Abstract
The androgen receptor (AR) plays a key role in progression to incurable androgen-ablation resistant prostate cancer (PCA). We have identified three novel AR splice variants lacking the ligand binding domain (designated as AR3, AR4 and AR5) in hormone insensitive PCA cells. AR3, one of the major splice variants expressed in human prostate tissues, is constitutively active and its transcriptional activity is not regulated by androgens or antiandrogens. Immunohistochemistry analysis on tissue microarrays containing 429 human prostate tissue samples shows that AR3 is significantly upregulated during PCA progression and AR3 expression level is correlated with the risk of tumor recurrence after radical prostatectomy. Overexpression of AR3 confers ablation-independent growth of PCA cells while specific knock-down of AR3 expression (without altering AR level) in hormone resistant PCA cells attenuates their growth under androgen-depleted conditions in both cell culture and xenograft models, suggesting an indispensable role of AR3 in ablation-independent growth of PCA cells. Furthermore, AR3 may play a distinct yet essential role in ablation-independent growth through regulating a unique set of genes including AKT1, which are not regulated by the prototype AR. Our data suggest that aberrant expression of AR splice variants may be a novel mechanism underlying ablation-independence during PCA progression and AR3 may serve as a prognostic marker to predict patient outcome in response to hormonal therapy. Given that these novel AR splice variants are not inhibited by currently available anti-androgen drugs, development of new drugs targeting these AR isoforms may potentially be effective for treatment of ablation-resistant PCA.
INTRODUCTION
Androgen ablation therapy is one of the most common treatments for patients with advanced prostate cancer (PCA). However, the majority of PCA patients will eventually develop androgen-depletion-independent recurrent tumors that are resistant to currently available treatments (1). It has, therefore, become a focus of intensive study to understand the mechanisms underlying the transition to ablation-resistant PCA (2–4). The androgen receptor(AR) is primarily responsible for mediating the physiological effects of androgens by binding to specific DNA sequences, known as androgen-responsive-elements (AREs) (5, 6). Upon ligand binding, AR undergoes a conformational change and translocates into the nucleus, where it binds to specific AREs in the androgen-responsive genes and thereby modulates their expression (7, 8). Human AR gene is structurally composed of 8 exons and encodes a multi-domain protein including an N-terminal transactivation domain (NTD), a central DNA-binding domain (DBD), a hinge region and a C-terminal ligand-binding domain (LBD) (9, 10). The LBD appears to be dispensable for AR transcriptional activity as its deletion leads to constitutive activation of its transcriptional capability in reporter assays (11–14). However, it remains elusive whether such constitutively active AR isoform(s) are naturally expressed in the prostate gland. Several studies have been carried out to identify potential AR-regulated genes by microarray and ChIP-chip analyses and revealed a cell- and gene-specific transcriptional regulation by AR in prostate cells (15, 16).
A majority of ablation-resistant PCAs express AR and androgen-responsive genes, indicating that AR-signaling pathway is still functional under androgen-depleted conditions (17, 18). Several studies established an essential role for AR in both hormone-sensitive and ablation-resistant PCA (19, 20). The mechanisms underlying ablation-resistant AR-mediated signaling have yet to be fully elucidated. Mutation and amplification of AR gene, alterations in protein kinases, growth factors, nuclear receptor coactivators and steroid metabolism enzymes have all been proposed to modulate AR signaling and may, therefore, contribute to androgen-ablation resistance of prostate cancer (3, 18, 21–29). Another plausible hypothesis for activation of AR in the absence of hormones was proposed by Tepper et al (30). In their study, a mutant AR was identified in hormone-insensitive PCA cell line CWR22Rv1 that contains an in-frame tandem duplication of the exon 3 encoding the second zinc finger of the DNA-binding domain and this insertional mutation renders AR susceptible to the protease cleavage and generates a constitutively active form around 80kD. A recent report showed that two AR splice variants may possibly be involved in ablation-resistance in 22Rv1 (31). However, the clinical significance of these AR isoforms remains elusive.
In this study, we have identified three novel AR splice variants (designated as AR3, AR4 and AR5) in androgen-insensitive PCA cells. Our data suggest that aberrant expression of AR alternative splicing variants may be a novel mechanism underlying androgen-ablation-independence during PCA progression.
MATERIALS AND METHODS
Cell Culture
LNCaP, PC-3, 22Rv1 and COS-1 cells were purchased from the American Type Culture Collection. PCA cell lines C-81, C4-2, C4-2B and CWR-R1 were kindly provided by Drs. MF Lin (32), D. Tindall and E. Wilson (33), respectively. The cells were transfected with FuGENE 6(Roche) or LipofectAMINE2000(Invitrogen) following the manufacturer’s instruction.
Cloning and constructs
The primer corresponding to the shARc(5′-AGAGTCGCGACTACTACAACTTTCCA-3′) was used to amplify the 3′ end of the AR transcripts using the 5′/3′-Rapid Amplification of cDNA Ends (RACE) Kit (Roche) according to the manufacturer’s instructions.
The shRNAs specific for human AR and AKT1 were purchased from Sigma. The shRNAs specific for AR3 (shAR3) were constructed as described previously (26). The oligo sequences used were as following: shAR3-1, 5′-TGTAATAGTGGTTACCACTCTTCAAGAGAGAGTGGTAACC ACTATTACTTTTTTTTC-3′, 5′-TCGAGAAAAAAAAGTAATAGTGGTTACCACTCTCTCTTG AAGAGTGGTAACCACTATTACA-3′; shAR3-2, 5′-TAGGCTAATGAGGTTT ATTTCTCAAGAG AAAATAAACCTCATTAGCCTTTTTTTTTC-3′, 5′-TCGAGAAAAAAAAAGGCTAATGAGGT TTATTTTCTCTTGAGAAATAAACCTCATTAGCCTA-3′; shAR3-sc (AR3 scrambled control): 5′-TAAGAAACAGTCCGACTCAATTCAAGAGATTGAGTCGGACTGTTTCTTTCTTTTTTC-3′, 5′-TCGAGAAAAAAGAAAGAAACAGTCCGACTCAATCTCTTGAATTGAGTCGGACTGTTTCTT A-3′.
Quantitative Real-time PCR
Quantitative realtime PCR was performed as described previously (34). The primer sequences used for AR isoforms are: AR sense: 5′-CTACTCCGGACCTTACGGGGACATGCG -3′; antisense: 5′-GGGCTGACATTCATAGCCTTCAATGTGTGAC-3′; AR3 sense: 5′-CTACTCCGGA CCTTACGGGGACATGCG -3′; antisense: 5′-TGCCAACCCGGAATTTTTCTCCC-3′; AR4: sense: 5′-CTACTCCGGACCTTACGGGGACATGCG -3′; antisense: 5′-GATTCTTTCAGAAACAACA ACAGCTGCT-3′; AR5: sense: 5′-CTACTCCGGACCTTACGGGGACATGCG -3′; antisense: 5′-CTTTTAATTTGTTCATTCTGAAAAATCCTC-3′; 18S sense: 5′-TTGACGGAAGGGCA CCACCAG-3′ and antisense: 5′-GCACCACCACCCACGGAATCG-3′. The relative abundance of each AR isoform transcript was quantified by using the comparative ΔΔCt with 18S as an internal control. The primer sequences used for human AKT1 were sense: 5′-TCTATGGCGCTGAGATTGTG-3′ and antisense: 5′-CTTAATGTGCCCGTCCTTGT-3′. Human ACTIN and PSA primers are described previously (26). The relative expression levels of AKT1 and PSA transcript was quantified by using the comparative ΔΔCt with ACTIN as an internal control.
