New crystal structures of the well studied protein-folding model system villin headpiece in a new space group provide insights into the conformations available in the folded state.
Keywords: villin-type headpiece domain, protein folding, native state
Abstract
The villin headpiece domain (HP67) is the C-terminal F-actin-binding motif that confers F-actin-bundling activity to villin, a component of the actin bundles that support the brush-border microvilli. It has been investigated extensively by both experimental and theoretical measurements. Our laboratory, for example, has determined both its NMR and its crystal structures. This study presents the structures of HP67 and its pH-stabilized mutant (H41Y) in a different crystal form and space group. For both constructs, two molecules are found in each asymmetric unit in the new space group P61. While one of the two structures (Mol A) is structurally similar to our previously determined structure (Mol X), the other (Mol B) has significant deviations, especially in the N-terminal subdomain, where lattice contacts do not appear to contribute to the difference. In addition, the structurally most different crystal structure, Mol B, is actually closer to the averaged NMR structure. Harmonic motions, as suggested by the B-factor profiles, differ between these crystal structures; crystal structures from the same space group share a similar pattern. Thus, heterogeneity and dynamics are observed in different crystal structures of the same protein even for a protein as small as villin headpiece.
1. Introduction
Protein structures are the underlying basis for protein functions. Two techniques have been widely used to determine protein structures: X-ray crystallography and nuclear magnetic resonance (NMR; Branden & Tooze, 1991 ▶). However, in some cases NMR and crystal structures do not exactly agree with each other (Fan & Mark, 2003 ▶), especially for mobile loops and surface residues (Billeter et al., 1992 ▶). Sometimes, divergence is even observed within crystal structures determined under slightly different conditions (Andrec et al., 2007 ▶), counselling caution against over-interpreting structural data (DePristo et al., 2004 ▶).
In addition to the structure, protein dynamics or flexibility, especially adjacent to the active sites, is essential for protein function (Mittermaier & Kay, 2006 ▶). Dynamics can be measured rigorously by NMR relaxation experiments (Kay et al., 1989 ▶; Palmer et al., 2001 ▶) and approximated by molecular-dynamics (MD) simulations. In the absence of NMR order parameters or MD simulation data, the structures themselves may shed light on protein mobility. For crystal structures, elevated thermal factors (B factors) give indications of the inherent mobility of the protein (Halle, 2002 ▶; Best et al., 2006 ▶).
We have previously determined the crystal structures of villin headpiece (HP67) and its pH-stabilized mutant H41Y using microseeding as the optimization step in crystallization (Meng & McKnight, 2008 ▶). Both HP67 and H41Y crystallized in space group P212121 and only one molecule was found in each asymmetric unit. Despite the differences in pH, thermal and chemical stability (Grey et al., 2006 ▶; Tang et al., 2006 ▶; Meng & McKnight, 2008 ▶), HP67 and H41Y have almost identical structures. The structure is composed of three α-helices within the C-terminal subdomain and several short α-helices or 310-helices from the N-terminal subdomain connected by loops and turns.
In this study, we present a new crystal form of villin headpiece and its pH-stabilized mutant H41Y. Two molecules are found in each asymmetric unit in the new space group P61. While similar to our previously published structure overall (Meng & McKnight, 2008 ▶; Meng et al., 2005 ▶), substantial deviations are observed in the N-terminal subdomain in one crystal form, where lattice interactions play a minor role in ordering the structure. We propose that this new crystal form traps substates within the native structure ensemble that are not favored under the previously identified condition (Meng & McKnight, 2008 ▶; Meng et al., 2005 ▶). Interestingly, this crystal form is more easily obtained for the more stable point mutant H41Y, which has been reported to exhibit decreased exchange with the aqueous environment (Meng & McKnight, 2008 ▶; Tang et al., 2006 ▶; Grey et al., 2006 ▶). Thus, slight modifications to the kinetics of the protein are likely to shift the equilibrium towards otherwise disfavored substates within the native structure ensemble.
