Abstract
The COP9 signalosome (CSN) is a highly conserved protein complex that was originally described as a repressor of light-dependent growth and transcription in Arabidopsis. The most studied CSN function is the regulation of protein degradation, which occurs primarily through the removal of the ubiquitin-like modifier Nedd8 from cullin-based E3 ubiquitin ligases. This activity can regulate transcription-factor stability and, therefore, transcriptional activity. Recent data suggest that the CSN also regulates transcription on the chromatin by mechanisms that are not yet clearly understood. Furthermore, the CSN subunits CSN5 and CSN2 seem to act as transcriptional coactivators and corepressors, respectively. Here, I re-evaluate the mechanisms by which the CSN acts as a transcriptional regulator, and suggest that they could extend beyond the regulation of protein stability.
Keywords: Alien, gene expression, Jab1, proteasome, transcriptional regulation
Glossary
AP-1 activator protein 1
β-TrcP β-transducin repeat containing protein
Cab chlorophyll A/B binding protein
Ccnd2 gene encoding cyclin D2
CDK cyclin-dependent kinase
Cdkn1a gene encoding p21
ChIP chromatin immunoprecipitation
CSA Cockayne syndrome WD repeat protein
Cul1 cullin 1
DAX1 dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome protein 1
DDB damaged DNA binding protein
E2f1 transcription factor E2f1
HEK human embryonic kidney
IκB inhibitor of κB
Jab1 Jun activation domain-binding protein 1/COP9 signalosome 5
JAMM Jab1/MPN/MOV34 domain-containing metalloisopeptidase
MPR1p multistep phosphorelay protein 1
MPN MPR1p and PAD1p amino-terminal
NAP-1 nucleosome assembly protein 1
Nedd8 neural precursor cell expressed, developmentally down-regulated 8
NF-κB nuclear factor-κB
PAD1p phenylacrylic acid decarboxylase protein
PCI proteasome-COP9-eukaryotic initiation factor 3
PCNA proliferating cell nuclear antigen
Pdcd4 programmed cell death 4
Polα DNA polymerase α-catalytic subunit
rbcS ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit
Rbf retinoblastoma family
RFX-1 MHC class II regulatory factor X, 1
SAP130 Sin3A-associated protein
SCF Skp1–cullin–F-box
Skp2 S-phase kinase associated 2
Slimb supernumerary limbs
TCR T-cell receptor
TIM timeless
Ubp12/USP15 ubiquitin specific protease
Introduction: the many functions of the CSN
The COP9 signalosome (CSN) is a highly conserved protein complex that consists of eight subunits known as CSN1 to CSN8 in higher eukaryotes (Fig 1; Deng et al, 2000). The complex was originally discovered as a repressor of light-dependent growth in Arabidopsis (Chamovitz et al, 1996; Wei et al, 1994). Subsequent work has identified and characterized the CSN in mammals, Drosophila, budding and fission yeast, fungi, Dictyostelium and Caenorhabditis elegans, highlighting its role as a general modulator of diverse cellular and developmental processes (reviewed in Wei et al, 2008).
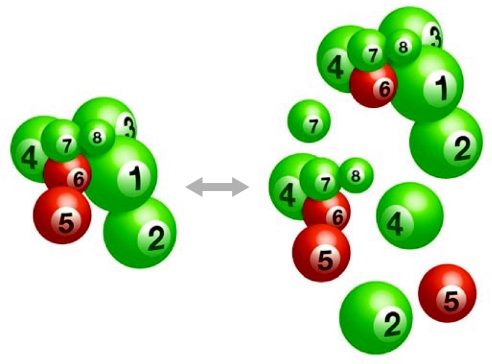
Figure 1.
Dynamic equilibrium among the eight-subunit CSN core complex, partial complexes and subunit monomers. Not all partial complexes or monomers are known or shown, and the composition of the partial complexes is probably both tissue and organism specific. The CSN4–CSN8 complex was described in human cells as the JAC (Tomoda et al, 2002). The size of each ball is proportional to the relative protein size. The subunit arrangement is shown according to Sharon et al (2009). The green balls represent PCI-containing subunits and the red balls represent MPN-containing subunits. CSN, COP9 signalosome; JAC, Jab1-associated complex; MPN, MPR1p and PAD1p amino-terminal; PCI, proteasome-COP9-eukaryotic initiation factor 3.