Antibodies
The antibodies used in this study include mouse monoclonal anti-Akt1(2H10) (Cell Signaling), mouse monoclonal anti-AR(441), anti-actin(C2) and rabbit polyclonal anti-AR(H-280) and anti-AR(C-19) (Santa Cruz). The anti-AR3 was developed by immunizing the rabbits with a synthetic peptide corresponding to the C-terminal 16-unique-amino-acids of AR3(EKFRVGNCKHLKMTRP) and antisera were affinity purified against the immobilized immunogen.
Luciferase Reporter Assay
Luciferase assay was carried out as described previously (26). Briefly, at 24h post transfection, cells were incubated with phenol-red free medium containing 5% charcoal-stripped FBS. Dual-Luciferase assays were performed according to the manufacturer instructions (Promega). The results are presented as the relative changes of luciferase activity to the untreated control.
Immunohistochemical Analysis
Immunohistochemical staining was carried out with anti-AR3 or anti-AR following a procedure as described previously (26). Immunoreactivity of prostatic epithelial cells was evaluated manually by pathologists (J.M. and X.K.) and graded using a two-score system based on intensity score (IS) and proportion score (PS) as described previously (26, 35). Intensity was scored on a scale of: 0, negative; 1, weak; 2, moderate; 3, strong. Distribution of immunopositive tumor cells was scored on a scale of 0 (0%), 1 (0.1–1%), 2 (2–10%), 3 (11–33%), 4 (34–66%) and 5 (67–100%). The immunoreactivity score was the sum of IS and PS. Additional information on TMAs is available in the Supplementary Material.
Chromatin-Immunoprecipitation
Chromatin-immunoprecipitation was performed as described previously (26). The PCR primers were as follows: P1-ARE, sense 5′-CCACAGAGCACCTCAGCAGTCC-3′, antisense, 5′-GAGCAGGGCAC CCTCTCATGG-3′; P2-ARE sense, 5′-GCTCCTCACTGACGGACTTGTCTG-3′ and antisense, 5′-CCCCTGGTGACAGATGGCC-3′; P3-ARE sense, 5′-GTGCATTTGAGAGAAGCCACGCTG -3′ and antisense, 5′-CACATTGCGCATAGCTGCAGAAG -3′. The PSA Enhancer ARE(PSA-E) and promoter ARE(PSA-P) detection primers were used as previously (26).
In vitro cell growth assay and In vivo tumor growth in xenograft models
The tumor growth of LNCaP, 22Rv1 and CWR-R1 in the xenograft models was carried out as described previously (26). Briefly, at 48h-postinfection, 106 cells were mixed with 100 μl of Matrigel and then subcutaneously(s.c.) injected in the flanks of the castrated SCID/nude mice. Tumor volumes (=0.5236 × r12 × r2 (r1<r2)) were measured weekly and calculated. The differences in tumor sizes formed on both flanks were compared by the paired t-test.
RESULTS
Cloning of AR splice variants
Expression of AR in a panel of PCA cell lines was examined using an antibody recognizing the N-terminus of AR. In addition to the well-characterized 110-kD AR protein, we detected one 80-kD band in the LNCaP derivative C-81, CWR-R1 and 22Rv1 cells which are known to grow in the androgen-depleted medium (Fig. 1A). This short-form AR(ARs) appeared to correspond to the truncated AR previously reported in CWR-R1 (36) and 22Rv1 cells (11, 30). On the other hand, ARs was barely detectable in the androgen-dependent LAPC4 and LNCaP cells. These data implied an inverse correlation between ARs expression and androgen-dependency of these cell lines. To confirm that ARs was indeed derived from the AR gene, we treated CWR-R1 with a panel of shRNAs targeting distinct regions of the AR gene. These shRNAs appeared to differentially knock down AR and ARs (Fig. 1B), suggesting that AR and ARs may be translated from more than one transcript. Similar effects were also observed in 22Rv1 cells (Suppl Fig 17A). These findings prompted us to clone possible alternative splice variants of AR by 3′ RACE using a primer corresponding to the shARc target sequence. As shown in Fig 1C, multiple PCR products resulted from the 3′ RACE were detected. Subsequent cloning and sequencing analysis revealed that the major band around 2 kb turned out to be the 110-kD prototype AR. The other bands were found to be resulted from alternative splicing through various mechanisms including exon skipping, cryptic splicing donor or acceptor usage, cryptic exon inclusion, etc. More than 20 splicing variants have been identified so far. Among them, three variants (designated as AR3, AR4 and AR5) were predicted to encode a protein around 80 kD (Suppl. Figs 1–3). The schematic structures of these AR variants are shown in Fig. 1D. They contain the intact NTD and the DBD but lack the hinge region and the LBD. Instead, they contain 16–53 unique amino acids at their C-termini respectively.
Figure 1.
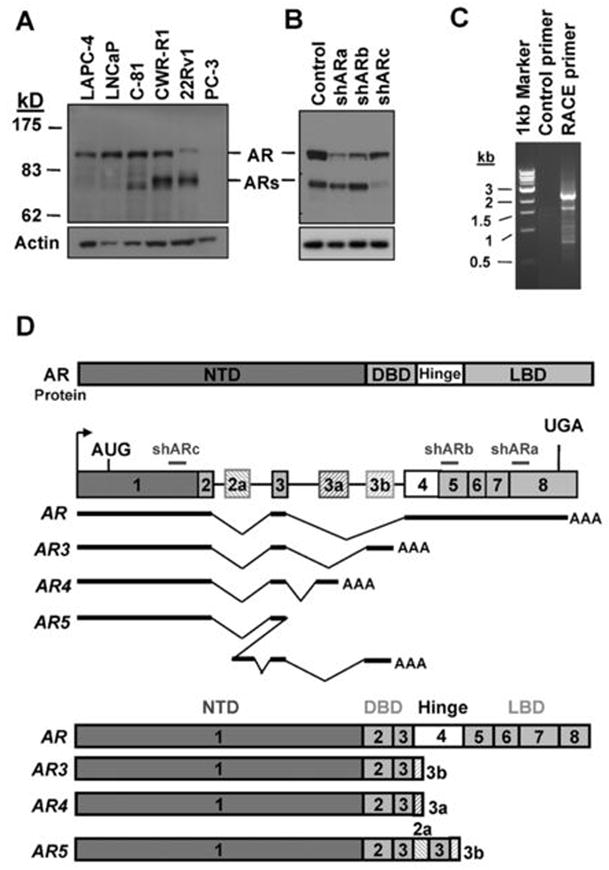
Cloning of novel alternative splice AR isoforms. (A) Cell lysates of different PCA cells were blotted with anti-AR(upper) and anti-Actin(lower). (B) CWR-R1 were infected with the lentivirus encoding the GFPshRNA(Control), ARshRNAa, ARshRNAb, and ARshRNAc(shARa, shARb and shARc) targeting different exons of AR as indicated in (D). At 48h-postinfection, cell lysates were subjected to western blot with anti-AR and anti-Actin, respectively. (C) Total RNA was isolated from CWR-R1 cells and reverse transcribed. The primer derived from AR shRNAc sequence or a control primer was used to perform 3′-RACE. (D) Schematic structure of the human AR splice variants. Hatched cassettes, cryptic exons; Solid thick lines, transcribed sequences.