2. Materials and methods
2.1. Crystallization
Wild-type HP67 and the H41Y mutant were expressed and purified as described previously (Meng & McKnight, 2008 ▶). Initial crystallization conditions were identified using a sparse-matrix crystal screen (Crystal Screen, Hampton Research, Laguna Niguel, California, USA). Hanging drops consisting of 2 µl protein stock solution (30 mg ml−1 in pure water) and 2 µl reservoir solution were equilibrated against 0.5 ml reservoir solution consisting of 0.2 M ammonium sulfate, 0.1 M 2-morpholinoethanesulfonic acid pH 6.5 and 30% polyethylene glycol (PEG) 8000. Crystallization was optimized by using 4 µl protein stock solution (30 mg ml−1 in pure water) with 4 µl reservoir solution in the hanging drop. Conical or bullet-shaped crystals with six faces grew at room temperature to a maximum dimension of ∼200 µm in 2–3 d for H41Y and in 1–2 weeks for HP67. The current and previous crystallization conditions (Meng & McKnight, 2008 ▶) are listed and compared in Table 1 ▶.
Table 1. Crystallization conditions for villin headpiece constructs.
| Construct | PDB code | Molecules | Space group | Protein stock (mgml1) | Precipitant | Seeding | Matthews coefficient (3Da1) | Solvent content (%) |
|---|---|---|---|---|---|---|---|---|
| HP67 | 2rjy † | Mol X | P212121 | 15 | 25% PEG 8000 | Yes | 2.06 | 40.32 |
| 2rjx | Mol A, Mol B | P61 | 30 | 30% PEG 8000 | No | 2.68 | 54.17 | |
| H41Y | 2rjv † | Mol X | P212121 | 15 | 25% PEG 8000 | Yes | 2.08 | 40.89 |
| 2rjw | Mol A, Mol B | P61 | 30 | 30% PEG 8000 | No | 2.72 | 54.85 |
Structures from Meng McKnight (2008 ▶).
2.2. X-ray diffraction data collection and structure determination
The crystals were cryoprotected by soaking sequentially in reservoir solutions containing 10% glycerol followed by 20% glycerol for 3 min each. The crystals were then immediately frozen under N2 vapor at 95 K. Diffraction data were collected using an R-AXIS IV (Boston University School of Medicine). The data were processed with the HKL suite (HKL Research Inc.). The structures were determined by molecular replacement using the first crystal structure of HP67 (PDB code 1yu5; Meng et al., 2005 ▶), which was crystallized from high salt (not from high concentrations of PEG 8000 as discussed here), as the template. Electron-density maps were calculated at 1.7 and 1.55 Å for HP67 and H41Y, respectively. The structures were modelled using O (Jones et al., 1991 ▶) and refined using torsional dynamics and the maximum-likelihood target function in CNS (Brünger et al., 1998 ▶). The two molecules in the asymmetric unit were refined separately for both HP67 and H41Y. The structural statistics are summarized in Table 2 ▶.
Table 2. Statistics of HP67 and H41Y X-ray structure determination.
Values in parentheses are for the outer shell.
| HP67 | H41Y | |
|---|---|---|
| X-ray source | R-AXIS IV | R-AXIS IV |
| Space group | P61 | P61 |
| Unit-cell parameters (, ) | a = b = 64.278, c = 68.422, = = 90, = 120 | a = b = 65.408, c = 67.291, = = 90, = 120 |
| Resolution limit () | 1.70 | 1.55 |
| No. of reflections (total/unique) | 156325/17717 (15492/1761) | 206751/23793 (20027/2374) |
| Redundancy | 8.8 (8.8) | 8.7 (8.4) |
| R merge † (%) | 3.8 (28.9) | 4.7 (31.4) |
| Completeness (%) | 99.7 (100) | 99.8 (100) |
| I/(I) | 59.0 (8.5) | 47.2 (7.1) |
| Parameters for molecular replacement | ||
| Correlation coefficient | 0.349 | 0.352 |
| R factor (%) | 24.9 | 23.7 |
| Statistics of model refinement | ||
| Non-H atoms | ||
| Protein atoms | 1051 | 1078 |
| Waters | 111 | 175 |
| R cryst ‡ (%) | 21.5 | 20.2 |
| R free § (%) | 24.0 | 22.9 |
| R.m.s. deviations | ||
| Bond lengths () | 0.008 | 0.008 |
| Bond angles () | 1.25 | 1.30 |
| Averaged B values (2) | ||
| Main chain | 27.12 | 20.88 |
| Side chain | 29.43 | 23.47 |
| Waters | 34.37 | 29.79 |
| Ramachandran plot (%) | ||
| Most favored | 96.5 | 96.6 |
| Additionally allowed | 3.5 | 3.4 |
| Disallowed | 0 | 0 |
R
merge = 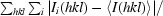
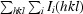 for the intensity (I) of i observations of reflection hkl.
for the intensity (I) of i observations of reflection hkl.
