Abstract
The nuclear factor-kappaB (NF-κB) transcription factor system is a crucial component that controls several important biological functions, thus raising the need for mechanisms that ensure the correct termination of its activity. Here, we identify a new phosphorylation/ubiquitination switch in the NF-κB network that controls the stability of the transactivating p65 subunit. Tumour necrosis factor-induced phosphorylation of p65 at Ser468 allows binding of COMMD1 and cullin 2, components of a multimeric ubiquitin ligase complex mediating p65 ubiquitination. Mutation of p65 at Ser468 largely prevents p65 ubiquitination and proteasomal degradation. Inducible p65 elimination is restricted to a subset of NF-κB target genes such as Icam1. Accordingly, chromatin immunoprecipitation experiments reveal the selective recruitment of Ser468-phosphorylated p65 and COMMD1 to the Icam1 promoter. Phosphorylation of p65 at Ser468 leads to ubiquitin/proteasome-dependent removal of chromatin-bound p65, thus contributing to the selective termination of NF-κB-dependent gene expression.
Keywords: NF-κB, phosphorylation, ubiquitination, transcription, ubiquitin ligase
Introduction
Nuclear factor-kappaB (NF-κB) is a dimeric transcription factor that controls the expression of a wide variety of genes involved not only in innate and adaptive immunity, but also in apoptosis and proliferation (Li & Verma, 2002; Wietek & O'Neill, 2007). The DNA-binding capacity of NF-κB dimers is primarily controlled by its interaction with inhibitory IκB proteins (Hacker & Karin, 2006). Canonical NF-κB activation by a wide variety of signals, including lipopolysaccharide (LPS), interleukin-1 (IL-1) or tumour necrosis factor (TNF), triggers a signalling cascade that leads to the phosphorylation of IκB, and its subsequent ubiquitination and proteasomal elimination (Hayden & Ghosh, 2008). In addition, several mechanisms mediate further layers of NF-κB regulation, including the post-translational modification of the DNA-binding subunits by phosphorylation, acetylation and ubiquitination. An increasing number of p65 phosphorylation sites have been identified such as Ser468, which is contained in a carboxy-terminal transactivation domain. Basal Ser468 phosphorylation is regulated by glycogen synthase kinase 3 (GSK3; Buss et al, 2004), whereas stimulus-induced modification is mediated by IκB kinase (IKK)-β and IKKɛ (Schwabe & Sakurai, 2005; Mattioli et al, 2006). Both the p50 and p65 subunits were found to be ubiquitinated and degraded in the nucleus, thus allowing termination of the nuclear NF-κB response (Ryo et al, 2003; Saccani et al, 2004; Carmody et al, 2007). Ubiquitination of p65 is controlled by various ubiquitin E3 ligases, including suppressor of cytokine signalling (SOCS1), PDZ and LIM domain 2 (PDLIM2) and COMM domain-containing 1 (COMMD1; Ryo et al, 2003; Maine et al, 2007; Tanaka et al, 2007). The p65-binding PDLIM2 protein is responsible for LPS-induced polyubiquitination of p65 and sequesters this transcription factor in subnuclear domains for proteasomal degradation. Stimulation of TNF enhances p65 interaction with COMMD1 and further components of this ubiquitin E3 ligase complex, including SOCS1 and cullin 2 (CUL2), thus resulting in inducible ubiquitination and destabilization of p65. Immunoblotting experiments show that proteolytic elimination occurs only for a limited subfraction of the p65 protein, suggesting that the turnover of p65 must be tightly controlled by regulatory mechanisms. Here, we identify the phosphorylation of p65 at Ser468 as a regulatory switch for ubiquitination of this transcription factor.