AR3 is one of the major AR splice variants in PCA
To determine the relative abundance of these AR splice variants, we designed the isoform specific primers recognizing the unique junction sequence present in each isoform. These isoforms were detected in a panel of human prostate tissues by RT-PCR (Suppl. Fig. 5). Fig. 2A shows that AR3 appeared to be one of the most frequently and abundantly expressed isoforms detected in all three hormone-insensitive cell lines. Consistent with the Western Blot data in Fig 1A, AR3 expression is significantly increased in the high-passage LNCaP androgen-insensitive derivative C-81, compared with the parental androgen-sensitive LNCaP cells (Fig. 2A middle panel). We also detected a dramatic increase of AR3 expression in castration-resistant CWR22 xenografts compared to their hormone naïve counterparts (Fig. 2A right panel), suggesting a role of AR3 in developing androgen-ablation resistance. All three AR isoforms were able to induce androgen-independent activation of the ARR2 reporter in COS-1 cells and AR3 appeared to be more active compared to the other two splice variants (Fig 2B). In addition, the AR3 activity was increased in a dose-dependent manner, however, unlike the prototype AR whose activity was dramatically stimulated by DHT, AR3 activity is independent of androgen (Fig 2C). We also overexpressed AR3 in LNCaP cells and examined whether its activity could be modulated by AR, androgen or anti-androgen. Fig. 2D(left panel) shows that inhibition of AR either by the specific shRNA or casodex did not affect AR3 activity regardless of DHT treatment while the activity of endogenous AR or exogenous codon-switched wild-type AR (ARcs, as described previously (26)) was induced by DHT and blocked by casodex as expected. Furthermore, overexpression of AR3 in LNCaP cultured in androgen-depleted medium induced a moderate increase of endogenous PSA expression and such change was not inhibited by casodex (Fig 2D right panel). Thus, AR3 activity is not controlled by DHT, casodex or AR, suggesting that AR3 may be a true androgen-independent transcription factor.
Figure 2.
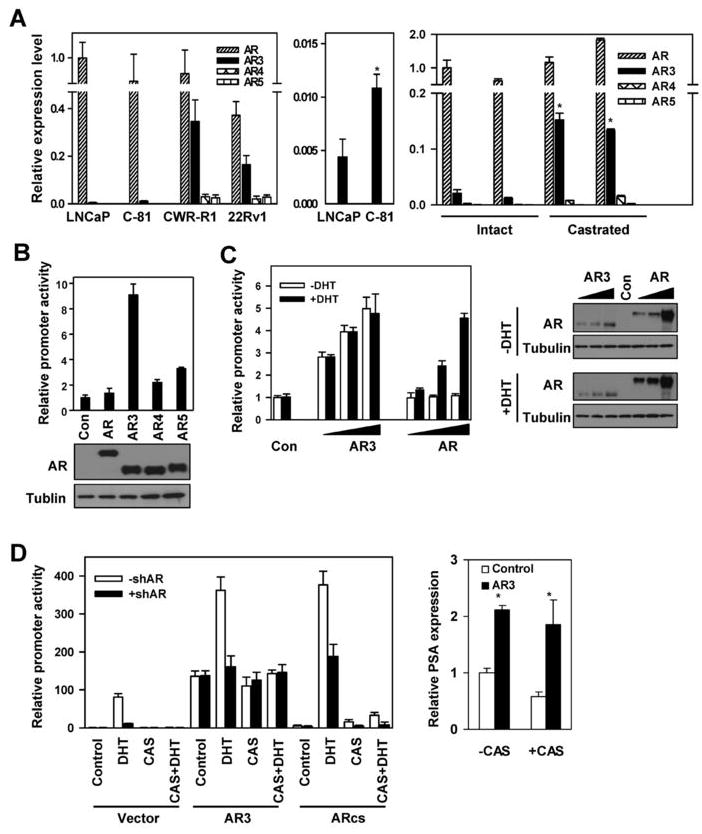
Expression of AR isoforms in PCA cells. (A) The relative expression levels of AR, AR3, AR4 and AR5 were quantified using real-time PCR(left panel). The AR level in LNCaP was arbitrarily set as 1. AR3 expression in LNCaP and C-81 were further plotted with a higher resolution. *p<0.05(middle panel). Their expression in two pairs of CW22R xenograft tumors derived from the intact and castrated male mice were also quantified(right panel). *p<0.05. (B) Transcriptional activity of AR isoforms. COS-1 were transfected with ARR2-luciferase reporter together with the indicated expression vector. At 24h-posttransfection, the luciferase activity was measured. Cell lysates were blotted with anti-AR and anti-Tubulin, respectively (bottom). (C) COS-1 were transfected with ARR2-Luciferase reporter along with increasing doses of AR3 or AR expressing vector. At 24h-posttransfection, cells were treated with or without 10nM DHT for 24h and luciferase activities were measured. Cell lysates were blotted with anti-AR and anti-Tubulin, respectively (right panel). (D) LNCaP were infected with (+) or without(−) the lentivirus encoding ARshRNA (shAR) as described previously (26). At 6h-postinfection, cells were transfected with ARR2-Luciferase reporter along with AR3 or the codon-switched wild-type AR(ARcs). At 24h-posttransfection, cells were treated with DHT and casodex(CAS) as indicated for 24h before luciferase activity was measured (left panel). Cell lysates were blotted with anti-AR and anti-Tubulin (Suppl. Fig. 15). Right panel, LNCaP were transfected with AR3 vector or control. At 24h-posttransfection, cells were treated with or without Casodex (+CAS or −CAS), the relative PSA levels were quantified using real-time PCR. The PSA level in the control LNCaP was arbitrarily set as 1. *p<0.05.