The most studied function of the CSN is the regulation of protein degradation, which is carried out by several mechanisms (Table 1). The CSN removes Nedd8—a ubiquitin-like modifier—from cullin-based E3 ubiquitin ligases (Lyapina et al, 2001), thereby regulating ligase activity. The deneddylation activity resides in the JAMM/MPN+ domain of CSN5 (Cope et al, 2002; Maytal-Kivity et al, 2002), and the intact complex is necessary and sufficient for deneddylation (Sharon et al, 2009). This activity of the CSN is well established, although not all cullin-based E3 ligase activities are sensitive to CSN-mediated deneddylation. For example, the signal-dependent degradation of IκB—which is mediated by SCFβ-TrcP—seems to be unaffected in mammalian cells after the knockdown of different CSN subunits (Harari-Steinberg et al, 2007; Menon et al, 2007; Panattoni et al, 2008; Schweitzer et al, 2007). Surprisingly, the TCR-stimulated downregulation of p27—mediated by SCFSkp2—is also unaffected in CSN8-deficient T cells (Menon et al, 2007). Similarly, the signal-dependent degradation of the IκB homologue Cactus and the clock protein TIM—which is mediated by SCFSlimb—is unaffected in Drosophila csn5null mutants (Harari-Steinberg et al, 2007; Knowles et al, 2009). Several excellent reviews have elaborated on the role of the CSN as a deneddylase, which is therefore not covered here (Cope & Deshaies, 2003; Wei & Deng, 2003; Wolf et al, 2003).
Table 1.
Activities ascribed to the CSN or CSN subunits
| Activity | References |
|---|---|
| Deneddylation | Lyapina et al, 2001 |
| Deubiquitination | Wu et al, 2006; Zhou et al, 2003 |
| Kinase activity | Naumann et al, 1999; Seeger et al, 1998 |
| Regulation of protein subcellular localization | Chamovitz et al, 1996; Tomoda et al, 2002; Wang et al, 2009 |
| Transcriptional coactivation/corepression | Claret et al, 1996; Dressel et al, 1999 |
CSN, COP9 signalosome.
However, CSN5-mediated deneddylation seems to be only one—albeit an extremely important one—of the CSN functions (Table 1). Indeed, if CSN5-mediated deneddylation were the only activity of the CSN, one would expect that loss of CSN5 function would be phenotypically equal to loss of the entire complex. This is indeed the case for null mutations in Arabidopsis and mice (Dohmann et al, 2005; Gusmaroli et al, 2007; Lykke-Andersen et al, 2003; Tomoda et al, 2004; Yan et al, 2003), whereas it is not the case in Drosophila and Schizosaccharomyces pombe. Although mutations in Drosophila CSN4, CSN5 and CSN8 all result in larval lethality, the mutants are characterized by overlapping—but unique—morphological phenotypes (Cope et al, 2002; Doronkin et al, 2003; Oren-Giladi et al, 2008; Oron et al, 2002). At a molecular level, the loss of the entire complex in Drosophila csn4null mutants leads to more severe transcriptome changes than the loss of only CSN5 in csn5null mutants, which maintain a complex that lacks CSN5 (Oron et al, 2007). Although this difference in transcriptomes might be explained by differences in the persistence of maternally contributed subunits, a similar picture arises from genetic studies in S. pombe. All the CSN subunit-null mutants of S. pombe that have been tested have impaired deneddylation of different cullins; however, only csn1-d and csn2-d mutants—and not csn5-d mutants—have obvious phenotypes (Mundt et al, 2002; Zhou et al, 2001). In addition, partial loss-of-function mutants of Arabidopsis csn1 and csn5 also show different growth phenotypes, as opposed to the null mutants of each subunit, which are phenotypically equivalent (Gusmaroli et al, 2007; Wang et al, 2002, 2003). In summary, although CSN5-mediated deneddylation is clearly dependent on the entire CSN—and is essential in most organisms—the loss of the entire complex can have different consequences than the loss of only CSN5.
One of the first functions of the CSN to be identified in mammalian cells was regulation of the phosphorylation of ubiquitin-proteasome pathway substrates through the activity of CSN-associated kinases (Naumann et al, 1999; Seeger et al, 1998). These kinases act on several crucial transcriptional regulators and much of the effect of the CSN on transcription can be ascribed to this activity (reviewed in Harari-Steinberg & Chamovitz, 2004).
A second CSN-associated activity—deubiquitination—has also garnered much attention (Hetfeld et al, 2005; Zhou et al, 2003). This activity—mediated by the CSN-associated Ubp12/USP15—also influences protein stability and has been proposed to stabilize E3 ligase subunits by preventing their promiscuous autoubiquitination (reviewed in Wu et al, 2006). This was recently shown to extend to E3 ligase substrates, the stability of which is dependent on the CSN. For example, following signal-dependent degradation of IκBα and the subsequent nuclear localization of NF-κB, CSN-dependent USP15-mediated deubiquitination stabilizes newly synthesized IκBα, thereby attenuating NF-κB activity (Schweitzer et al, 2007).
The CSN also has poorly defined roles in regulating the subcellular localization of crucial signalling molecules, including the COP1 E3 ubiquitin ligase (Chamovitz et al, 1996; Wang et al, 2009), the cell-cycle regulator p27 (Tomoda et al, 2002), p53 (Oh et al, 2006) and the IκB homologue Cactus (Harari-Steinberg et al, 2007).