Results And Discussion
TNF-triggered p65 degradation depends on phosphorylation
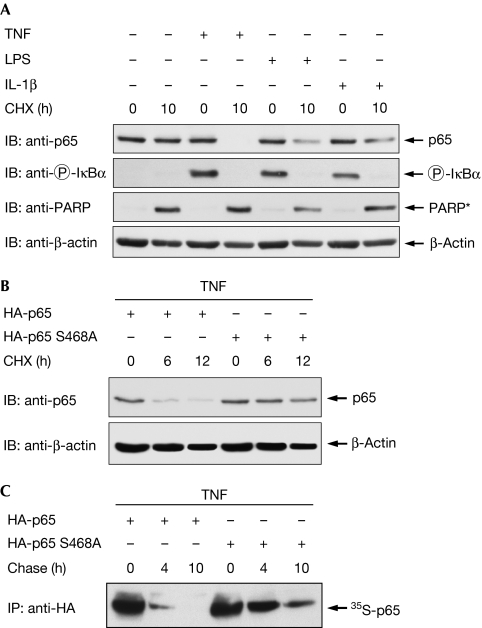
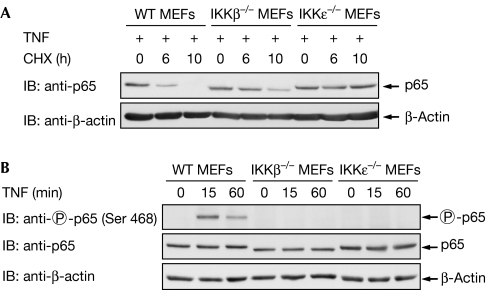
To find a signalling pathway suitable for the study of p65 degradation in mouse embryonic fibroblasts (MEFs), we compared the NF-κB-inducing stimuli LPS, IL-1 and TNF for their ability to mediate p65 decay. As only a fraction of p65 undergoes proteasomal degradation, MEFs were stimulated with a pulse of TNF, LPS or IL-1 to induce a single wave of NF-κB activation, followed by the administration of cycloheximide (CHX) to prevent resynthesis of the eliminated protein and to allow the convenient detection of p65 diminishment. Western blot experiments revealed that TNF had the highest efficacy for the degradation of p65 (Fig 1A). Although treatment with CHX alone triggered caspase activity (as revealed by the occurrence of the caspase-cleaved, truncated form of poly (ADP-ribose) polymerase), induction of TNF signalling was necessary to trigger p65 decay. As LPS can trigger the degradation of p65 in RAW264.7 macrophage cells (Tanaka et al, 2007), our data imply that LPS controls p65 stability in a cell type-specific manner. TNF-induced elimination of p65 was also detectable in other cell types such as HeLa cells (supplementary Fig 1 online). Given the need for a tightly controlled regulation of p65 stability, we investigated the occurrence of phosphorylation-dependent destabilization, as seen, for example, for IκBα (Yaron et al, 1998). To investigate this question, p65-deficient MEFs were retransfected with wild-type p65 or a p65 mutant in which the phosphorylated Ser468 is replaced by an alanine (p65 S468A). TNF-induced degradation of the wild-type p65 protein was detectable after 6 h, whereas the unphosphorylatable p65 mutant remained largely stable (Fig 1B). Accordingly, TNF-induced phosphorylation of p65 at Ser468 occurred only for the wild type, but not for the point-mutated p65 protein (supplementary Fig 2 online). It has been shown previously that p65 degradation is more prominent in IκBα−/− cells in which the IκBα-dependent removal of NF-κB from its target genes is absent (Saccani et al, 2004). Thus, we compared the stabilities between the p65 wild-type and Ser468 mutant in IκBα−/− MEFs to observe potential differences in the absence of IκBα-mediated promoter clearance. In addition, this experimental setting revealed the importance of Ser468 for the control of p65 stability (supplementary Fig 3 online). The differential stabilities of p65 and its point-mutated version were also shown by pulse-chase experiments, which showed a clearly increased half-life for the mutant p65 protein at early time points (Fig 1C). The amount of the mutant p65 protein was significantly reduced at later time points, which suggests the occurrence of further degradation mechanisms such as caspase- or serine protease-mediated elimination of p65 (Franzoso et al, 1994; Ravi et al, 1998). Accordingly, prolonged treatment of cells with TNF and CHX also resulted in the destabilization of the p65 S468A mutant (supplementary Fig 4 online), suggesting that this phosphorylation site regulates early, but not late, degradation of p65. As phosphorylation of p65 at Ser468 can be mediated either by IKKβ (Schwabe & Sakurai, 2005) or IKKɛ (Mattioli et al, 2006), we compared TNF-induced p65 decay between wild-type MEFs and cells lacking either IKKβ or IKKɛ (Fig 2A). A comparative analysis of TNF-induced p65 degradation showed that the protein was completely degraded in wild-type cells 10 h after the TNF pulse, whereas cells lacking IKKβ showed incomplete p65 decay at this time point. The residual p65 degradation at later time points might be attributable to other degradation pathways using Ser468 phosphorylation-independent mechanisms (Lawrence et al, 2005). By contrast, the p65 protein was largely stable in cells deficient for IKKɛ, revealing a crucial contribution of IKKɛ-mediated p65 phosphorylation for the control of its stability. NF-κB p65 phosphorylation was absent in the knock-out cells (Fig 2B), thus revealing another phosphorylation-dependent ubiquitination switch in the NF-κB system.