R
cryst = 
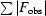 , where F
obs and F
calcare the observed and calculated structure-factor amplitudes, respectively.
, where F
obs and F
calcare the observed and calculated structure-factor amplitudes, respectively.
R free was calculated as R cryst but with 8% of amplitudes chosen randomly and omitted from the start of refinement.
3. Results
3.1. New crystal forms were obtained for both HP67 and H41Y
By omitting microseeds from the hanging drop (Meng & McKnight, 2008 ▶), we crystallized HP67 and its pH-stabilized mutant H41Y in a new space group, P61. Two molecules were found in each asymmetric unit and most of the interface contained water molecules (Fig. 1 ▶). These two molecules are not a true dimer because only monomers were found in the solution state by gel-filtration chromatography for both HP67 and H41Y (data not shown). We define these two molecules as molecule A (Mol A) and molecule B (Mol B) in order to distinguish them from the published structures, which we term molecule X (Mol X). A difference was also observed in the crystallization rate between the two constructs: H41Y crystallized much faster than HP67 under the current conditions, whereas the reverse was true in the presence of microseeds (Meng & McKnight, 2008 ▶).
Figure 1.
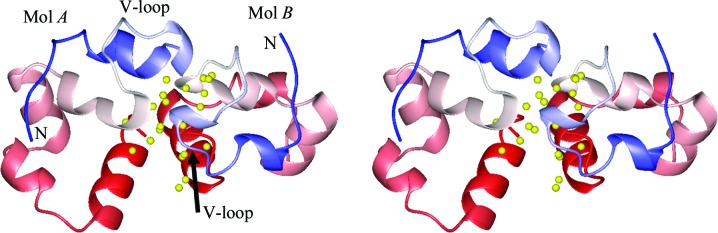
Stereoview of the two molecules of HP67 in the asymmetric unit of space group P61 crystals. Both Mol A (left) and Mol B (right) are shown in a backbone ribbon representation for the HP67 structure. The backbone is colored from blue at the respective N-termini though white in the center of the sequence to red at the C-termini. Selected water molecules at the interface between Mol A and Mol B are shown as yellow spheres. The graphic was created by MOLMOL (Koradi et al., 1996 ▶).
3.2. Overall structural features of Mol A and Mol B
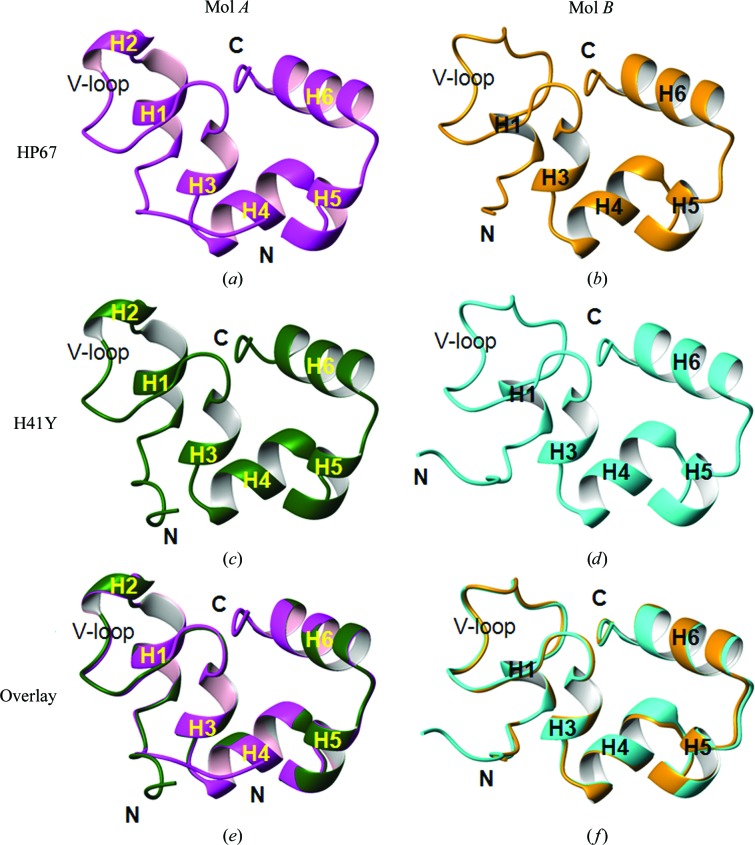
Since Mol A and Mol B are not a true dimer, they are drawn separately to characterize their major structural features (Fig. 2 ▶). The secondary structures shown in Fig. 2 ▶ were calculated using STRIDE (Heinig & Frishman, 2004 ▶). Similar to our published structures (Mol X; Fig. 3 ▶ a; Meng & McKnight, 2008 ▶), Mol A of both HP67 (Fig. 2 ▶ a) and H41Y (Fig. 2 ▶ c) contains three long α-helices in the C-terminal subdomain and several short α-helices or 310-helices in the N-terminal subdomain. While similarly structured in the C-terminal subdomain, Mol B of both HP67 (Fig. 2 ▶ b) and H41Y (Fig. 2 ▶ d) has less canonical secondary structure in the N-terminal subdomain. Specifically, the 310-helix (H2) is lost and the α-helix (H1) is converted into a shorter 310-helix in both HP67 and H41Y.