Although the CSN was originally identified as a transcriptional repressor involved in Arabidopsis photomorphogenesis (Wei et al, 1994; Wei & Deng, 1992), much of the subsequent work has concentrated on its role as a regulator of protein degradation. Several recent studies have emphasized the need to re-evaluate the role of the CSN as a transcriptional regulator.
The CSN as a regulator of transcription
The CSN was originally described as a transcriptional repressor of a range of Arabidopsis genes that are normally repressed in the dark. As such, these ‘light-activated' genes are expressed in darkness in null mutants of CSN subunits (Wei & Deng, 1992). This repression was subsequently ascribed to the role of the CSN in regulating the stability of the transcription factors that are normally unstable and degraded in darkness, but are stabilized in csn mutants (reviewed in Serino & Deng, 2003). This paradigm for the role of CSN in regulating the stability—and therefore the activity—of transcription factors and other signalling proteins has been shown in many systems (see Table 3 in Wei et al, 2008). In addition to light-regulated genes (Ma et al, 2003), numerous others are misregulated in Arabidopsis csn mutants, including those regulated by the phytohormones auxin (Dohmann et al, 2008a) and jasmonate (Feng et al, 2003), and those involved in cell-cycle regulation (Dohmann et al, 2008b).
The mutation of different CSN subunits in Drosophila leads to a misregulation of approximately 20% of the transcriptome during larval development (Oron et al, 2007). It is not surprising that mutations in a protein complex that lead to severe phenotypes and death also induce many transcriptome changes as an indirect consequence. However, many of these changes are observed in early larval development (mid-second instar)—before the onset of visible mutant phenotypes, but at a stage where maternally contributed proteins have been depleted, rendering the larva true null mutants. This indicates that a primary effect of CSN perturbation is a change in gene-expression profiles. The most obvious among these changes in Drosophila is the achronic expression of numerous developmentally regulated genes (Oron et al, 2007), the transcription of many of which is usually limited to embryogenesis or metamorphosis. These results indicate that, in the absence of the CSN, the transcription of these genes is derepressed, similar to the derepression of light-activated genes in dark-grown mutants of the Arabidopsis CSN. However, although most of these early misregulated genes were derepressed, the transcription of another large set of genes was repressed in the mutants relative to the wild type (Oron et al, 2007), indicating a positive requirement of the CSN for gene expression.
This positive requirement is in agreement with the role of the CSN in regulating photomorphogenesis. Close re-examination of the early work on the cop9 (csn8) mutant in Arabidopsis (Wei & Deng, 1992) reveals that the light-activated genes cab and rbcS are not only derepressed in the dark, but that their light-induced expression levels are lower than those of wild-type plants (N. Wei, personal communication; see Fig 5 in Wei & Deng, 1992). Similarly, although the expression of many auxin-induced genes is derepressed in Arabidopsis csn mutants, the induction of an auxin-responsive reporter gene in the presence of auxin is also reduced (Dohmann et al, 2008a), further illustrating a role of the CSN not only as a transcriptional repressor, but also as a transcriptional activator. This effect is mirrored in mice; during T-cell development, CSN5 is required for T-cell receptor-driven signals that are involved in positive selection (Panattoni et al, 2008), and the specific deletion of Csn8 in T cells leads to an impairment of signal-induced expression of cell cycle-related genes (Menon et al, 2007). For example, in the absence of CSN8, the genes encoding cyclin E1, cyclin D2 and E2f1 are elevated relative to wild-type mice—consistent with the repressive role of the CSN. However, on stimulation for cell-cycle re-entry, the expression of these genes could not be upregulated to wild-type levels.
Despite these clear effects on transcription, a question remains as to whether the mechanism of this regulation is based exclusively on the CSN-dependent stability of transcription regulators, or whether other mechanisms are also involved. Some CSN functions are clearly cytoplasmic (Schweitzer et al, 2007; Tomoda et al, 2002); however, could the nuclear CSN (Chamovitz et al, 1996; Mundt et al, 2002; Tomoda et al, 2002) regulate transcription directly? An amino-terminal fragment of human CSN1 has been shown to translocate to the nucleus and inhibit gene expression from several specific promoters (Tsuge et al, 2001). Although the mechanism of this inhibition is still unclear, it might be related to the fact that this part of CSN1 interacts with SAP130—a member of the DDB1 protein family and an established component of the transcription machinery (Menon et al, 2008).
Is the CSN a chromatin-based transcriptional regulator?
Several recent studies have shown that CSN subunits can localize to chromatin, further suggesting a direct role for the CSN in transcriptional regulation. The first indication that the CSN might be associated with chromatin came from the work of Groisman and colleagues (2003), who detected it in purified chromatin fractions from HeLa cells. They found that the CSN is bound to chromatin in association with both the CSA and DDB2-containing E3 ligase complexes, where it participates in regulating cellular responses to DNA damage.