AR3 expression is increased in androgen-depletion-insensitive PCA cells and predicts PCA recurrence
To further characterize the endogenous AR3, we developed a polyclonal antibody specific for AR3 and two AR3 shRNAs specifically targeting the unique Exon 3b of AR3. Fig. 3A shows that the anti-AR3 antibody only detected the overexpressed AR3 but not AR. Knock-down of AR3 in 22Rv1 and CWR-R1 cells diminished the immunoreactivity of anti-AR3 as well as anti-AR with the 80-kD ARs, while it had little effect on anti-AR reactivity with the 110-kD AR (Fig 3B). The anti-AR3 antibody could efficiently and selectively immunoprecipitate the endogenous AR3 but not AR (Fig 3C). The specificity of anti-AR3 in immunohistochemical staining was further validated as shown in Supplementary Fig. 6. Immunofluorescence staining revealed that AR3 was present in both nucleus and cytoplasm in CWR-R1 and 22Rv1 cells (Suppl. Fig. 7). Western Blot analysis showed that AR3 is expressed in all tested AR-positive PCA lines. It is noteworthy that the level of AR3 in the androgen-insensitive LNCaP derivatives C81, C4-2 and C4-2B was significantly higher than that in the androgen-sensitive LNCaP (Fig 3D top panel). In addition, AR3 protein level was also dramatically increased in CWR22 xenograft tumor derived from the castrated mice compared to that from the intact animals (Fig 3D bottom panel). Immunohistochemistry analysis on human prostate tissue microarrays revealed a marked change in AR3 expression level and pattern in malignant prostate tissues compared to the benign counterparts (Fig 4A). In benign tissues, anti-AR3 mainly stained basal and stromal cells but most of luminal epithelial cells were barely stained (the mean epithelial cytoplasmic staining score =1.52 ± 0.34) (Fig. 4B and Suppl. Fig. 9). On the other hand, the majority of luminal cells in malignant glands showed stronger cytoplasmic AR3 staining (mean score = 4.74 ± 0.13). In addition, a significant redistribution of AR3 protein to the nucleus was observed in hormone-resistant tumor samples (44% nuclear positive) compared to hormone-naïve counterparts (9% nuclear positive). Thus, nuclear translocation of AR3 is significantly increased in hormone-resistant tumors. To assess whether AR3 could be used as a potential prognostic marker, clinical outcome analysis was performed on 224 PCA patient samples with clinicopathlogical information. Patients with elevated PSA levels following radical prostatectomy are at a high risk to develop distant metastases and die of PCA. Clinical failure was defined as a PSA elevation of greater than 0.2 ng/ml following radical prostatectomy with successive increasing PSA values. Kaplan-Meier analysis indicated that PCA patients who have higher cytoplasmic staining of AR3 (staining score >=6) have a greater risk for PSA recurrence after radical prostatectomy (Fig 4C, log-rank test, p < 0.0001). Furthermore, multivariable Cox Regression Analysis showed that AR3 is a significant predictor of PCA recurrence after adjusting for other important clinicopathological variables including: Gleason sum score, preoperative PSA, size of largest individual nodule of invasive cancer, surgical margin status (Fig 4D). As indicated in the model, AR3 was the strongest predictor with hazard ratio of 2.5 (95% CI: 1.3–4.6, p=0.004).
Figure 3.
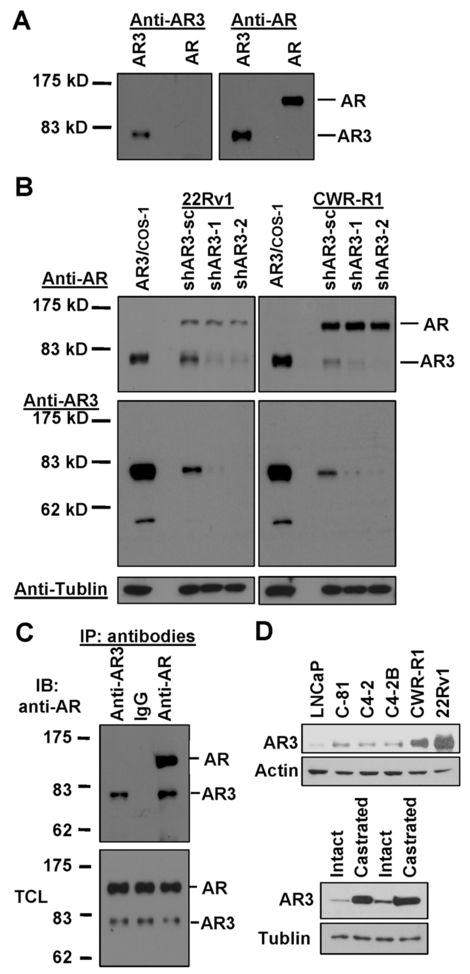
Detection of AR3 in hormone-insensitive PCA cells. (A) COS-1 were transfected with AR3 or AR vector. Total protein lysates were immunoblotted with anti-AR3 and anti-AR, respectively. (B) CWR-R1 and 22Rv1 were infected with lentivirus encoding AR3shRNAs (shAR3-1, −2) or the scrambled control (shAR3-sc). At 48h-postinfection, cell lysates were subjected to immunoblotting with anti-AR and anti-AR3, respectively. COS-1 overexpressing AR3 was used as a positive control (first lane). (C) CWR-R1 lysates were split into three equal aliquots and immnuoprecipitated with anti-AR3, control IgG and anti-AR, respectively. The resultant immunoprecipitates and the input total cell lysates (TCL) were immunoblotted with anti-AR. (D) Total cell lysates of a panel of PCA cells were blotted with anti-AR3 and anti-Actin, respectively (top panel). Bottom panel, extracts of two pairs of CW22R tumor xenografts derived from the intact and castrated male mice were blotted with anti-AR3 and anti-Tubulin, respectively.
Figure 4.
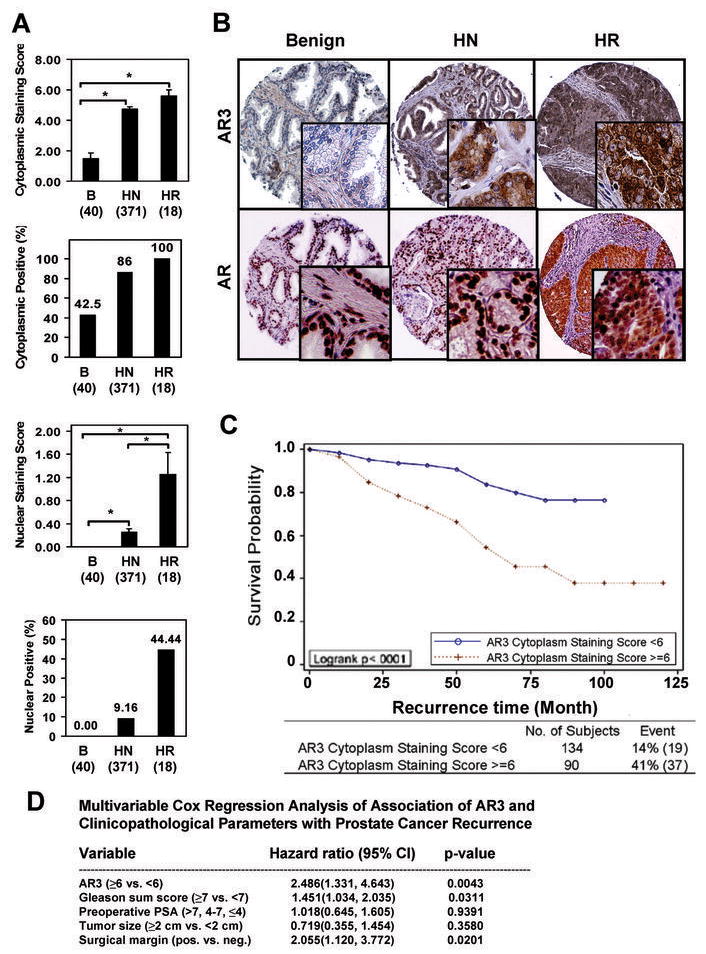
Increased AR3 expression in androgen-depletion-resistant PCA cells. (A) Human prostate tissue microarrays(TMAs) were stained with anti-AR3. The mean score of cytoplasmic and nuclear staining of the luminal cell as well as the positive rates for cytoplasmic and nuclear staining were shown. (Error bars indicate standard error, *p<0.01). Benign (B), hormone-naïve (HN) and hormone-resistant (HR). (B) The representative fields of TMAs stained with anti-AR3. The anti-AR stained arrays were included as a control. (C) Correlation of AR3 cytoplasmic staining with PSA recurrence after prostatectomy. (D) Multivariable Cox Regression Analysis.
