Abstract
Previously, we reported evidence of genetic heterogeneity in late-onset familial Alzheimer’s disease, based on sex of affected parent, demonstrating linkage to chromosome 10q, a region identified by other groups and implicated as a quantitative trait loci for Aβ levels, in families with an affected mother. Using functional magnetic resonance imaging and a memory encoding task, we now show differential brain activation patterns among asymptomatic offspring which correspond to the previous linkage finding. These results suggest the possibility that activation patterns may prove useful as a preclinical quantitative trait related to the putative familial late-onset AD gene in this chromosome 10 region.
Although several causative genetic mutations have been identified in early-onset Alzheimer’s disease (AD), no genetic mutation with a Mendelian pattern of transmission has yet been identified in late-onset AD, which represents more than 90% of AD cases. Recent genomewide scans in samples of late-onset familial AD cases have reported linkage for several chromosomal regions, with, in general, little agreement between studies.1–5 Lack of consistent findings is a problem that plagues the search for causative genes in many complex adult-onset disorders. These inconsistencies may reflect differences in the samples, study designs, or analytic techniques. They may also reflect heterogeneity within the samples or more complicated genetic mechanisms such as the interplay of mutation and epigenetic control. Our research team has focused on using phenotypic information to identify more homogeneous groups for linkage analysis in our families, a subset of the National Institute of Mental Health AD Genetics Initiative sample. Recently, we examined parental affection status and reported evidence of linkage to chromosome 10, approaching genomewide significance (nonparametric logarithm of odds score = 3.27), when only families with affected mothers were analyzed (N = 49). This maternal effect was shown to be statistically significant via multiple random stratifications of the overall data set. In contrast, families with affected fathers were not linked to this region.6 This chromosome 10 (c10) region, close to the centromere, is linked to late-onset AD in several studies and implicated as a quantitative trait loci (QTL) for Aβ levels.7,8
Because the disease starts perhaps decades before the onset of clinical symptoms, functional brain imaging, especially of the offspring of AD cases, provides a method of studying brain changes among those at-risk but as yet unaffected. A few studies of asymptomatic carriers of the early-onset AD PS1 and APP mutations have shown altered patterns of metabolic and hemodynamic response on positron emission tomography (PET) and single-proton emission computed tomography (SPECT) imaging distinct from those seen in diagnosed cases.9 –11 Johnson and colleagues examined a large extended family whose affected members carry the 280 PS1 mutation.11 The asymptomatic carriers displayed decreased perfusion compared with noncarriers, with decreases noted in the hippocampal complex, cingulate, posterior parietal, and superior frontal lobes. In contrast, the family members who had developed AD showed reduced perfusion only in the posterior parietal and superior frontal cortex. Wahlund and Perani each examined a family whose affected members carried mutations of the APP gene. In asymptomatic carriers of the 670/671 mutation, there was reduced glucose metabolism in the right temporal lobe when compared with the noncarriers,10 whereas, asymptomatic carriers from a family with the 717 mutation showed reductions in the thalamus and frontal lobe,9 potentially indicating differential patterns for particular mutations.
Changes in brain activation in these gene-positive asymptomatic individuals are perhaps not surprising given that the disease starts decades before the appearance of clinical symptoms. However, it is unknown whether such changes are evident in individuals from late-onset families with significant linkage findings. We were interested in knowing whether unaffected offspring of autopsy-confirmed cases, drawn from these families, would exhibit phenotypic differences based on the heterogeneity indicated in our linkage findings. Thus, we used functional magnetic resonance imaging (fMRI) to explore differences in brain activation patterns during memory encoding, a task compromised early in AD, contrasting those offspring from families linked to chromosome 10q with those from families unlinked to this region. Such information could be extremely valuable as we try to understand the biological significance of this putative AD gene on chromosome 10.
Participants included female offspring of autopsy-confirmed familial AD cases, nine from families linked to the 10q region and six from families unlinked to this region. All were apolipoprotein E (ApoE) ε4-negative, right-handed, and free of memory complaints and performed within the normal range on cognitive testing. The two groups did not differ in age (60 ± 8 vs 67 ± 5), estimated IQ (111 ± 6 vs 105 ± 10) or educational achievement (16 ± 4 vs 14 ± 4). These individuals were on average 11.3 years standard deviation, 7.8) from the onset age of their parent. The study was approved by the Johns Hopkins University Institutional Review Board, and all participants provided informed consent. Individuals were administered a verbal word-pair memory test both before and after fMRI scanning.