Figure 2.
The alternative conformation of HP67 and H41Y in space group P61. Mol A (a, c) and Mol B (b, d) of HP67 (a, b) and H41Y (c, d) are shown as ribbon representations, with the structural elements labelled. Mol A of HP67 and H41Y are overlaid in (e) from residues 13 to 76, omitting the mutated residue 41; Mol B of HP67 and H41Y are overlaid in (f). Magenta, HP67 Mol A; orange, HP67 Mol B; green, H41Y Mol A; cyan, H41Y Mol B. Graphics were created by MOLMOL (Koradi et al., 1996 ▶).
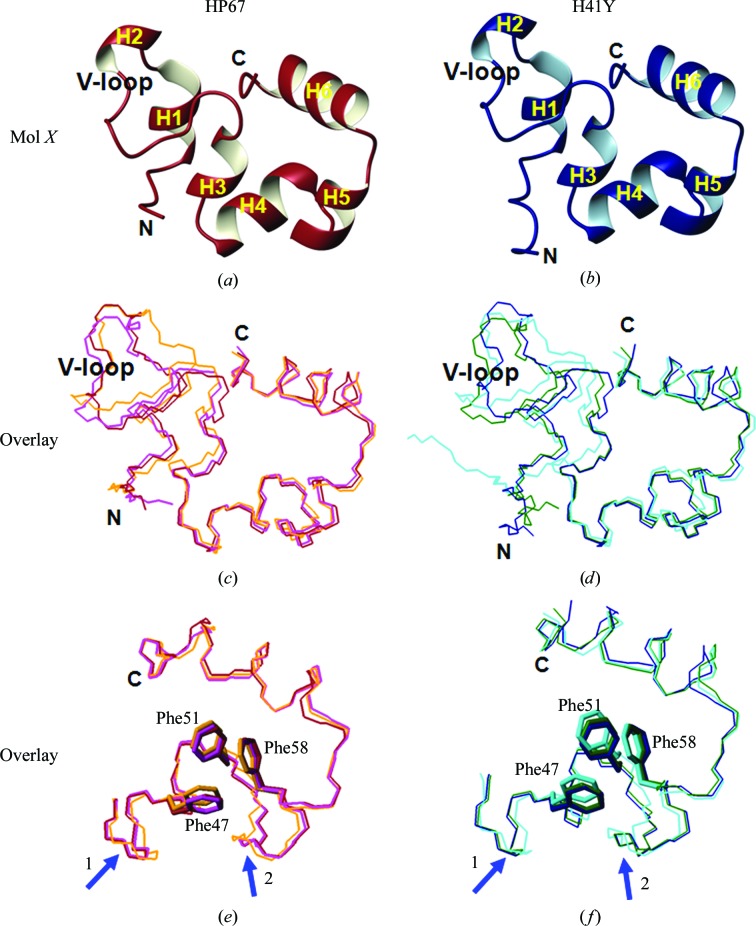
Figure 3.
Comparisons of crystal structures within the same construct. Ribbon views of our previously determined crystal structures (Mol X) of HP67 (red) and H41Y (blue) are shown in (a) and (b). The three crystal structures of HP67 are overlaid for the entire sequence in (c) and for the C-terminal subdomain in (e); the three crystal structures of H41Y are overlaid for the entire sequence in (d) and for the C-terminal subdomain in (f), with Phe47, Phe51 and Phe58 labelled. Blue arrows indicate variations in the alignment. Red, HP67 Mol X; magenta, HP67 Mol A; orange, HP67 Mol B; blue, H41Y Mol X; green, H41Y Mol A; cyan, H41Y Mol B. Graphics were created by MOLMOL (Koradi et al., 1996 ▶).