Figure 1.
Phosphorylation of p65 at Ser468 controls TNF-induced p65 degradation. (A) NF-κB p65 degradation in response to various stimuli. MEFs were stimulated with TNF (15 min), LPS (30 min) or IL-1β (30 min) as shown. After washout of the stimuli, the protein synthesis blocker cycloheximide (CHX; 10 μg/ml) was added for 10 h. Equal amounts of protein contained in the total cell extracts were analysed by immunoblotting (IB) for the occurrence of p65, the caspase-cleaved PARP fragment (indicated as PARP*) and β-actin as shown. Phosphorylation of IκBα was shown by a phospho-specific antibody. (B) p65−/− MEFs were transiently transfected to express HA-p65 or the HA-p65 S468A mutant as indicated. After 36 h, cells were stimulated with a 15-min pulse of TNF, followed by the addition of cycloheximide for the indicated periods. Total cell extracts were tested by immunoblotting for p65 and β-actin protein levels. (C) HEK 293 cells transfected to express p65 or p65 S468A were metabolically labelled with 35S-methionine and cysteine. After the addition of TNF and unlabelled cysteine and methionine, lysates were prepared at the indicated time points and the p65 protein was immunoprecipitated by αHA antibodies. The p65 protein was further analysed by SDS–PAGE, and an autoradiography of a dried gel is shown. HA, haemagglutinin; HEK, human embryonic kidney; IL-1β, interleukin-1β; LPS, lipopolysaccharide; MEF, mouse embryonic fibroblast; NF-κB, nuclear factor-kappaB; PARP, poly (ADP-ribose) polymerase; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; TNF, tumour necrosis factor.
Figure 2.
IKKɛ is crucial for p65 decay. (A) Wild-type MEFs and MEFs deficient for IKKβ or IKKɛ were pulsed with TNF as shown, followed by the analysis of p65 stability with the protocol used in Fig 1A. (B) The wild-type and knockout MEFs were stimulated for 15 or 60 min with TNF, followed by the analysis of protein expression and p65 Ser 468 phosphorylation with specific antibodies. CHX, cycloheximide; IB, immunoblotting; IKK, IκB kinase; MEF, mouse embryonic fibroblast; TNF, tumour necrosis factor; WT, wild type.