A stable association between CSN subunits and chromatin was subsequently identified by ChIP experiments in both Drosophila (Ullah et al, 2007) and mammalian cells (Menon et al, 2007; Mori et al, 2008). Ullah and colleagues described a direct interaction between CSN4 and the Drosophila Rbf proteins, and showed that CSN4 could associate with Rbf-targeted promoters in both S2 cells and Drosophila embryos. CSN4 and Rbf simultaneously occupied the promoter region of the Polα and PCNA genes, suggesting that Rbf activity is regulated directly on the chromatin by the CSN, presumably through the modulation of Rbf stability.
The Groisman and Ullah papers are consistent with a role of the CSN in regulating transcription-factor stability and/or activity, albeit directly on the chromatin. The work of Menon et al (2007) further suggests that the CSN is a direct regulator of the transcription of cell cycle-related genes. By using Csn8-deficient T cells—which have impaired proliferation—the authors showed the normal, signal-induced degradation of several SCF substrates, including IκBα, p27 and Pdcd4. By contrast, the accumulation of G1 cyclins and CDKs was defective in these mutant cells, suggesting that the turnover of these proteins is affected by the loss of the CSN. Interestingly, treatment with the proteasome inhibitor MG132 did not restore the normal accumulation of G1 cyclins and CDKs in these Csn8-deficient cells, leading the authors to speculate that CSN-mediated transcriptional regulation—rather than direct regulation of protein turnover—might lead to the reduced levels of these proteins. Indeed, the expression of the genes encoding E2F1 and the G1 cyclins E1 and D2 could not be upregulated after TCR stimulation in the absence of Csn8, mirroring the corresponding protein-accumulation patterns. Both Csn1 and Csn8 were detected bound to the Ccnd2, Cdk4 and Cdkn1a promoters by ChIP assays; the localization of two CSN subunits—and therefore probably the entire CSN complex—at the promoter regions of genes encoding cell-cycle regulators suggests that the CSN has the ability to regulate transcription directly, and this might be—at least partly—protease independent.
The caveat to this hypothesis is that it is partly based on negative results—the lack of MG132 rescue of G1 cyclin and CDK protein levels. Therefore, the mechanism by which the CSN regulates transcription on chromatin—apart from the regulation of transcription-factor stability—remains to be identified.
Most of these studies describe chromatin-associated roles for the CSN in the context of cell-cycle regulation, although they are probably not limited to this process. Indeed, Ullah et al (2007) claimed that CSN4 immunoprecipitated with the promoters of other important Drosophila transcription factors, such as the dorsal/ventral patterning gene zerknullt, the Gap gene tailless and the segmentation gene fushi tarazu, although the data were not shown. Therefore, the CSN probably associates with genomic regions that regulate many developmental processes.
Is the PCI domain a DNA-binding motif?
The crystal structure of CSN7, which was recently published, could shed new light on the role of the CSN in transcriptional regulation (Dessau et al, 2008). CSN7, similar to five other CSN subunits (CSN1, CSN2, CSN3, CSN4 and CSN8), contains a PCI domain, which is also common to six subunits of the proteasome lid and six subunits of eIF3. This domain is thought to mediate and stabilize protein–protein interactions within the complexes (Halimi & Chamovitz, 2008; Hofmann & Bucher, 1998).
The PCI domain is comprised of two subdomains, an N-terminal helical bundle and a carboxy-terminal winged helix, which are intimately connected through a central helix (Dessau et al, 2008). Importantly for the CSN-mediated transcription discussed here, the winged helix subdomain comprises a canonical helix-turn-helix that has a structure and electrostatic potential similar to those of winged helix nucleic acid-binding proteins, such as RFX-1 (Gajiwala et al, 2000). This suggests that an intact CSN would have six winged helix domains with the potential to bind to nucleic acids. Does the presence of multiple winged helix domains in the CSN imply that it has direct involvement—either as a complex or as individual subunits—in nucleotide binding? Although multiple CSN subunits have been shown to associate with chromatin, there is currently no evidence of direct nucleotide binding. However, two additional lines of evidence suggest that this possibility is worthy of further study. First, the CSN has been shown to bind to heparin, which is a highly sulphated glycosaminoglycan with the ability to bind to nucleic acid-binding proteins (Chamovitz et al, 1996). Second, theoretical modelling shows that the winged helix domain of CSN7 can dock with nucleic acids (see Supplementary Fig 11 in Dessau et al, 2008). Nonetheless, whether such a direct interaction occurs in vivo remains to be determined by further experimentation.
Which forms of the CSN regulate transcription?