AR3 promotes androgen-depletion-independent growth
Overexpression of AR3 in LNCaP cell promoted growth in androgen-depleted medium (Fig 5A). This was accompanied by the increase of DNA synthesis measured by EdU incorporation (Suppl. Fig 14A), suggesting that overexpression of AR3 stimulates LNCaP proliferation in androgen-depleted medium. Such growth enhancement was also observed in the castrated SCID mice xenograft models (Fig 5B). We further investigated whether expression of AR3 in hormone-insensitive PCA cells is required for their growth by specifically knocking down AR3. As shown in Fig 5C and 5D, treatment of both 22Rv1 and CWR-R1 cells with the shRNA specific for AR3 attenuated their growth in the androgen-depleted medium as well as in the castrated nude mice, suggesting that AR3 activity is required for PCA cell growth under androgen-depleted conditions. We also examined the effects of AR3 knock-down on DNA synthesis and apoptosis in CWR-R1 cells in parallel with AR knock-down. We found that AR3 knock-down significantly reduced EdU incorporation (Suppl. Fig. 14B), but had little effects on apoptosis (Suppl. Fig. 12). Meanwhile, AR knock-down appeared to induce a marked increase in the number of apoptotic cells and reduce the number of proliferating cells. It should be noted that knock-down of AR3 in these cells did not alter the expression of AR, therefore, AR3 may play an indispensable role in promoting prostate cancer cell proliferation, possibly through regulating a different set of target genes.
Figure 5.
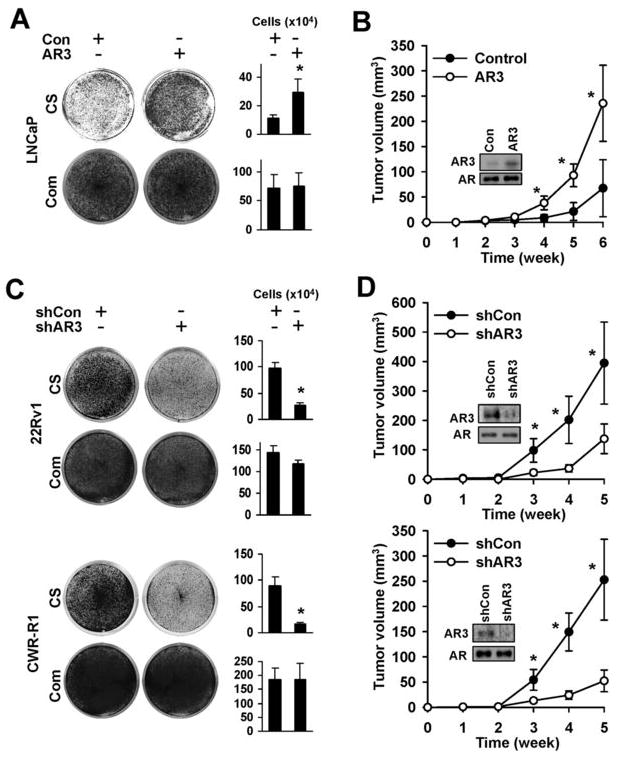
AR3 promotes PCA cell growth under androgen-depleted conditions. LNCaP were infected with lentivirus encoding AR3 or the control vector. After 2-week culture in the androgen-depleted (CS) or the complete(Com) medium, cells were visualized by Coomassie Blue staining. Under the same experimental conditions, cell numbers were quantified and plotted as a bargraph (right panel) (*p<0.05) (A). At 48h-postinfection, cells were injected into the castrated SCID mice and tumor growth were monitored weekly. The result represents the mean tumor volume ±SE (n = 5 mice/group), *p<0.05. Inset, Western blots of anti-AR3 and anti-AR of LNCaP xenograft tumor lysates (B). CWR-R1 and 22Rv1 cells were infected with lentivirus encoding AR3shRNA-1 (shAR3) or control shRNA (shCon). Cell growth was visualized and quantified as in Fig 5A (C). Tumor growth was monitored as in Fig 5B (D). Inset, Western blots of anti-AR3 and anti-AR of the CWR-R1 and 22Rv1 xenograft tumor lysates.
AR3 regulates AKT1 expression in PCA cells
To identify potential AR3 regulated genes, we selectively knocked down AR3 or AR by the specific shRNAs in CWR-R1 and 22Rv1. The differential gene expression resulted from AR3 or AR knockdown were determined by microarray analysis. The differential expression of a set of 188 genes was consistently detected in both cell lines when AR3 was specifically knocked down while the expression of 412 genes was altered in both cell lines when AR was specifically inhibited (Fig 6A). Among them, 71 genes are commonly regulated by both AR and AR3. A partial list of these genes is summarized in Supplementary Table 1. Several known AR regulated genes such as IGFBP3 and FKBP5 are also regulated by AR3. However, many classical AR regulated genes such as CLU, TMEPAI, KLK3 (PSA) and CLDN4 were not affected by AR3 knock-down under our experimental conditions (Supplementary Table 2). Among the 117 genes that are preferentially regulated by AR3 (Supplementary Table 3), there are a number of genes, such as MAP4K4, HOXB7 and ELK1, have been found to be upregulated in hormone-resistant or metastatic prostate cancers in previous gene profiling studies (19, 37, 38). Interestingly, the serine/threonine kinase AKT1, which has been implicated in PCA development and progression, appeared to be preferentially regulated by AR3 as well. As shown in Fig 6B(left panel), the level of AKT1 transcript in AR3-knockdown cells is significantly less than that in cells treated with the scrambled shRNA or the shRNA specific for AR. The protein level of AKT1 was also reduced accordingly in AR3-knockdown CWR-R1 cells (Fig 6B middle panel). In addition, we examined AKT1 protein level in some of the xenograft tumors described in Fig. 4. Consistent with the results in cell lines, AKT1 protein level is increased in the xenograft tumor of LNCaP overexpressing AR3 and decreased in the xenograft tumor of 22Rv1 with AR3 knock-down (Fig 6B right panel). Concurrent with the change of AKT1 expression level in these cells, the phosphorylation status of the AKT substrate GSK3β was also altered accordingly. To test whether AKT1 is essential for PCA growth under androgen-depleted conditions, AKT1 in CWR-R1 cells was knocked down by the specific shRNAs. Consistent with the previous report on the mouse model (39), even a 50% reduction of AKT1 expression diminished PCA cell growth (Fig 6C). Furthermore, we identified at least two putative ARE sites in the AKT1 regulatory region and showed that AR3 but not AR was able to bind to these ARE sites determined by the chromatin-immunoprecipitation (ChIP) assays (Fig 6D), suggesting that AR3 may directly regulate AKT1 transcription. Meanwhile, AR3 failed to bind to the ARE site located at the enhancer region of PSA gene. Taken together, our data suggest that AR3 and AR may play an overlapping but yet distinct role in prostate cells by regulating their respective target genes.