p65 Ser468 phosphorylation enables ubiquitination
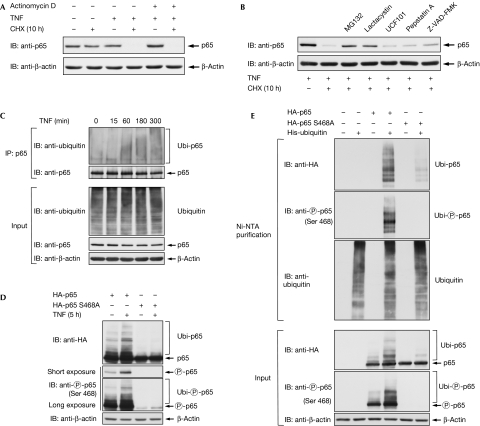
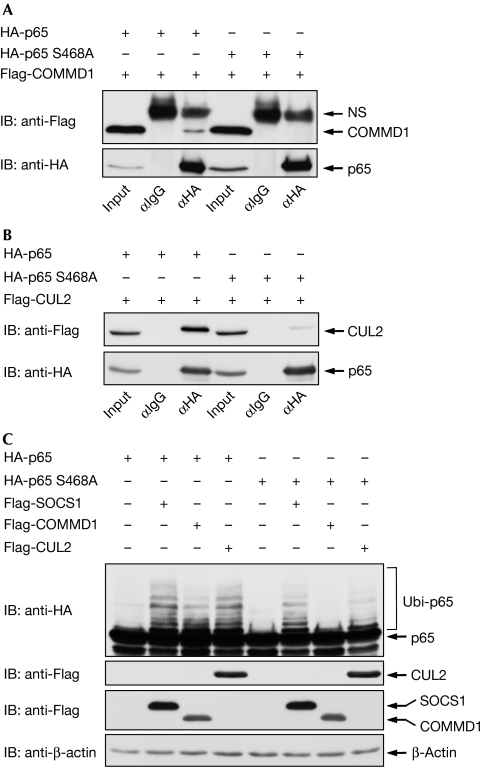
Many proteins that mediate the termination of the NF-κB response are themselves target genes of this transcription factor (Hayden & Ghosh, 2008). To investigate whether gene expression is required for p65 destabilization, transcription was blocked by actinomycin D before TNF treatment and the analysis of p65 stability. These experiments showed full degradation of p65 in the absence of transcription (Fig 3A), revealing that this process does not depend on de novo gene expression. Similarly, preincubation with CHX also allowed the occurrence of p65 degradation (supplementary Fig 5 online). The stability of p65 can be controlled by caspases (Ravi et al, 1998), serine proteases (Franzoso et al, 1994) and the proteasome (Saccani et al, 2004). To reveal the pathway involved in TNF-induced elimination of p65, cells were preincubated with various inhibitors targeting distinct pathways. Inhibition of various NF-κB-regulating kinases did not have any impact on p65 stability (supplementary Fig 6 online). The test of various protease inhibitors showed only a slight stabilization of p65 by the caspase inhibitor Z-VAD-FMK, whereas inducible degradation of p65 was largely blocked by the proteasome inhibitors, MG132 and lactacystin (Fig 3B). Given the relevance of the proteasome for the control of p65 stability, we next compared p65 ubiquitination between unstimulated and TNF-treated cells. Although ubiquitination of p65 was already detectable in unstimulated cells, it was significantly increased by TNF treatment (Fig 3C). We then compared ubiquitination between the wild-type transcription factor and the point mutant defective in Ser468 phosphorylation. Cells were transfected with the various p65 expression plasmids and either left untreated or stimulated for 5 h with TNF. Cell extracts were analysed by immunoblotting for p65 phosphorylation and ubiquitination. Basal p65 ubiquitination was enhanced in TNF-stimulated cells, whereas the p65 S468A mutant showed a strongly impaired ubiquitination of p65 (Fig 3D). To explore p65 ubiquitination using another approach, the transfected cells were lysed and His-tagged ubiquitin was enriched on nickel-nitrilotriacetic acid (Ni-NTA) columns. Subsequent immunoblotting clearly identified the upshifted bands as both ubiquitinated and Ser468 phosphorylated p65, and also confirmed the strongly diminished ubiquitination in the Ser468-mutated p65 protein (Fig 3E). A steadily growing number of enzymes from the ubiquitination machinery have been identified in the NF-κB system (Krappmann & Scheidereit, 2005; Wullaert et al, 2006). Many of these modifiers show functional redundancy, as, for example betaTrCP—the receptor subunit of a Skp1–Cul1–F-box (SCF)-type ubiquitin E3 ligase—controls the ubiquitination of IκB and also the p50 precursor p105 (Yaron et al, 1998; Lang et al, 2003). Thus, we screened a large set of ubiquitin-modifying proteins with relevance to NF-κB regulation for binding to p65. Coimmunoprecipitation experiments under buffer conditions maintaining ubiquitination confirmed published interactions and also revealed several new p65 interaction partners (data not shown). All p65 interacting proteins were then systematically compared for potential differences in binding to p65 wild-type or the p65 S468A mutant. Most interactors showed no differences in binding affinities, as exemplified