The exact functional configuration of the CSN subunits is not always clear. Some of the eight subunits are detected only in a ‘core complex' of approximately 500 kDa (also known as the COP9 signalosome), whereas others—such as CSN2, CSN4, CSN5, CSN6 and CSN7—are also detected independently of the core complex. The distribution of these subunit conformations might be dependent on the organism, tissue or specific conditions analysed, and several studies have suggested various equilibria between these different subunit forms (Fukumoto et al, 2005; Gusmaroli et al, 2007; Oron et al, 2002; Tsuge et al, 2001).
Deneddylation is clearly dependent on CSN5 within the context of the entire eight-subunit core complex (Cope et al, 2002; Sharon et al, 2009). A recombinant human CSN assembled in Escherichia coli shows deneddylation activity against Cul1, whereas a CSN complex that lacks CSN5 has no such activity (Sharon et al, 2009), although the complex can be readily detected in vivo and in vitro (Dohmann et al, 2005; Oron et al, 2002; Sharon et al, 2009).
Two subunits in particular—CSN2 and CSN5—seem to be stable in the absence of the CSN (Gusmaroli et al, 2007). In the model proposed by Sharon and colleagues (2009), these two subunits protrude from the core of the CSN and are each connected to it by a single interaction—CSN5 to CSN6 and CSN2 to CSN1—such that the loss of either subunit is not expected to affect the overall complex stability (Fig 1). CSN5 and CSN2 are the two subunits for which the most evidence exists regarding their roles independent of the CSN core complex. CSN5—also known as Jab1—was originally identified as a coactivator (with Jun1) of AP-1 transcription factor-binding sites (Claret et al, 1996), and has since been shown to interact with numerous proteins affecting diverse signalling pathways, both cytoplasmic and nuclear (Chamovitz & Segal, 2001; Richardson & Zundel, 2005; Zhang et al, 2008). CSN2—also known as Alien—was originally identified as a corepressor of steroid hormone signalling (Dressel et al, 1999). Alien interacts with a subset of nuclear hormone receptors—such as the human thyroid and DAX1 receptors and the Drosophila ecdysone and seven-up receptors—in the absence of hormone. The addition of the appropriate hormones leads to the dissociation of Alien from the receptor, allowing for hormone-mediated transcription (Altincicek et al, 2000; Dressel et al, 1999). Indeed, many of the physical phenotypes and transcriptome alterations identified in Drosophila csn mutants could be ascribed to the impaired function of the ecdysone receptor (Oren-Giladi et al, 2008; Oron et al, 2002, 2007). However, characterizing the function of CSN2/Alien in mammals is not as straightforward because CSN2 and Alien are two different splicing variants of the same locus. CSN2 is the full-length gene product, whereas Alien refers to the N-terminal 300 amino-acid residues of CSN2, which interestingly lack part of the PCI domain (Tenbaum et al, 2003). Whether the Alien form of CSN2 assembles into the CSN is not yet clear. Alien has also been recently shown to function directly on the chromatin, where it enhances NAP-1-mediated nucleosome assembly, thereby participating in gene silencing (Eckey et al, 2007). Although Alien was found to associate with chromatin in HEK293 cells in this study, both CSN8 and CSN2 were detected in the cytoplasm. The discrepancy between these results, and the nuclear localization of CSN8 and other subunits found in other studies and discussed above, remains to be resolved.
Outlook and perspectives
Seventeen years after the initial description of COP9 as a transcriptional repressor (Wei & Deng, 1992), the mechanisms by which the CSN regulates transcription are beginning to be understood. Recent studies have shed light on the roles of the CSN in regulating transcription directly on the chromatin, although many important questions remain regarding how this is achieved on a mechanistic level (Sidebar A). In contemplating a chromatin-based role, we must keep in mind that the CSN is not the only component of the ubiquitin-proteasome system to be implicated in transcriptional control on the chromatin. For example, the entire yeast proteasome is able to associate with regulatory sequences directly on the chromatin (Sikder et al, 2006). Therefore, it is conceivable that the CSN could participate in a chromatin-localized supercomplex with other components of the ubiquitin-proteasome system. Interestingly, although the proteasome obviously has a proteolytic role in regulating transcription-factor stability directly on the chromatin (Saccani et al, 2004), it is also thought to regulate transcription by nonproteolytic mechanisms (Lassot et al, 2007). Therefore, although solid positive data are still lacking, a nonproteolytic role for the CSN—or at least for some CSN subunits—remains a possibility. Future work is expected to unravel the complex regulatory mechanisms of the CSN and its individual subunits in regulating gene expression.
Sidebar A | In need of answers.
Is the COP9 signalosome (CSN) a transcriptional regulator? The activity of the CSN definitely influences both the repression and the activation of a range of genes. This activity was originally ascribed to CSN-mediated stability of transcription factors and other regulatory proteins. Recent work indicates that the CSN can associate with specific promoters to regulate transcription, although whether this interaction is direct—or mediated by other proteins—and the mechanism by which the CSN affects transcription on the chromatin are not known.
