- 5.Kay M S, Baldwin R L. Nat Struct Biol. 1996;3:439–445. doi: 10.1038/nsb0596-439. [DOI] [PubMed] [Google Scholar]
- 6.Barrick D, Baldwin R L. Protein Sci. 1993;2:869–876. doi: 10.1002/pro.5560020601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weaver D. Biopolymers. 1992 May;32:477–490. doi: 10.1002/bip.360320504. [DOI] [PubMed] [Google Scholar]
- 8.Richmond T J, Richards F M. J Mol Biol. 1978;119:537–555. doi: 10.1016/0022-2836(78)90201-2. [DOI] [PubMed] [Google Scholar]
- 9.Lesk A M, Chothia C. J Mol Biol. 1980;136:225–270. doi: 10.1016/0022-2836(80)90373-3. [DOI] [PubMed] [Google Scholar]
- 10.Lee B, Richards F M. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 11.Takano T. J Mol Biol. 1977;110:537–568. doi: 10.1016/s0022-2836(77)80111-3. [DOI] [PubMed] [Google Scholar]
- 12.Loh S N, Kay M S, Baldwin R L. Proc Natl Acad Sci USA. 1995;92:5446–5450. doi: 10.1073/pnas.92.12.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Springer B A, Sligar S G. Proc Natl Acad Sci USA. 1987;84:8961–8965. doi: 10.1073/pnas.84.24.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santoro M M, Bolen D W. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 15.Luo Y, Kay M S, Baldwin R L. Nat Struct Biol. 1997;4:925–930. doi: 10.1038/nsb1197-925. [DOI] [PubMed] [Google Scholar]
- 16.Serrano L, Kellis J T, Jr, Cann P, Matouschek A, Fersht A R. J Mol Biol. 1992;224:783–804. doi: 10.1016/0022-2836(92)90562-x. [DOI] [PubMed] [Google Scholar]
- 17.Hubbard S J, Thornton J M. naccess. University College, London: Department of Biochemistry and Molecular Biology; 1993. [Google Scholar]
- 18.Hughson F M, Baldwin R L. Biochemistry. 1989;28:4415–4422. doi: 10.1021/bi00436a044. [DOI] [PubMed] [Google Scholar]
- 19.Chakrabartty A, Baldwin R L. Adv Protein Chem. 1995;46:141–176. [PubMed] [Google Scholar]
- 20.Marmorino J L, Lehti M, Pielak G J. J Mol Biol. 1998;275:379–388. doi: 10.1006/jmbi.1997.1450. [DOI] [PubMed] [Google Scholar]
- 21.Wu L C, Kim P S. J Mol Biol. 1998;280:175–182. doi: 10.1006/jmbi.1998.1825. [DOI] [PubMed] [Google Scholar]
- 22.Song J, Bai P, Luo L, Peng Z Y. J Mol Biol. 1998;280:167–174. doi: 10.1006/jmbi.1998.1826. [DOI] [PubMed] [Google Scholar]
- 23.Colon W, Elove G A, Wakem L P, Sherman F, Roder H. Biochemistry. 1996;35:5538–5549. doi: 10.1021/bi960052u. [DOI] [PubMed] [Google Scholar]