When aligned, the same types of structures from the two different constructs, i.e. Mol A of HP67 and H41Y or Mol B of HP67 and H41Y, overlay with each other well (Figs. 2 ▶ e and 2 ▶ f). Omitting the mutated residue, the root-mean-square deviations (r.m.s.d.s) for backbone heavy atoms are only 0.328 Å for Mol A between HP67 and H41Y and 0.371 Å for Mol B between HP67 and H41Y (Table 3 ▶). We have previously shown that Mol X of H41Y is very similar to Mol X of HP67 (Meng & McKnight, 2008 ▶), with a backbone r.m.s.d. of 0.405 Å (Table 3 ▶). Thus, the same types of structures, Mol A, Mol B or Mol X, are nearly identical between the two constructs.
Table 3. Pairwise backbone r.m.s.d. comparison between the HP67 and H41Y crystal structures.
R.m.s.d.s are for the backbone heavy atoms of the residues indicated in parentheses. Values above the diagonal are for the entire HP67 [HP67 (1276)]. Values below the diagonal are for the HP35 subdomain [HP35 (4276)].
| HP67 | H41Y | HP67 | ||||||
|---|---|---|---|---|---|---|---|---|
| Mol X | Mol A | Mol B | Mol X | Mol A | Mol B | NMR | ||
| HP67 | Mol X | 0.790 | 1.044 | 0.405 | 0.751 | 1.224 | 1.777 | |
| Mol A | 0.456 | 1.225 | 0.869 | 0.328 | 1.378 | 1.736 | ||
| Mol B | 0.587 | 0.563 | 1.165 | 1.201 | 0.371 | 1.218 | ||
| H41Y | Mol X | 0.202 | 0.482 | 0.644 | 0.798 | 1.321 | 1.915 | |
| Mol A | 0.479 | 0.120 | 0.601 | 0.479 | 1.343 | 1.766 | ||
| Mol B | 0.697 | 0.557 | 0.292 | 0.742 | 0.620 | 1.218 | ||
| HP67 | NMR | 1.247 | 0.950 | 1.030 | 1.224 | 0.981 | 0.896 | |
3.3. Mol B is the most different of the three crystal structures
When compared with Mol X, our previously determined crystal structure (Figs. 3 ▶ a and 3 ▶ b; Meng & McKnight, 2008 ▶), the overall traces are quite similar (Figs. 3 ▶ c and 3 ▶ d). However, significant differences are seen in the N-terminal subdomain. Correspondingly, the r.m.s.d.s between Mol B and Mol A/Mol X are higher (Table 3 ▶). Differences are also observed in the more conserved C-terminal subdomain (Figs. 3 ▶ e and 3 ▶ f, Table 3 ▶), as reflected in the reorganization of one or more of the three key aromatic residues (Frank et al., 2002 ▶), Phe47, Phe51 and Phe58, in Mol B. Thus, Mol B is the most distinctive structure among the three crystal structures within the same construct.
3.4. Lattice contacts do not account for the different structural types
Lattice contacts were mapped for each structure (Fig. 4 ▶) to address whether the differences observed in the crystal structures arise from crystal packing. Substantial deviations are observed between Mol B of HP67 and H41Y (Fig. 4 ▶). In addition, all three structures of H41Y have more contacts in the extreme N-terminus than those of HP67 (Fig. 4 ▶). The fact that Mol A and Mol B crystallized in the same space group under identical conditions further indicates that lattice interaction is not the driving force in ordering the structures. Thus, Mol A and Mol B are likely to represent substates inherent in the native structure ensemble in solution that are selectively favoured by the current crystallization condition.