Where in the cell does the CSN regulate transcription? Probably both in the cytoplasm and in the nucleus. Individual subunits have been detected in various subcellular compartments, including the cell membrane, cytoplasm and nucleus. However, as outlined in the text, some of the CSN functions seem to be mediated in the nucleus directly on the chromatin.
Is CSN-mediated transcriptional control achieved through its regulation of transcription-factor stability? The CSN definitely has a crucial role in regulating the stability of specific transcription factors. However, some roles of CSN subunits in transcriptional regulation might be protease independent.
Does the entire CSN, or do individual CSN subunits, regulate transcription? Probably both, although we do not completely understand the relationship between individual subunits and the CSN complex.
What is unique about CSN5 and CSN2? Both CSN5 and CSN2 have established functions that are independent of the CSN complex. Interestingly, CSN5 is a coactivator of transcription, whereas CSN2 is a corepressor. Could these subunits be the basis of CSN-mediated activation and repression? Both subunits have the weakest physical connection to the complex, and it is tempting to speculate that the CSN releases and rebinds to these proteins to regulate transcription differentially. Answering this question will require further study.
What regulates CSN-mediated transcription? We do not know how, which or even whether signals directly impinge on CSN activity. Certain CSN subunits can be phosphorylated (Fang et al, 2008; Harari-Steinberg & Chamovitz, 2004); however, we do not understand the implications of this modification for functionality.
What determines CSN specificity in transcriptional control? We do not have the answer to this question, although there are obviously tissue and environmental specificities that need to be considered both when analysing the primary literature and in planning future experiments.

Daniel A. Chamovitz
Acknowledgments
I thank N. Wei for helpful discussion and unpublished ideas, M. Sharon for providing her manuscript before publication, and M. Glickman, A. Yahalom and E. Oron for critical reading of the manuscript. Our research on the CSN is supported by the Israel Academy of Sciences, The United States–Israel Binational Agricultural Research and Development Fund, and the Israel Ministry of Sciences, Cooperation with Taiwan Grant.
References
- Altincicek B, Tenbaum SP, Dressel U, Thormeyer D, Renkawitz R, Baniahmad A (2000) Interaction of the corepressor Alien with DAX-1 is abrogated by mutations of DAX-1 involved in adrenal hypoplasia congenita. J Biol Chem 275: 7662–7667 [DOI] [PubMed] [Google Scholar]
- Chamovitz DA, Segal D (2001) JAB1/CSN5 and the COP9 signalosome. A complex situation. EMBO Rep 2: 96–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng XW (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86: 115–121 [DOI] [PubMed] [Google Scholar]
- Claret FX, Hibi M, Dhut S, Toda T, Karin M (1996) A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature 383: 453–457 [DOI] [PubMed] [Google Scholar]
- Cope G, Deshaies RJ (2003) COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114: 663–671 [DOI] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of NEDD8 from CUL1. Science 15: 15. [DOI] [PubMed] [Google Scholar]
- Deng XW et al. (2000) Unified nomenclature for the COP9 signalosome and its subunits: an essential regulator of development Trends Genet 16: 202–203 [DOI] [PubMed] [Google Scholar]
- Dessau M, Halimi Y, Erez T, Chomsky-Hecht O, Chamovitz DA, Hirsch JA (2008) The Arabidopsis COP9 signalosome subunit 7 is a model PCI domain protein with subdomains involved in COP9 signalosome assembly. Plant Cell 20: 2815–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann EM, Kuhnle C, Schwechheimer C (2005) Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 17: 1967–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann EM, Levesque MP, Isono E, Schmid M, Schwechheimer C (2008a) Auxin responses in mutants of the Arabidopsis Cop9 signalosome. Plant Physiol 147: 1369–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann EM, Levesque MP, De Veylder L, Reichardt I, Jurgens G, Schmid M, Schwechheimer C (2008b) The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development 135: 2013–2022 [DOI] [PubMed] [Google Scholar]
- Doronkin S, Djagaeva I, Beckendorf SK (2003) The COP9 signalosome promotes degradation of cyclin E during early Drosophila oogenesis. Dev Cell 4: 699–710 [DOI] [PubMed] [Google Scholar]