The fMRI paradigm was a modified block design with six repetitions of intervals in which seven word pairs were presented for memorization (encoding), followed by presentation of the first word of the word pair with the subject told to internally speak the second word of the pair, followed by a rest period (baseline). The 6-minute and 10-second paradigm was repeated twice using two distinct word-pair sets. Scanning was performed on a Tesla Phillips Gyroscan ACS-NT scanner (Phillips Medical Systems, Endhoven, The Netherlands), acquiring two functional scans using echo planar imaging (EPI) and a blood oxygenation level dependent (BOLD) technique with TR, 1,000 milliseconds; TE, 39 milliseconds; flip angle, 90 degrees, field-of-view, 230mm, and matrix size, 64 × 64. Eighteen coronal slices were acquired with a 4.5mm thickness and an interslice gap of 0.5mm, oriented perpendicular to the anterior commissure–posterior commissure line.
fMRI data were preprocessed and analyzed using Statistical Parametric Mapping (SPM) 99 within the framework of the General Linear Model. The individual time series was fit to each subject producing SPM[t] maps. Activation during encoding was examined in each group using one sample Student’s t tests. Group comparisons were conducted using two sample Student’s t tests to examine differences in brain regions that are more active when memorizing word pairs. Statistical testing was conducted using an uncorrected height threshold of p value less than 0.01 and a minimum cluster size of 20 voxels.
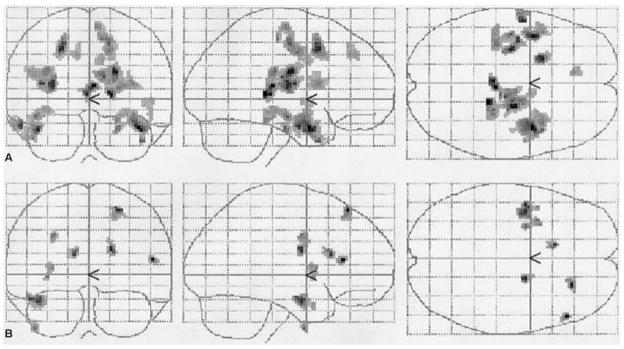
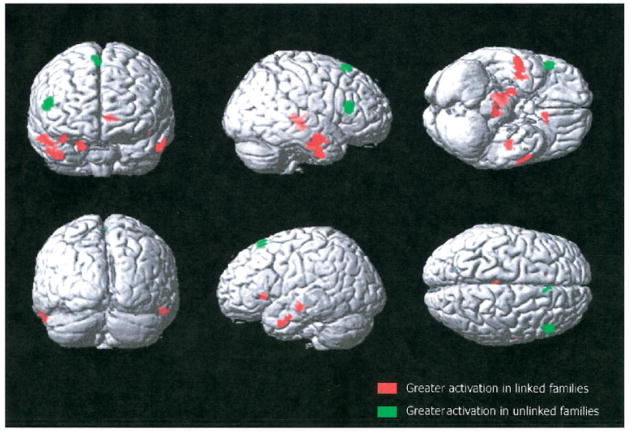
Although performance on word-pair recall both before scanning (12.5 ± 2.7 vs 10.8 ± 4.1) and immediately after scanning (3.6 ± 1.6 vs 2.8 ± 1.8) did not differ significantly, the fMRI results did show significantly different patterns of activation during encoding (Fig 1). Single group maps show that individuals from linked families had more extensive and bilateral temporal and frontal activation, whereas offspring from unlinked families showed only unilateral frontal and unilateral temporal activation. Group comparisons using a two-sample t test showed statistically significant differences in activation between the groups. More specifically, the thalamus and the temporal gyri as well as the right parahippocampal gyrus were more activated in the those from linked families (Table and Fig 2, red), whereas increased activation was found only for the right frontal lobe within the offspring from the unlinked families (see Table and Fig 2, green).
Fig 1.
Group activation maps for verbal encoding. The three images on top (A) represent the pattern of activation during verbal encoding in offspring from linked families, whereas the three images on the bottom (B) represent the activation in offspring from unlinked families.
Table.