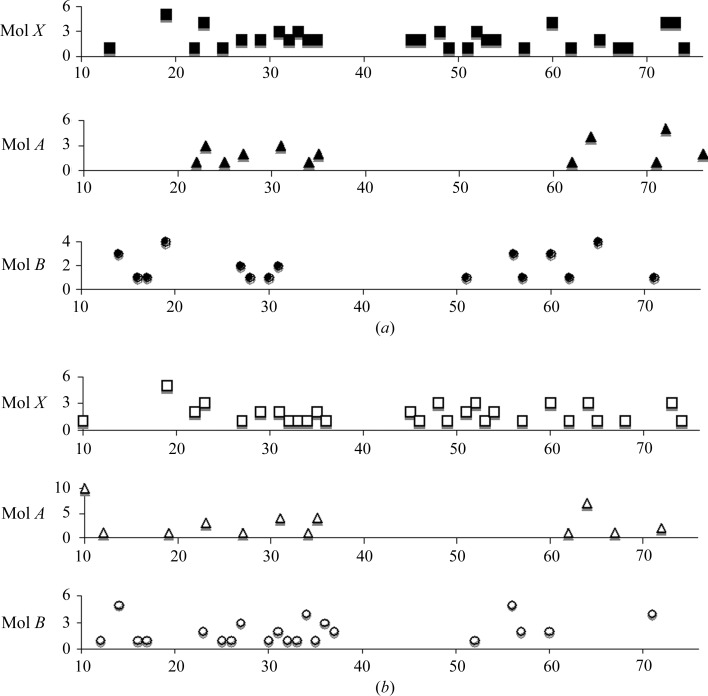
Figure 4.
Lattice contacts in the crystal structures of HP67 and H41Y. Lattice contacts for Mol X (filled squares), Mol A (filled triangles) and Mol B (filled circles) of HP67 are shown in (a) and lattice contacts for Mol X (open squares), Mol A (open triangles) and Mol B (open circles) of H41Y are shown in (b). A distance of 3.5 Å was used as the cutoff in calculating lattice contacts.
3.5. Mol B is closest to the NMR structure
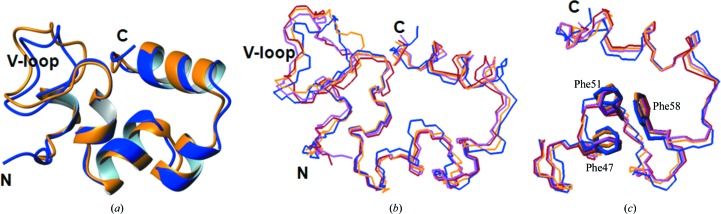
When compared with the solution NMR structure (Vardar et al., 1999 ▶), Mol B overlays best with the averaged NMR structure (Figs. 5 ▶ a–5 ▶ c; Table 3 ▶). Thus, Mol B is the closest to the NMR structure of the three crystal forms.
Figure 5.
Comparisons of the crystal structures with the NMR structure of HP67. Mol B of HP67 (orange) is overlaid with the averaged NMR structure (blue) in (a) in ribbon representation. The backbones of Mol X (red), Mol A (magenta), Mol B (orange) and the averaged NMR structure (blue) are aligned in (b) for the entire sequence and in (c) for the C-terminal subdomain, with Phe47, Phe51 and Phe58 labelled. Graphics were created by MOLMOL (Koradi et al., 1996 ▶).
3.6. Comparison of the thermal factors of the crystal structures
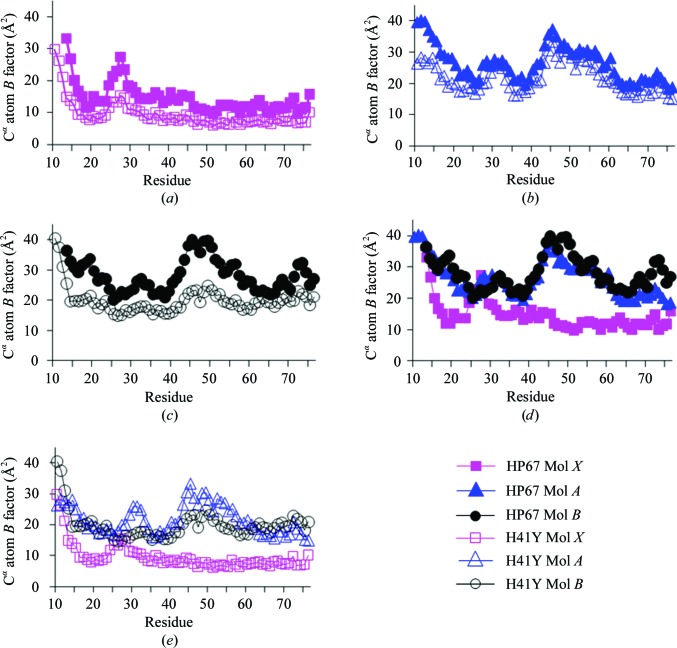
The B factors of crystal structures shed light on harmonic motions of the protein (Halle, 2002 ▶). To address whether the different structures represent substates with different dynamic profiles, we compared the B factors of these crystal structures (Fig. 6 ▶). The B-factor profiles are consistent within the same types of structures (Figs. 6 ▶ a, 6 ▶ b and 6 ▶ c). In addition, the B-factor profiles are quite similar between Mol A and Mol B for each construct (Figs. 6 ▶ d and 6 ▶ e), which have distinctive lattice contacts within the same space group, indicating that crystal-packing forces only play a minimal role in B-factor profiling. Thus, the B-factor profiles of these crystal structures are likely to represent true motions within the crystalline environment.