- Dressel U, Thormeyer D, Altincicek B, Paululat A, Eggert M, Schneider S, Tenbaum SP, Renkawitz R, Baniahmad A (1999) Alien, a highly conserved protein with characteristics of a corepressor for members of the nuclear hormone receptor superfamily. Mol Cell Biol 19: 3383–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckey M, Hong W, Papaioannou M, Baniahmad A (2007) The nucleosome assembly activity of NAP1 is enhanced by Alien. Mol Cell Biol 27: 3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Wang X, Yamoah K, Chen P-l, Pan Z-Q, Huang L (2008) Characterization of the human COP9 signalosome complex using affinity purification and mass spectrometry. J Proteome Res 7: 4914–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Ma L, Wang X, Xie D, Dinesh-Kumar SP, Wei N, Deng XW (2003) The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 15: 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto A, Tomoda K, Kubota M, Kato JY, Yoneda-Kato N (2005) Small Jab1-containing subcomplex is regulated in an anchorage- and cell cycle-dependent manner, which is abrogated by ras transformation. FEBS Lett 579: 1047–1054 [DOI] [PubMed] [Google Scholar]
- Gajiwala KS, Chen H, Cornille F, Roques BP, Reith W, Mach B, Burley SK (2000) Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 403: 916–921 [DOI] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113: 357–367 [DOI] [PubMed] [Google Scholar]
- Gusmaroli G, Figueroa P, Serino G, Deng XW (2007) Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant Cell 19: 564–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halimi Y, Chamovitz DA (2008) The PCI complexes and ubiquitin protesome system (UPS) in plant development. In Intracellular Signaling in Plants, Z Yang (Ed), Vol 33, pp 273–306. Oxford, UK: Blackwell [Google Scholar]
- Harari-Steinberg O, Chamovitz DA (2004) The COP9 signalosome: mediating between kinase signaling and protein degradation. Curr Protein Pept Sci 5: 185–189 [DOI] [PubMed] [Google Scholar]
- Harari-Steinberg O, Cantera R, Denti S, Bianchi E, Oron E, Segal D, Chamovitz DA (2007) COP9 signalosome subunit 5 (CSN5/Jab1) regulates the development of the Drosophila immune system: effects on Cactus, Dorsal and hematopoiesis. Genes Cells 12: 183–195 [DOI] [PubMed] [Google Scholar]
- Hetfeld BK, Helfrich A, Kapelari B, Scheel H, Hofmann K, Guterman A, Glickman M, Schade R, Kloetzel PM, Dubiel W (2005) The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr Biol 15: 1217–1221 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P (1998) The PCI domain: A common theme in three multi-protein complexes. Trends Biochem Sci 23: 204–205 [DOI] [PubMed] [Google Scholar]
- Knowles A, Koh K, Wu JT, Chien CT, Chamovitz DA, Blau J (2009) The COP9 signalosome is required for light-dependent TIM degradation and Drosophila clock resetting. J Neurosci 29: 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassot I, Latreille D, Rousset E, Sourisseau M, Linares LK, Chable-Bessia C, Coux O, Benkirane M, Kiernan RE (2007) The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol Cell 25: 369–383 [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Deshaies RJ (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen K, Schaefer L, Menon S, Deng X-W, Miller JB, Wei N (2003) Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol Cell Biol 23: 6790–6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhao H, Deng XW (2003) Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 130: 969–981 [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V, Reis N, Hofmann K, Glickman MH (2002) MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem 3: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Chi H, Zhang H, Deng XW, Flavell RA, Wei N (2007) COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat Immunol 8: 1236–1245 [DOI] [PubMed] [Google Scholar]
- Menon S, Tsuge T, Dohmae N, Takio K, Wei N (2008) Association of SAP130/SF3b-3 with Cullin-RING ubiquitin ligase complexes and its regulation by the COP9 signalosome. BMC Biochem 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Yoneda-Kato N, Yoshida A, Kato J-y (2008) Stable form of JAB1 enhances proliferation and maintenance of hematopoietic progenitors. J Biol Chem 283: 29011–29021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt KE, Liu C, Carr AM (2002) Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol Biol Cell 13: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M, Bech-Otschir D, Huang X, Ferrell K, Dubiel W (1999) COP9 signalosome-directed c-Jun activation/stabilization is independent of JNK. J Biol Chem 274: 35297–35300 [DOI] [PubMed] [Google Scholar]
- Oh W, Lee EW, Sung YH, Yang MR, Ghim J, Lee HW, Song J (2006) Jab1 induces the cytoplasmic localization and degradation of p53 in coordination with Hdm2. J Biol Chem 281: 17457–17465 [DOI] [PubMed] [Google Scholar]
- Oren-Giladi P, Krieger O, Edgar BA, Chamovitz DA, Segal D (2008) Cop9 signalosome subunit 8 (CSN8) is essential for Drosophila development. Genes Cells 13: 221–231 [DOI] [PubMed] [Google Scholar]
- Oron E, Mannervik M, Rencus S, Harari-Steinberg O, Neuman-Silberberg S, Segal D, Chamovitz DA (2002) COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 129: 4399–4409 [DOI] [PubMed] [Google Scholar]