Areas of Significant Activation Differences during Memory Encoding
| # | Talairach Coordinates |
||||||
|---|---|---|---|---|---|---|---|
| Cluster Size | Voxel T | x | y | z | L/R | Anatomical Label | Brodmann Area |
| c10 Linked > c10 Unlinked | |||||||
| 319 | 4.75 | 10 | −25 | 1 | R | Thalamus | — |
| 3.59 | 12 | −20 | −6 | R | Substantia nigra | — | |
| 3.58 | 14 | −31 | 7 | R | Thalamus: pulvinar | — | |
| 3.58 | 14 | −24 | −4 | R | Substantia nigra | — | |
| 3.47 | 16 | −33 | 5 | R | Thalamus: pulvinar | — | |
| 3.21 | −4 | −25 | 5 | L | Thalamus: pulvinar | — | |
| 3.12 | −6 | −23 | 1 | L | Thalamus | — | |
| 2.74 | 0 | −23 | 7 | L | Thalamus | — | |
| 135 | 3.83 | 50 | −3 | −30 | R | Inferior temporal gyrus | 20 |
| 3.59 | 57 | −9 | −18 | R | Inferior temporal gyrus | 21 | |
| 3.41 | 40 | −7 | −33 | R | Middle temporal gyrus | 21 | |
| 3.03 | 48 | −9 | −21 | R | Inferior temporal gyrus | 20 | |
| 2.79 | 46 | −7 | −25 | R | Fusiform gyrus | 20 | |
| 48 | 4.80 | 20 | −15 | −23 | R | Parahippocampal gyrus | 28 |
| 46 | 3.51 | −44 | −18 | −9 | L | Superior temporal gyrus | 22 |
| 35 | 3.50 | −8 | 21 | 1 | L | Caudate head | — |
| 2.84 | −16 | 23 | −3 | L | Caudate head | — | |
| 24 | 3.19 | −53 | 4 | −27 | L | Middle temporal gyrus | 21 |
| 3.18 | −55 | 3 | −25 | L | Middle temporal gyrus | 21 | |
| 3.05 | −57 | −1 | −25 | L | Middle temporal gyrus | 21 | |
| 2.95 | −61 | −7 | −18 | L | Inferior temporal gyrus | 21 | |
| 2.92 | −359 | −3 | −23 | L | Middle temporal gyrus | 21 | |
|
| |||||||
| c10 Unlinked > c10 Linked | |||||||
| 84 | 4.63 | 53 | 28 | 12 | R | Inferior frontal gyrus | 46 |
| 26 | 4.05 | 2 | 26 | 58 | R | Superior frontal gyrus | 6 |
| 3.73 | 4 | 28 | 54 | R | Superior frontal gyrus | 6 | |
| 3.01 | 8 | 24 | 60 | R | Superior frontal gyrus | 6 | |
Fig 2.
This is a representation of two contrasts of the functional magnetic resonance imaging activation during verbal encoding. There is increased activation in linked families in temporal areas (shown in red), whereas unlinked families show a very different pattern of activation, mainly localized in the frontal lobes (shown in green).
Our results show that when attempting to encode or learn nonrelated word pairs, the brain regions recruited for this effort differ drastically between offspring of families linked to a region on chromosome 10q and offspring of families unlinked to this region. Most significantly, offspring from c10-linked families, in addition to having more extensive activation, showed increased bilateral activation in the thalamus and temporal lobe, brain regions related to episodic memory encoding,12,13 whereas offspring from c10-unlinked families showed increased activation only in the right frontal lobe. These differential activation patterns were seen despite similar memory performance on these paired-associate tasks, both before and after scanning.
Several confounding factors, such as sex, handedness, or genetic predisposition at another genomic location, which might explain differences in fMRI activation patterns, have been controlled in this study. The sample was restricted to right-handed female offspring, with no ε4 alleles at the APOE locus. This exclusion addresses a previous report that ε4/ε4 homozygotes evidencing no memory problems showed decreased metabolism on PET in temporal/parietal regions similar to that seen in AD cases.14 Therefore, these potential confounders cannot explain the fMRI differences between these groups.
Previous imaging findings from asymptomatic early-onset mutation carriers have encouraged the concept that particular mutations may create a preclinical imaging signature. For example, individuals with the double APP mutation 670/6710 and those with the APP 717 mutations9 showed altered but different patterns of glucose metabolism on PET scanning. By analogy, our use of imaging phenotypes among asymptomatic at-risk offspring from late-onset families may also identify imaging signatures that correspond to the effects of particular genetic causes. With this in mind, genetic heterogeneity at the linkage level has been further documented at the preclinical imaging level, supporting the existence of actual genetic heterogeneity in these families and the importance of identifying the responsible gene in this region of c10.