Figure 6.
Comparisons of the B factors of the crystal structures. The B factors of Cα atoms for Mol X, Mol A and Mol B are shown in (a), (b) and (c), respectively (filled symbols for HP67 and open symbols for H41Y). The B factors of Cα atoms for Mol X, Mol A and Mol B are shown in (d) for HP67 and (e) for H41Y (squares, Mol X; triangles, Mol A; circles, Mol B).
What is consistent among all these crystal structures is the extreme N-terminus, for which all data sets suggest high mobility. This mobility is also seen in NMR spin-relaxation studies, where the extreme N-terminus (residues 10–16) and residues in the V-loop exhibit enhanced flexibility in wild-type HP67 and the H41Y mutant (Grey et al., 2006 ▶).
4. Discussion
Proteins are dynamic and heterogeneous in nature, whether in the solution state or in the crystalline environment (Frauenfelder et al., 1991 ▶; DePristo et al., 2004 ▶). Heterogeneity is not usually captured in crystal structures because the diffraction patterns of most proteins do not reach a sufficiently high resolution to identify and model heterogeneity (DePristo et al., 2004 ▶). Alternative crystal conformations from different crystallization conditions, on the other hand, partially ameliorate this deficiency (DePristo et al., 2004 ▶). By omitting microseeding from the optimization step (Meng & McKnight, 2008 ▶; Meng et al., 2005 ▶), we were able to crystallize HP67 and its point mutant H41Y in a different crystal form in a new space group, P61, and thus to address the conformational heterogeneity of villin headpiece.
Unlike in our previously identified conditions, two molecules, Mol A and Mol B, were found in each asymmetric unit in the structures reported here. These structures were compared with the structures of HP67 and H41Y in space group P212121 (Mol X; Meng & McKnight, 2008 ▶) to identify similarities and differences within these structures. Consistent with our previous findings, high structural similarity is observed between HP67 and H41Y within the same structural type, thus confirming that the H41Y substitution has little impact on the structure. Although Mol A is similar to Mol X, Mol B is structurally more different, especially in the N-terminal subdomain. The basic question is whether these different structures reflect heterogeneity inherent in the native structure ensemble or are merely artefacts or byproducts of crystal packing.
Lattice interactions do not appear to play a key role in ordering these structures. Except for the extreme N- and C-termini, Mol A (from space group P61) makes far fewer contacts than Mol X (from space group P212121), yet Mol A adopts a conformation similar to Mol X. The lattice contacts differ significantly between Mol B of HP67 and Mol B of H41Y, yet their final conformations are almost identical. Therefore, the different crystal structures are likely to represent substates within the native structure ensemble in solution that are selectively favored by specific crystallization conditions.
In addition to providing structural information, both the crystal and the NMR structures shed light on inherent mobility. The B factors of crystal structures, for example, give indications of the harmonic motions in the crystal lattice. Interestingly, the B-factor profiles for structures in the same space group are very similar despite differences in coordinates and lattice contacts: although Mol A is structurally more similar to Mol X, dynamically it is closer to Mol B. What is consistent within all structures are the elevated B factors in the N-terminus, indicating that it is flexible. The flexibility of the N-terminus has also been observed in solution using NMR-relaxation measurements (Grey et al., 2006 ▶) and in molecular-dynamics simulations (Khandogin et al., 2007 ▶).