- Oron E, Tuller T, Li L, Rozovsky N, Yekutieli D, Rencus-Lazar S, Segal D, Chor B, Edgar BA, Chamovitz DA (2007) Genomic analysis of COP9 signalosome function in Drosophila melanogaster reveals a role in temporal regulation of gene expression. Mol Syst Biol 3: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panattoni M, Sanvito F, Basso V, Doglioni C, Casorati G, Montini E, Bender JR, Mondino A, Pardi R (2008) Targeted inactivation of the COP9 signalosome impairs multiple stages of T cell development. J Exp Med 205: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson KS, Zundel W (2005) The emerging role of the COP9 signalosome in cancer. Mol Cancer Res 3: 645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Marazzi I, Beg AA, Natoli G (2004) Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J Exp Med 200: 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer K, Bozko PM, Dubiel W, Naumann M (2007) CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. EMBO J 26: 1532–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, Schade R, Gordon C, Naumann M, Dubiel W (1998) A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J 12: 469–478 [PubMed] [Google Scholar]
- Serino G, Deng X-W (2003) The COP9 signalosome: regulating plant development through control of proteolysis. Annu Rev Plant Biol 54: 165–182 [DOI] [PubMed] [Google Scholar]
- Sharon M, Mao H, Erba EB, Stephens E, Zheng N, Robinson CV (2009) Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure 17: 31–40 [DOI] [PubMed] [Google Scholar]
- Sikder D, Johnston SA, Kodadek T (2006) Widespread, but non-identical, association of proteasomal 19 and 20 S proteins with yeast chromatin. J Biol Chem 281: 27346–27355 [DOI] [PubMed] [Google Scholar]
- Tenbaum SP, Juenemann S, Schlitt T, Bernal J, Renkawitz R, Munoz A, Baniahmad A (2003) Alien/CSN2 gene expression is regulated by thyroid hormone in rat brain. Dev Biol 254: 149–160 [DOI] [PubMed] [Google Scholar]
- Tomoda K, Kubota Y, Arata Y, Mori S, Maeda M, Tanaka T, Yoshida M, Yoneda-Kato N, Kato JY (2002) The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem 277: 2302–2310 [DOI] [PubMed] [Google Scholar]
- Tomoda K, Yoneda-Kato N, Fukumoto A, Yamanaka S, Kato JY (2004) Multiple functions of Jab1 are required for early embryonic development and growth potential in mice. J Biol Chem 279: 43013–43018 [DOI] [PubMed] [Google Scholar]
- Tsuge T, Matsui M, Wei N (2001) The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J Mol Biol 305: 1–9 [DOI] [PubMed] [Google Scholar]
- Ullah Z, Buckley MS, Arnosti DN, Henry RW (2007) Retinoblastoma protein regulation by the COP9 signalosome. Mol Biol Cell 18: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kang D, Feng S, Serino G, Schwechheimer C, Wei N (2002) CSN1 N-terminal-dependent activity is required for Arabidopsis development but not for Rub1/Nedd8 deconjugation of cullins: a structure-function study of CSN1 subunit of COP9 signalosome. Mol Biol Cell 13: 646–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Feng S, Nakayama N, Crosby WL, Irish V, Deng XW, Wei N (2003) The COP9 signalosome interacts with SCF(UFO) and participates in Arabidopsis flower development. Plant Cell 15: 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li W, Piqueras R, Cao K, Deng XW, Wei N (2009) Regulation of COP1 nuclear localization by the COP9 signalosome via direct interaction with CSN1. Plant J [doi: 10.1111/j.1365-313X.2009.03805.x] [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW (1992) COP9: a new genetic locus involved in light-regulated development and gene expression in Arabidopsis. Plant Cell 4: 1507–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW (2003) The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286 [DOI] [PubMed] [Google Scholar]
- Wei N, Chamovitz DA, Deng XW (1994) Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78: 117–124 [DOI] [PubMed] [Google Scholar]
- Wei N, Serino G, Deng XW (2008) The COP9 signalosome: more than a protease. Trends Biochem Sci 33: 592–600 [DOI] [PubMed] [Google Scholar]
- Wolf DA, Zhou C, Wee S (2003) The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol 5: 1029–1033 [DOI] [PubMed]
- Wu JT, Chan YR, Chien CT (2006) Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol 6: 362–369 [DOI] [PubMed] [Google Scholar]
- Yan J, Walz K, Nakamura H, Carattini-Rivera S, Zhao Q, Vogel H, Wei N, Justice MJ, Bradley A, Lupski JR (2003) COP9 signalosome subunit 3 is essential for maintenance of cell proliferation in the mouse embryonic epiblast. Mol Cell Biol 23: 6798–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Chen J, Su CH, Yang HY, Lee MH (2008) Roles for CSN5 in control of p53/MDM2 activities. J Cell Biochem 103: 1219–1230 [DOI] [PubMed] [Google Scholar]
- Zhou C, Seibert V, Geyer R, Rhee E, Lyapina S, Cope G, Deshaies RJ, Wolf DA (2001) The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Wee S, Rhee E, Naumann M, Dubiel W, Wolf DA (2003) Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol Cell 11: 927–938 [DOI] [PubMed] [Google Scholar]