It is possible the differential activity pattern by region between these asymptomatic offspring groups reflects underlying differences in cognitive strategies between c10-linked and unlinked families. For example, the lateral and medial temporal regions showing increased activation in the linked families have been shown to be activated with language-based episodic memory tasks in healthy controls, whereas the frontal regions activated in the unlinked families are more often activated in working memory tasks.12 The underlying genetic cause may have led to different regional limitations and subsequent coping strategies over time, such that different areas are recruited for the same task during midlife.
In conclusion, these imaging results provide initial evidence of functional brain differences related to genetic linkage findings and suggest the possibility that activation patterns may prove useful as a quantitative trait to examine genetic heterogeneity in late-onset familial AD. Although this finding is exciting for the further elucidation of this specific c10 gene and is the first to correlate linkage heterogeneity with preclinical brain function, these imaging results are not interpretable as biomarkers for AD per se, because all offspring were considered at-risk individuals, rather than members of the general population.
Acknowledgments
This work was supported by grants from the NIH (National Institute on Aging, R01 AG16324, R01 AG21804, S.S.B.).
References
- 1.Myers A, Wavrant De-Vrieze F, Holmans P, et al. Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet. 2002;114:235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- 2.Kehoe P, Wavrant-De Vrieze F, Crook R, et al. A full genome scan for late onset Alzheimer’s disease. Hum Mol Genet. 1999;8:237–245. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- 3.Pericak-Vance MA, Bass MP, Yamaoka LH, et al. Complete genomic screen in late-onset familial Alzheimer disease. Evidence for a new locus on chromosome 12. JAMA. 1997;278:1237–1241. [PubMed] [Google Scholar]
- 4.Pericak-Vance MA, Bass ML, Yamaoka LH, et al. Complete genomic screen in late-onset familial Alzheimer’s disease. Neurobiol Aging. 1998;19:S39–S42. doi: 10.1016/s0197-4580(98)00037-2. [DOI] [PubMed] [Google Scholar]
- 5.Blacker D, Bertram L, Saunders AJ, et al. Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 6.Bassett SS, Avramopoulos D, Fallin D. Evidence for parent of origin effect in late-onset Alzheimer disease. Am J Med Genet. 2002;114:679–686. doi: 10.1002/ajmg.10648. [DOI] [PubMed] [Google Scholar]
- 7.Lendon C, Craddock N. Susceptibility gene(s) for Alzheimer’s disease on chromosome 10. Trends Neurosci. 2001;24:557–559. doi: 10.1016/s0166-2236(00)01912-3. [DOI] [PubMed] [Google Scholar]
- 8.Ertekin-Taner N, Graff-Radford N, Younkin LH, et al. Linkage of plasma Abeta42 to a quantitative locus on chromosome 10 in late-onset Alzheimer’s disease pedigrees. Science. 2000;290:2303–2304. doi: 10.1126/science.290.5500.2303. [DOI] [PubMed] [Google Scholar]
- 9.Perani D, Grassi F, Sorbi S. PET studies in subjects from two Italian FAD families with APP717 Val to Ileu mutation. Eur J Neurol. 1997;4:214–220. [Google Scholar]
- 10.Wahlund LO, Basun H, Almkvist O, et al. A follow-up study of the family with the Swedish APP 670/671 Alzheimer’s disease mutation. Dement Geriatr Cogn Disord. 1999;10:526–533. doi: 10.1159/000017200. [DOI] [PubMed] [Google Scholar]
- 11.Johnson KA, Lopera F, Jones K, et al. Presenilin-1-associated abnormalities in regional cerebral perfusion. Neurology. 2001;56:1545–1551. doi: 10.1212/wnl.56.11.1545. [DOI] [PubMed] [Google Scholar]
- 12.Cabeza R, Nyberg L. Imaging cognition II: an empirical review of 275 PET and fMRI studies. J Cogn Neurosci. 2000;12:1–47. doi: 10.1162/08989290051137585. [DOI] [PubMed] [Google Scholar]
- 13.Van der Werf YD, Jolles J, Witter MP, Uylings HB. Contributions of thalamic nuclei to declarative memory functioning. Cortex. 2003;39:1047–1062. doi: 10.1016/s0010-9452(08)70877-3. [DOI] [PubMed] [Google Scholar]
- 14.Reiman EM, et al. Preclinical evidence of Alzheimer’s disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N Engl J Med. 1996;334:752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]