A difference is observed in the crystallization rate and space-group preference for HP67 and H41Y. HP67 crystallizes within hours after seeding into crystals of space group P212121. However, it takes 1–2 weeks for HP67 to crystallize in the absence of seeds in space group P61 and high-quality crystals are rare. In contrast, diffraction-quality H41Y crystals form in 2–3 d without microseeding in space group P61. Microseeding results in P212121 space-group crystals, but crystallization slows to 1–2 weeks and there are fewer diffraction-quality crystals. Considering there is only one residue that differs between HP67 and H41Y, where the point mutation disrupts only localized interactions with residue 41 (Meng & McKnight, 2008 ▶), it is hard to imagine that the resulting increase in stability from the mutation would contribute to such a difference (Tang et al., 2006 ▶; Grey et al., 2006 ▶). It is more likely that the resulting increase in the kinetics of the N-terminal subdomain introduced by the mutation (Grey et al., 2006 ▶) enhances the interchange between substates within the native structure ensemble, favors alternative conformations with different dynamic properties and, if captured by the right crystallization condition, pushes the equilibrium toward the alternative conformations. In turn, these alternative conformations provide a more comprehensive and accurate view of the native structure ensemble and reveal the heterogeneous nature of native protein structures, even for proteins as small as villin headpiece.
Supplementary Material
PDB reference: HP67, 2rjx, r2rjxsf
PDB reference: H41Y, 2rjw, r2rjwsf
Acknowledgments
This work was supported by National Institutes of Health grant GM62886 to CJM.
References
- Andrec, M., Snyder, D. A., Zhou, Z., Young, J., Montelione, G. T. & Levy, R. M. (2007). Proteins, 69, 449–465. [DOI] [PubMed]
- Best, R. B., Lindorff-Larsen, K., DePristo, M. A. & Vendruscolo, M. (2006). Proc. Natl Acad. Sci. USA, 103, 10901–10906. [DOI] [PMC free article] [PubMed]
- Billeter, M., Vendrell, J., Wider, G., Aviles, F. X., Coll, M., Guasch, A., Huber, R. & Wuthrich, K. (1992). J. Biomol. NMR, 2, 1–10. [DOI] [PubMed]
- Branden, C. & Tooze, J. (1991). Introduction to Protein Structure. New York: Garland Publishing.
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- DePristo, M. A., de Bakker, P. I. & Blundell, T. L. (2004). Structure, 12, 831–838. [DOI] [PubMed]
- Fan, H. & Mark, A. E. (2003). Proteins, 53, 111–120. [DOI] [PubMed]
- Frank, B. S., Vardar, D., Buckley, D. A. & McKnight, C. J. (2002). Protein Sci. 11, 680–687. [DOI] [PMC free article] [PubMed]
- Frauenfelder, H., Sligar, S. G. & Wolynes, P. G. (1991). Science, 25, 1598–1603. [DOI] [PubMed]
- Grey, M. J., Tang, Y., Alexov, E., McKnight, C. J., Raleigh, D. P. & Palmer, A. G. III (2006). J. Mol. Biol. 355, 1078–1094. [DOI] [PubMed]
- Halle, B. (2002). Proc. Natl Acad. Sci. USA, 99, 1274–1279.
- Heinig, M. & Frishman, D. (2004). Nucleic Acids Res. 32, W500–W502. [DOI] [PMC free article] [PubMed]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed]
- Kay, L. E., Torchia, D. A. & Bax, A. (1989). Biochemistry, 28, 8972–8979. [DOI] [PubMed]
- Khandogin, J., Raleigh, D. P. & Brooks, C. L. III (2007). J. Am. Chem. Soc. 129, 3056–3057. [DOI] [PMC free article] [PubMed]
- Koradi, R., Billeter, M. & Wüthrich, K. (1996). J. Mol. Graph. 14, 51–55. [DOI] [PubMed]
- Meng, J. & McKnight, C. J. (2008). Biochemistry, 47, 4644–4650. [DOI] [PubMed]
- Meng, J., Vardar, D., Wang, Y., Guo, H. C., Head, J. F. & McKnight, C. J. (2005). Biochemistry, 44, 11963–11973. [DOI] [PubMed]
- Mittermaier, A. & Kay, L. E. (2006). Science, 312, 224–228. [DOI] [PubMed]
- Palmer, A. G., 3rd, Kroenke, C. D. & Loria, J. P. (2001). Methods Enzymol. 339, 204–238. [DOI] [PubMed]
- Tang, Y., Grey, M. J., McKnight, J., Palmer, A. G., 3rd & Raleigh, D. P. (2006). J. Mol. Biol. 355, 1066–1077. [DOI] [PubMed]
- Vardar, D., Buckley, D. A., Frank, B. S. & McKnight, C. J. (1999). J. Mol. Biol. 294, 1299–1310. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: HP67, 2rjx, r2rjxsf
PDB reference: H41Y, 2rjw, r2rjwsf