by SOCS1 (supplementary Fig 7 online). By contrast, binding of COMMD1 and CUL2, which are both contained in a multimeric ubiquitin ligase complex targeting p65, was only detectable for the wild-type subunit (Fig 4A,B). Control experiments ensured the occurrence of p65 phosphorylation at under these conditions (supplementary Fig 8 online). Accordingly, expression of COMMD1 triggered p65 ubiquitination, whereas ubiquitination of p65 S468A was largely unaffected. In addition, CUL2-induced p65 ubiquitination was strongly diminished for the Ser468 mutant (Fig 4C). How can phosphorylation of a single amino acid control the binding to a multimeric ubiquitin ligase complex? It is tempting to speculate that the phosphorylation of Ser468 affects the p65 conformation. A previous study revealed intramolecular binding between the C- and amino-terminal p65 portions (closed conformation) that could be disrupted by Ser276 phosphorylation (open conformation; Zhong et al, 1998). This model implicates that relief of intramolecular masking by Ser468 phosphorylation allows for binding of the COMMD1 complex and subsequent ubiquitination.
Figure 3.
Ubiquitin/proteasome-dependent degradation of p65. (A) IκBα−/− MEFs were pre-incubated with actinomycin D (1 μg/ml) for 1 h and pulsed for 15 min with TNF. After washout of this stimulus, cells were further incubated for 10 h in the presence of CHX. The levels of p65 and β-actin were analysed by immunoblotting (IB) as indicated. (B) TNF-induced elimination of the endogenous p65 protein was triggered in IκBα−/− MEFs, as described in Fig 1A, but cells were pretreated for 1 h with the following inhibitors: the proteasome inhibitors MG132 (10 μM) and lactacystin (10 μM), the Omi/Htr A protease inhibitor UCF101 (10 μM), aspartic protease inhibitor pepstatin A (20 μM) and the broad-spectrum caspase inhibitor Z-VAD-FMK (5 μM) as shown. Cell extracts were further analysed by immunoblotting using the indicated antibodies. (C) IκBα−/− MEFs were stimulated with TNF for the indicated periods, followed by lysis in a buffer inhibiting ubiquitin isopeptidases and thus maintaining ubiquitination. After immunoprecipitation (IP) of p65, its ubiquitination was shown by immunoblotting. The lower panels show the input material. (D) HEK 293 cells were transfected to express either HA-p65 or S468A mutant. After 36 h, cells were stimulated with TNF for 5 h. After denaturing lysis, ubiquitination of p65 was shown by immunoblotting with the indicated antibodies. The short exposure time (upper panel) of the Western blot is more suitable for the detection of p65 Ser468 phosphorylation, whereas the long exposure time (lower panel) also allows phosphorylation of the ubiquitinated p65 protein to be observed. (E) HEK 293T cells were transfected with expression vectors for HA-p65, HA-p65 S468A and His-tagged ubiquitin as shown. An aliquot thereof was lysed under denaturing conditions, followed by purification of His-tagged ubiquitin on Ni-NTA agarose columns (Qiagen, Hilden, Germany). Upper panel: the eluted proteins were shown by immunoblotting using the indicated antibodies. Lower panel: another aliquot was tested by Western blotting as in (D). CHX, cycloheximide; HA, haemagglutinin; HEK, human embryonic kidney; MEF, mouse embryonic fibroblast; Ni-NTA, nickel-nitrilotriacetic acid; Omi/Htr, Omi stress-regulated endoprotease/5-hydroxytryptamine receptor; TNF, tumour necrosis factor; Ubi, ubiquitinated p65; UCF101, furfurylidine-thiobarbituric acid.