Figure 6.
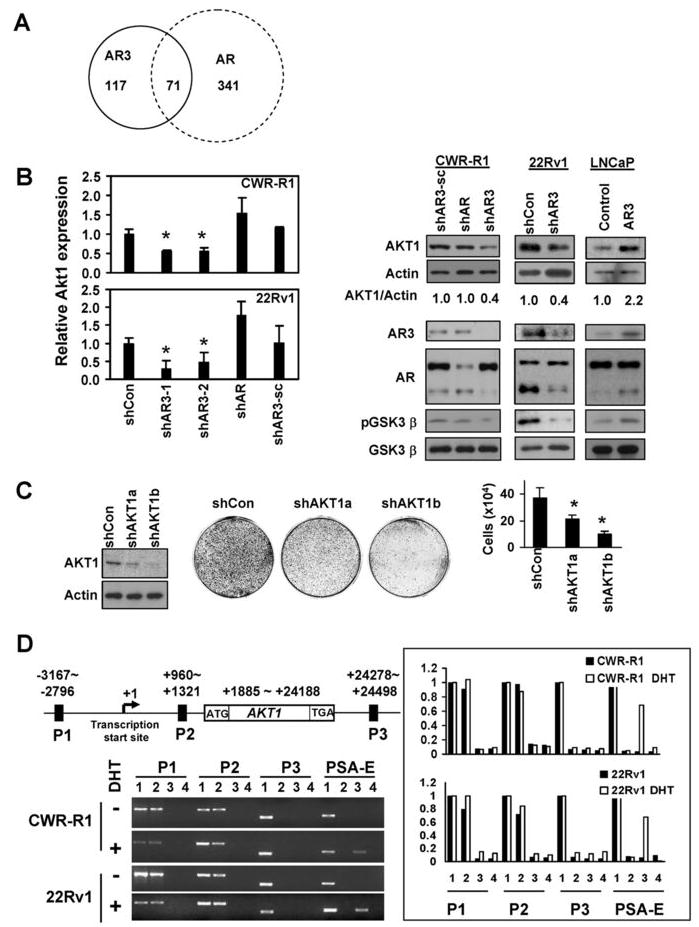
AKT1 is a target gene regulated by AR3. (A) Schematic representation of AR3 and AR target genes. (B) The effects of AR3 on AKT1 transcription. CWR-R1 and 22Rv1 were infected with lentivirus encoding the control shRNA(shCon), AR3shRNAs(shAR3-1 or shAR3-2), ARshRNA(shAR) or the scrambled control(shAR3-sc). At 48h-postinfection, the relative expression levels of AKT1 transcripts compared to the shCon was quantified by real-time PCR (*p<0.05) (Left Panel). The protein levels of AKT1, Actin, AR3, AR, pGSK3β and GSK3β were also detected by immunoblotting. The levels of AKT1 from the immunoblots were normalized by calculating the ratios of AKT1/actin. The changes in fold compared to the control were shown (bottom). Right Panels, The lysates of LNCaP and 22Rv1 xenograft tumors from Figure 5A and 5B were subjected to immunoblotting as described above. (C) The effects of AKT1 knockdown on PCA cell growth. CWR-R1 cells were infected with the lentivirus encoding two independent AKT1 shRNAs (shAKT1a and shAKT1b) respectively. After 2-week culture in androgen-depleted medium, cells were visualized and quantified as described in Fig 5A. (D) CWR-R1 and 22Rv1 were treated with or without DHT(10nM) for 1h. Binding of AR3 or AR to the putative ARE sites (P1, P2 and P3) of human AKT1 gene was analyzed by ChIP assays. The ARE at the PSA enhancer region (PSA-E) was used as a positive control for AR. PCR products from input(1), immunoprecipitation with anti-AR3(2), anti-AR(3) or the control antibody(4), were resolved on agarose gels(Left panel). The PCR products were quantified by using the software Quantity One(Right panel).
DISCUSSION
Mechanisms underlying prostate cancer progression to androgen-ablation-resistance are complicated and many factors may be involved. In this report, we demonstrated that alternative splicing of human AR gene may be one of the means to diversify its signaling and confer androgen-independent activation of AR in prostate cells. We showed that several AR splice variants are constitutively active in transcription and their activity is not expected to be affected by either androgens or anti-androgen drugs. Among these variants, AR3 appears to be one of the most abundantly and ubiquitously expressed isoforms in our screening of a panel of human prostate cancer cell lines and tissues. In normal prostate tissues, AR3 appears to be mainly expressed in the basal and stromal cells but virtually no or only weak AR3 expression was detected in the luminal epithelial cells. This is consistent with the previous reports that the basal and stromal compartments are insensitive to androgen-ablation (40–43). Our data suggested that AR3 may play a role in androgen-insensitive regulation of normal prostate gland homeostasis. However, in malignant glands, a marked increase of cytoplasmic AR3 expression is detected in carcinoma cells in 86% of the cases examined in this study. Although the cytoplasmic AR3 may not be transcriptionally active prior to hormonal therapy, AR3 may translocate into the nucleus later on during disease progression (e.g., when Src activity is up-regulated upon androgen ablation) and exerts its transcriptional activity under androgen-depleted conditions. This is consistent with our observation that PCA patients with higher cytoplasmic AR3 protein level have a greater risk for tumor recurrence after prostatectomy. Therefore, the expression of AR3 may potentially be used to predict patient outcome in response to hormonal therapy. It is possible that AR3 transcriptional activity is tightly regulated by its subcellular localization like the prototype AR. However, the underlying mechanisms have yet to be determined. We previously showed that Src kinase may regulate AR nuclear translocation under androgen-depleted conditions, possibly through phosphorylating Y534 in the NTD. Most of Src-induced phosphorylation sites are present in AR3 and therefore Src kinase family kinases may likely be involved.
It is possible that tumor cells hijack the active AR splice variants lacking the LBD to escape from the hormonal therapy and aberrant expression of the constitutively active AR3 may contribute to ablation-independent growth. This is supported by our observations that overexpression of AR3 in LNCaP promoted tumor growth and knockdown of AR3 in CWR-R1 and 22Rv1 attenuated their growth under androgen-depleted conditions. Our gene expression profiling in CWR-R1 and 22Rv1 cells revealed that AR3 shares some overlapping target genes with AR despite of its lack of the AF2 domain(LBD). However, a large subset of classical androgen-responsive genes including KLK3(PSA) is preferentially regulated by AR under our experimental conditions. It should be noted that these cells were maintained in normal growth medium and under such conditions both AR and AR3 are believed to be active. Although AR3 knock-down did not alter PSA expression under this condition as the active AR may be the main driver for controlling PSA expression, overexpression of AR3 in LNCaP cultured in the androgen-depleted medium did induce a moderate increase in PSA transcription, suggesting that AR3 is able to compensate, at least in part, for AR activity under androgen-depleted conditions. This is supported by our observation that AR3 was able to bind to the proximal ARE site though it did not bind to the distal ARE site of the PSA gene (Figure 6D and Supplementary Figure 11).
Most importantly, microarray analysis allowed us to uncover a subset of genes that are preferentially regulated by AR3. These genes are involved in regulation of diverse biological processes including signal transduction, posttranslational modifications, transcription, chromatin remodeling, ion transportation and metabolism, suggesting that AR3 may play a critical role in homeostasis maintenance of its target cells though AR3 is relatively less abundant compared to the prototype AR. This is supported by our observation that knockdown of AR3 attenuated PCA cell growth. We have confirmed that AKT1 is one of AR3 preferred target genes in PTEN-positive CWR-R1 and 22Rv1 cells by real-time PCR and Western Blot. AKT1 was reported to be overexpressed in the primary epithelial cultures derived from human prostate tumors (44), suggesting that AKT signaling may be elevated in the epithelial compartment through a transcriptional mechanism. An increase of AKT1 expression in PCA patients is associated with PSA relapse (45). Thus, AR3 may contribute to upregulation of AKT1 signaling at the transcriptional level during PCA progression. Although the magnitude of changes of AKT1 is moderate(about 2–3 fold), such change may have a substantial impact on prostate cell growth as demonstrated in Fig 6C. This is supported by a previous study showing that haplodeficiency of Akt1 dramatically inhibits prostate tumor development in Pten+/− mice (39). Therefore, the increase of AR3 in luminal epithelial cells may be sufficient to confer a growth advantage, at least in part, through increasing AKT1 expression. Taken together, our data suggest that AR3 may have a distinct biological activity despite a partial overlapping biological function with AR. AR3 may primarily play a role in regulation of androgen-independent biological processes and maintain homeostasis of the prostate gland in concert with AR.
Although we showed that AR3 can function as a transcriptional factor independent of AR, it is still possible that AR3 may bind to a subset of ARE sites (e.g. the proximal PSA-P site) in complex with AR. However, so far we have not yet been able to detect such complex using our AR3 specific antibody. Future study should be carried out to examine whether they may synergistically function together to regulate a subset of ARE-containing promoters.
We also identified another splice variant AR6 lacking the second zinc finger in the DBD (Suppl. Fig 4). AR6 did not display detectable transcriptional activity on the PSA and ARR2 reporters in COS-1 cells, and therefore, was not characterized in this study. During preparation of this manuscript, two AR variants expressed in 22Rv1 cells were reported recently (31). Notably, they appear to be different from the ones identified in the present study. We propose that technical approaches (cDNA preparation, PCR amplification, cloning, etc.) might account for our differing observations. This possibility is supported by that the sequences of our splice variants have longer 3′ UTRs and contain the conserved AATAAA polyadenylation signals. Although their variants appear to share some sequence homology with AR5 and AR6, the unique coding sequences at the C-termini are quite different. Future study on human tissue samples will be necessary to resolve these discrepancies.
Nevertheless, these studies suggest that aberrant expression of AR splicing variants may be a mechanism underlying prostate cancer progression. Given that these AR isoforms are not inhibited by currently available anti-androgens, development of new drugs targeting these AR variants may potentially be effective for ablation-resistant PCA.
Supplementary Material
Acknowledgments
This work was supported by the NIH grants (CA106504), DOD grants (W81XWH-06-1-0199, W81XWH-08-1-0126) to Y.Q. and the NIH grant (NCI UO1- CA86772) to J.M., DOD Postdoctoral Fellowship (W81XWH-04-1-0015) to Z.G and Pre-doctoral Fellowship (W81XWH-08-1-0068) to D.L.
References
- 1.Denmeade SR, Isaacs JT. A history of prostate cancer treatment. Nat Rev Cancer. 2002;2:389–396. doi: 10.1038/nrc801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Litvinov IV, Antony L, Isaacs JT. Molecular characterization of an improved vector for evaluation of the tumor suppressor versus oncogene abilities of the androgen receptor. Prostate. 2004;61:299–304. doi: 10.1002/pros.20187. [DOI] [PubMed] [Google Scholar]
- 3.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 4.Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. N Engl J Med. 2004;351:1488–1490. doi: 10.1056/NEJMp048178. [DOI] [PubMed] [Google Scholar]
- 5.Chang CS, Kokontis J, Liao ST. Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science. 1988;240:324–326. doi: 10.1126/science.3353726. [DOI] [PubMed] [Google Scholar]
- 6.Lubahn DB, Joseph DR, Sar M, et al. The human androgen receptor: complementary deoxyribonucleic acid cloning, sequence analysis and gene expression in prostate. Mol Endocrinol. 1988;2:1265–1275. doi: 10.1210/mend-2-12-1265. [DOI] [PubMed] [Google Scholar]
- 7.McPhaul MJ. Factors that mediate and modulate androgen action. J Investig Dermatol Symp Proc. 2003;8:1–5. doi: 10.1046/j.1523-1747.2003.12163.x. [DOI] [PubMed] [Google Scholar]
- 8.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 9.Kuiper GG, Faber PW, van Rooij HC, et al. Structural organization of the human androgen receptor gene. J Mol Endocrinol. 1989;2:R1–4. doi: 10.1677/jme.0.002r001. [DOI] [PubMed] [Google Scholar]