Figure 4.
p65 Ser468 controls binding to COMMD1 and CUL2. HEK 293T cells were transiently transfected with HA-tagged p65 proteins either alone or together with (A) Flag-tagged COMMD1 or (B) CUL2 as shown. Following immunoprecipitation (IP) with either HA or control IgG antibodies, precipitated proteins were shown by immunoblotting (IB). (C) Cells were transfected to express limited amounts of the indicated HA-p65 proteins along with different components of the multimeric COMMD1 ubiquitin ligase complex as shown. After denaturing lysis, p65 ubiquitination and protein expression were shown by immunoblotting. COMMD1, COMM domain-containing 1; CUL2, cullin 2; HA, haemagglutinin; HEK, human embryonic kidney; IgG, immunoglobulin G; NS, non specific; SOCS1, suppressor of cytokine signalling; Ubi, ubiquitinated p65.
p65 degradation occurs at selective promoters
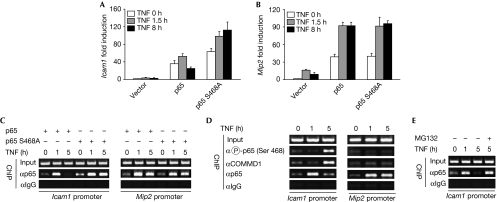
At which NF-κB target genes does elimination of p65 occur? To investigate this question, p65-deficient MEFs retransfected to express p65 wild-type or the S468A point mutant were left untreated or stimulated with TNF for various time points. Gene expression was monitored by a low-density microarray covering highly regulated inflammatory genes (Thiefes et al, 2005). Analysis of the 16 p65-dependent genes on this array revealed that the gene-regulating capacity of the p65 S468A mutant was strictly dependent on the individual target gene (supplementary Fig 9 online). As shown for the intercellular adhesion molecule 1 (Icam1) gene, real-time PCR experiments revealed that p65-dependent gene expression was already diminished 8 h after the stimulation of TNF (Fig 5A). By contrast, p65 S468A-induced transcription was not impaired under similar conditions. Other genes such as macrophage inflammatory protein 2 (Mip2) did not show any difference between p65 and the phosphorylation mutant (Fig 5B). Consistent with the observed selectivity for NF-κB target genes, we found that only a small subfraction of total p65 was phosphorylated at Ser468 (supplementary Fig 10 online). These gene expression data also imply that ubiquitin/proteasome-dependent degradation of p65 from its cognate promoters does not target the immediate early events in gene expression. Chromatin immunoprecipitation (ChIP) assays were performed to investigate whether these differences are reflected by p65 promoter occupancy. Expression of p65 was sufficient to cause basal association of p65 to the Icam1 promoter, which was further enhanced after 1 h of TNF treatment. At 5 h after TNF addition, the amount of promoter-associated p65 was strongly diminished (Fig 5C). By contrast, the p65 S468A mutant protein remained associated with its binding site even after prolonged periods of TNF treatment. Further ChIP experiments revealed the recruitment of Ser468 phosphorylated p65 and COMMD1 to the Icam1 but not to the Mip2 promoter (Fig 5D). Phosphorylated p65 is detected only at later time points and not at early phases when Icam1 transcription is fully active, but the mechanisms ensuring its retarded recruitment are unknown. Consistently, Ser468 phosphorylation can be detected even 5 h after the stimulation of TNF for expressed (supplementary Fig 2 online) and endogenous p65 (supplementary Fig 11 online). Accordingly, a previous report showed increased Icam1 transcription after knockdown of COMMD1 (Maine et al, 2007). To reveal the importance of ubiquitin/proteasome-mediated events for the removal of p65 from its cognate binding site at the Icam1 promoter, we repeated the ChIP experiments in the presence of MG132. The induced removal of p65 after 5 h of TNF stimulation was fully inhibited by the proteasome inhibitor (Fig 5E).
Figure 5.
Phosphorylation-induced p65 degradation occurs at selective NF-κB target genes. (A,B) p65−/− MEFs were transfected as shown. The next day, cells were left untreated or stimulated with TNF for 1.5 and 8 h. Gene expression of Icam1 and Mip2 was assessed by real-time PCR and normalized for β-actin expression. Fold activation relative to unstimulated cells transfected with empty expression vector is shown. Experiments were performed in triplicate and error bars show standard deviations. (C) p65−/− MEFs were transfected as shown, followed by stimulation of TNF for the indicated time points, and ChIP analysis using the indicated specific and control antibodies. Immunoprecipitates from each sample were analysed by PCR with specific primers. Input represents the PCR product from chromatin obtained before immunoprecipitation. PCR products were separated by agarose gel electrophoresis, and an ethidium bromide-stained gel is shown. (D) MEFs were stimulated with TNF for the indicated periods and analysed by ChIP for the recruitment of p65, phospho-p65 and COMMD1 as shown. (E) The experiment was performed as in (D) with the exception that cells were pretreated for 1 h with MG132 (10 μM) as shown. ChIP, chromatin immunoprecipitation; COMMD1, COMM domain-containing 1; ICAM1, intercellular adhesion molecule 1; MEF, mouse embryonic fibroblast; MIP2, macrophage inflammatory protein 2; NF-κB, nuclear factor-kappaB; TNF, tumour necrosis factor.
In summary, these data reveal a new negative feedback loop in the NF-κB system that contributes to the promoter-specific termination of the NF-κB response. Similar to the other feedback loops, this event occurs with a characteristic time delay, thereby allowing full NF-κB function during the interim period (Renner & Schmitz, 2009). The relative contribution of the various mechanisms used for NF-κB feedback inhibition will be a relevant point of future research.
Methods
Microarray and ChIP assays. Total RNA was isolated using the RNeasy kit (Qiagen, Hilden, Germany). Microarray experiments were performed using the first version of the mouse inflammation microarray (OciChip; Winzen et al, 2007). ChIP experiments were performed as described previously (Wolter et al, 2008) using specific antibodies and the respective IgG control antibody (Rockland, Gilbertsville, PA, USA) according to the recommendations of the manufacturer. Immunoprecipitated DNA was detected by 35 cycles of PCR using Taq DNA Polymerase (Fermentas, St Leon-Rot, Germany).
Further methods are available as supplementary information.
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Supplementary Material
Supplementary Materials and Methods
Acknowledgments
We thank all colleagues mentioned in the online section of the ‘Methods' who provided valuable reagents to make this study possible. Our study was supported by grants from the Deutsche Forschungsgemeinschaft projects SCHM 1417/4-1, SCHM 1417/5-1, SFB 547 and the ECCPS (Excellence Cluster Cardio-Pulmonary System).
Footnotes
The authors declare that they have no conflict of interest.