- 10.Lubahn DB, Brown TR, Simental JA, et al. Sequence of the intron/exon junctions of the coding region of the human androgen receptor gene and identification of a point mutation in a family with complete androgen insensitivity. Proc Natl Acad Sci U S A. 1989;86:9534–9538. doi: 10.1073/pnas.86.23.9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Libertini SJ, Tepper CG, Rodriguez V, Asmuth DM, Kung HJ, Mudryj M. Evidence for calpain-mediated androgen receptor cleavage as a mechanism for androgen independence. Cancer Res. 2007;67:9001–9005. doi: 10.1158/0008-5472.CAN-07-1072. [DOI] [PubMed] [Google Scholar]
- 12.Simental JA, Sar M, Lane MV, French FS, Wilson EM. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J Biol Chem. 1991;266:510–518. [PubMed] [Google Scholar]
- 13.Jenster G, van der Korput HA, van Vroonhoven C, van der Kwast TH, Trapman J, Brinkmann AO. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol. 1991;5:1396–1404. doi: 10.1210/mend-5-10-1396. [DOI] [PubMed] [Google Scholar]
- 14.Rundlett SE, Wu XP, Miesfeld RL. Functional characterizations of the androgen receptor confirm that the molecular basis of androgen action is transcriptional regulation. Mol Endocrinol. 1990;4:708–714. doi: 10.1210/mend-4-5-708. [DOI] [PubMed] [Google Scholar]
- 15.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Li W, Liu XS, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcelli M, Cunningham GR. Hormonal signaling in prostatic hyperplasia and neoplasia. J Clin Endocrinol Metab. 1999;84:3463–3468. doi: 10.1210/jcem.84.10.6083. [DOI] [PubMed] [Google Scholar]
- 18.Culig Z, Klocker H, Bartsch G, Hobisch A. Androgen receptors in prostate cancer. Endocr Relat Cancer. 2002;9:155–170. doi: 10.1677/erc.0.0090155. [DOI] [PubMed] [Google Scholar]
- 19.Chen CD, Welsbie DS, Tran C, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 20.Zegarra-Moro OL, Schmidt LJ, Huang H, Tindall DJ. Disruption of androgen receptor function inhibits proliferation of androgen-refractory prostate cancer cells. Cancer Res. 2002;62:1008–1013. [PubMed] [Google Scholar]
- 21.Yeh S, Lin HK, Kang HY, Thin TH, Lin MF, Chang C. From HER2/Neu signal cascade to androgen receptor and its coactivators: a novel pathway by induction of androgen target genes through MAP kinase in prostate cancer cells. Proc Natl Acad Sci U S A. 1999;96:5458–5463. doi: 10.1073/pnas.96.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling MT, Chan KW, Choo CK. Androgen induces differentiation of a human papillomavirus 16 E6/E7 immortalized prostate epithelial cell line. J Endocrinol. 2001;170:287–296. doi: 10.1677/joe.0.1700287. [DOI] [PubMed] [Google Scholar]
- 23.Lin HK, Hu YC, Yang L, et al. Suppression versus induction of androgen receptor functions by the phosphatidylinositol 3-kinase/Akt pathway in prostate cancer LNCaP cells with different passage numbers. J Biol Chem. 2003;278:50902–50907. doi: 10.1074/jbc.M300676200. [DOI] [PubMed] [Google Scholar]
- 24.Gregory CW, Fei X, Ponguta LA, et al. Epidermal growth factor increases coactivation of the androgen receptor in recurrent prostate cancer. J Biol Chem. 2004;279:7119–7130. doi: 10.1074/jbc.M307649200. [DOI] [PubMed] [Google Scholar]
- 25.Ueda T, Mawji NR, Bruchovsky N, Sadar MD. Ligand-independent activation of the androgen receptor by interleukin-6 and the role of steroid receptor coactivator-1 in prostate cancer cells. J Biol Chem. 2002;277:38087–38094. doi: 10.1074/jbc.M203313200. [DOI] [PubMed] [Google Scholar]
- 26.Guo Z, Dai B, Jiang T, et al. Regulation of androgen receptor activity by tyrosine phosphorylation. Cancer Cell. 2006;10:309–319. doi: 10.1016/j.ccr.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 27.Mahajan NP, Liu Y, Majumder S, et al. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proc Natl Acad Sci U S A. 2007;104:8438–8443. doi: 10.1073/pnas.0700420104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Titus MA, Schell MJ, Lih FB, Tomer KB, Mohler JL. Testosterone and dihydrotestosterone tissue levels in recurrent prostate cancer. Clin Cancer Res. 2005;11:4653–4657. doi: 10.1158/1078-0432.CCR-05-0525. [DOI] [PubMed] [Google Scholar]
- 30.Tepper CG, Boucher DL, Ryan PE, et al. Characterization of a novel androgen receptor mutation in a relapsed CWR22 prostate cancer xenograft and cell line. Cancer Res. 2002;62:6606–6614. [PubMed] [Google Scholar]
- 31.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a Novel Androgen Receptor Exon Generates a Constitutively Active Androgen Receptor that Mediates Prostate Cancer Therapy Resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Igawa T, Lin FF, Lee MS, Karan D, Batra SK, Lin MF. Establishment and characterization of androgen-independent human prostate cancer LNCaP cell model. Prostate. 2002;50:222–235. doi: 10.1002/pros.10054. [DOI] [PubMed] [Google Scholar]
- 33.Gregory CW, Johnson RT, Jr, Mohler JL, French FS, Wilson EM. Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Res. 2001;61:2892–2898. [PubMed] [Google Scholar]
- 34.Zhang Y, Wang XW, Jelovac D, et al. The ErbB3-binding protein Ebp1 suppresses androgen receptor-mediated gene transcription and tumorigenesis of prostate cancer cells. Proc Natl Acad Sci U S A. 2005;102:9890–9895. doi: 10.1073/pnas.0503829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 36.Gregory CW, He B, Wilson EM. The putative androgen receptor-A form results from in vitro proteolysis. J Mol Endocrinol. 2001;27:309–319. doi: 10.1677/jme.0.0270309. [DOI] [PubMed] [Google Scholar]
- 37.Best CJ, Gillespie JW, Yi Y, et al. Molecular alterations in primary prostate cancer after androgen ablation therapy. Clin Cancer Res. 2005;11:6823–6834. doi: 10.1158/1078-0432.CCR-05-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Chen ML, Xu PZ, Peng XD, et al. The deficiency of Akt1 is sufficient to suppress tumor development in Pten+/− mice. Genes Dev. 2006;20:1569–1574. doi: 10.1101/gad.1395006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johansson A, Jones J, Pietras K, et al. A stroma targeted therapy enhances castration effects in a transplantable rat prostate cancer model. Prostate. 2007;67:1664–1676. doi: 10.1002/pros.20657. [DOI] [PubMed] [Google Scholar]
- 41.Cunha GR, Cooke PS, Kurita T. Role of stromal-epithelial interactions in hormonal responses. Arch Histol Cytol. 2004;67:417–434. doi: 10.1679/aohc.67.417. [DOI] [PubMed] [Google Scholar]
- 42.Kurita T, Wang YZ, Donjacour AA, et al. Paracrine regulation of apoptosis by steroid hormones in the male and female reproductive system. Cell Death Differ. 2001;8:192–200. doi: 10.1038/sj.cdd.4400797. [DOI] [PubMed] [Google Scholar]
- 43.English HF, Santen RJ, Isaacs JT. Response of glandular versus basal rat ventral prostatic epithelial cells to androgen withdrawal and replacement. Prostate. 1987;11:229–242. doi: 10.1002/pros.2990110304. [DOI] [PubMed] [Google Scholar]
- 44.Nanni S, Priolo C, Grasselli A, et al. Epithelial-restricted gene profile of primary cultures from human prostate tumors: a molecular approach to predict clinical behavior of prostate cancer. Mol Cancer Res. 2006;4:79–92. doi: 10.1158/1541-7786.MCR-05-0098. [DOI] [PubMed] [Google Scholar]
- 45.Le Page C, Koumakpayi IH, Alam-Fahmy M, Mes-Masson AM, Saad F. Expression and localisation of Akt-1, Akt-2 and Akt-3 correlate with clinical outcome of prostate cancer patients. Br J Cancer. 2006;94:1906–1912. doi: 10.1038/sj.bjc.6603184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