References
- Buss H, Dorrie A, Schmitz ML, Frank R, Livingstone M, Resch K, Kracht M (2004) Phosphorylation of serine 468 by GSK-3β negatively regulates basal p65 NF-κB activity. J Biol Chem 279: 49571–49574 [DOI] [PubMed] [Google Scholar]
- Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH (2007) Negative regulation of toll-like receptor signaling by NF-κB p50 ubiquitination blockade. Science 317: 675–678 [DOI] [PubMed] [Google Scholar]
- Franzoso G, Biswas P, Poli G, Carlson LM, Brown KD, Tomita-Yamaguchi M, Fauci AS, Siebenlist UK (1994) A family of serine proteases expressed exclusively in myelo-monocytic cells specifically processes the nuclear factor-κ B subunit p65 in vitro and may impair human immunodeficiency virus replication in these cells. J Exp Med 180: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker H, Karin M (2006) Regulation and function of IKK and IKK-related kinases. Sci STKE 2006: 13. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S (2008) Shared principles in NF-κB signaling. Cell 132: 344–362 [DOI] [PubMed] [Google Scholar]
- Krappmann D, Scheidereit C (2005) A pervasive role of ubiquitin conjugation in activation and termination of IκB kinase pathways. EMBO Rep 6: 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang V, Janzen J, Fischer GZ, Soneji Y, Beinke S, Salmeron A, Allen H, Hay RT, Ben-Neriah Y, Ley SC (2003) TrCP-mediated proteolysis of NF-κB1 p105 requires phosphorylation of p105 serines 927 and 932. Mol Cell Biol 23: 402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, Bebien M, Liu GY, Nizet V, Karin M (2005) IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature 434: 1138–1143 [DOI] [PubMed] [Google Scholar]
- Li Q, Verma IM (2002) NF-κB regulation in the immune system. Nat Rev Immunol 2: 725–734 [DOI] [PubMed] [Google Scholar]
- Maine GN, Mao X, Komarck CM, Burstein E (2007) COMMD1 promotes the ubiquitination of NF-κB subunits through a cullin-containing ubiquitin ligase. EMBO J 26: 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli I, Geng H, Sebald A, Hodel M, Bucher C, Kracht M, Schmitz ML (2006) Inducible phosphorylation of NF-κ B p65 at serine 468 by T cell costimulation is mediated by IKK ζ. J Biol Chem 281: 6175–6183 [DOI] [PubMed] [Google Scholar]
- Ravi R, Bedi A, Fuchs EJ (1998) CD95 (Fas)-induced caspase-mediated proteolysis of NF-κB. Cancer Res 58: 882–886 [PubMed] [Google Scholar]
- Renner F, Schmitz ML (2009) Autoregulatory feedback loops terminating the NF-κB response. Trends Biochem Sci (in press) [DOI] [PubMed] [Google Scholar]
- Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP (2003) Regulation of NF-κB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell 12: 1413–1426 [DOI] [PubMed] [Google Scholar]
- Saccani S, Marazzi I, Beg AA, Natoli G (2004) Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor κB response. J Exp Med 200: 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe RF, Sakurai H (2005) IKKβa phosphorylates p65 at S468 in transactivation domain 2. FASEB J 19: 1758–1760 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Grusby MJ, Kaisho T (2007) PDLIM2-mediated termination of transcription factor NF-κB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol 8: 584–591 [DOI] [PubMed] [Google Scholar]
- Thiefes A, Wolter S, Mushinski JF, Hoffmann E, Dittrich-Breiholz O, Graue N, Dorrie A, Schneider H, Wirth D, Luckow B, Resch K, Kracht M (2005) Simultaneous blockade of NFκB, JNK, and p38 MAPK by a kinase-inactive mutant of the protein kinase TAK1 sensitizes cells to apoptosis and affects a distinct spectrum of tumor necrosis factor [corrected] target genes. J Biol Chem 280: 27728–27741 [DOI] [PubMed] [Google Scholar]
- Wietek C, O'Neill LA (2007) Diversity and regulation in the NF-κB system. Trends Biochem Sci 32: 311–319 [DOI] [PubMed] [Google Scholar]
- Winzen R, Thakur BK, Dittrich-Breiholz O, Shah M, Redich N, Dhamija S, Kracht M, Holtmann H (2007) Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol Cell Biol 27: 8388–8400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter S, Doerrie A, Weber A, Schneider H, Hoffmann E, von der Ohe J, Bakiri L, Wagner EF, Resch K, Kracht M (2008) c-Jun controls histone modifications, NF-κB recruitment, and RNA polymerase II function to activate the ccl2 gene. Mol Cell Biol 28: 4407–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullaert A, Heyninck K, Janssens S, Beyaert R (2006) Ubiquitin: tool and target for intracellular NF-κB inhibitors. Trends Immunol 27: 533–540 [DOI] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y (1998) Identification of the receptor component of the IκBα-ubiquitin ligase. Nature 396: 590–594 [DOI] [PubMed] [Google Scholar]
- Zhong H, Voll RE, Ghosh S (1998) Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell 1: 661–671 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods