Abstract
Cancer genome projects are now being expanded in an attempt to provide complete landscapes of the mutations that exist in tumours. Although the importance of cataloguing genome variations is well recognized, there are obvious difficulties in bridging the gaps between high-throughput resequencing information and the molecular mechanisms of cancer evolution. Here, we describe the current status of the high-throughput genomic technologies, and the current limitations of the associated computational analysis and experimental validation of cancer genetic variants. We emphasize how the current cancer-evolution models will be influenced by the high-throughput approaches, in particular through efforts devoted to monitoring tumour progression, and how, in turn, the integration of data and models will be translated into mechanistic knowledge and clinical applications.
Keywords: cancer bioinformatics, cancer genome, cancer models, driver/passenger, genetic variants
Introduction
Cancers result from the accumulation of genetic changes (Vogelstein & Kinzler, 2004), and the identification of gene variants involved in tumour development and progression has been a central goal of cancer research for years (Sidebar A). Projects such as the Human Cancer Genome Project, The Cancer Genome Atlas and the International Cancer Genome Consortium aim to decipher the spectrum of genetic variants in different cancer types. The goals of these high-throughput resequencing (HTR) studies are fourfold: to identify genetic changes associated with tumour phenotypes; to discover molecular biomarkers that might be used for early detection, more accurate diagnosis or prognosis; to determine the molecular events of tumorigenesis; and, ultimately, to use this knowledge to develop strategies for targeted therapy (Chin & Gray, 2008; Wood et al, 2007).
However, there is an intense debate about the extent to which large-scale variation data will help us to understand the molecular mechanisms of tumour evolution. It is fair to say that, so far, the first genome-wide cancer HTR projects have had limited impact on molecular cancer research. These studies are rarely quoted as a starting point for further experiments (supplementary Table S1), although it is clear that more time is needed to translate gene discovery into mechanistic understanding. Technical, cultural and scientific issues can be responsible for the gap between genomic data and outcomes in terms of the molecular understanding of tumorigenesis. In the first place, the current methods for the organization of genomic data are evolving along with sequencing developments and constitute a real handicap for the use of the information. Second, high-throughput technologies unavoidably generate noise; the computational and statistical methods used to filter out genomic data—on which the reliability of the observations provided to the community ultimately depend—are not exempt from complications. Third, the core of the scientific challenge lies in the difficulty of linking genomic data to the molecular processes that underlie cancer evolution, as discussed in the final section of this review. It is therefore not surprising that cancer genome initiatives have generated substantial criticism, as many biologists are used to (and favour) more targeted approaches (Chng et al, 2007; Loeb & Bielas, 2007; Strauss, 2007).
The mutational landscape of tumours
Many types of genetic variant contribute to cancer: small structural changes (such as point mutations or small insertions), major structural rearrangements (such as translocations), numerical changes and epigenetic changes (supplementary Table S2). Alterations in the control of aneuploidy could also have a role (Duesberg, 2007). Mutations can occur spontaneously in cancer cells—through cytosine deamination, for example—after exposure to carcinogens or as the result of a mutator phenotype caused by mutations in polymerases and/or in mismatch-repair genes, which can lead to chromosomal instability (Loeb et al, 2008). In principle, all genes that harbour modifications are candidate cancer genes.
Genetic variants can be transmitted through the germline or can arise through somatic mutation. Germline variants are present in all the cells of an individual and contribute to inherited cancer susceptibility. One particular case of germline variants are the single-nucleotide polymorphisms (SNPs), the most common genetic variants, which are, by definition, present in at least 1% of the population (Collins et al, 1998; The International HapMap Consortium, 2003). Germline variants are typically identified through resequencing, and their involvement in cancer is shown using linkage or association studies (supplementary Table S2). Somatic mutations arise in the genomes of dividing cells and, in fact, all adult organisms are probably mosaics of somatically mutated cells. Somatic changes are typically identified through the resequencing of candidate genes, by analysing chromosomal rearrangements, or by quantifying losses or gains in gene-copy numbers using a range of techniques—such as microsatellite analysis or the quantitative polymerase chain reaction (qPCR; supplementary Table S2). Evidence for epigenetic silencing and downregulation of expression provides further support for the identification of tumour-suppressor genes, whereas increased expression can provide evidence for oncogene identification. The experimental validation of the biochemical and/or biological effects of a given alteration is often considered as proof of mechanistic involvement. In this context, RNA interference provides an additional method to study the involvement of gene variants in tumorigenesis. It has, for example, been recently used to validate mouse tumour-suppressor candidates (Zender et al, 2008).
The detection of point mutations has generally been carried out with small-scale sequencing from one to a few genes; >25,000 mutations identified in the well-known tumour-suppressor TP53 (Soussi et al, 2000) have been collected using this approach.
In the past decade, technical advances have provided the opportunity to use high-throughput methods for the identification of candidate cancer genes. Functional genomics approaches—such as microarray or methylation studies—have also been used, as well as association analyses and, more recently, tumour HTR screenings to determine the genes responsible for the initiation and progression of cancer (supplementary Table S2).
Large-scale resequencing studies
HTR studies can detect point mutations and short insertions or deletions (Bardelli et al, 2003); the introduction of ‘next-generation sequencing' technologies (Mardis, 2008) has not only produced massive amounts of data, but also allows the quantitative identification of individual gene variants and the detection of abnormal transcripts (Campbell et al, 2008). So far, HTR studies have followed two approaches, focusing either on genes or on tumour types (Table 1). In the first approach, a subset of genes—such as those that encode protein kinases (Greenman et al, 2007)—is sequenced in a relatively large number of samples. This approach allows the identification of genes that are mutated at low frequencies, but also requires an a priori selection of genes. The second approach analyses the coding sequences of whole genomes in a smaller number of tumour samples, and has been applied to colon and breast tumours (Sjöblom et al, 2006; Wood et al, 2007), pancreas adenocarcinomas (Jones et al, 2008) and glioblastoma (Parsons et al, 2008). This approach allows for the identification of the most-frequently mutated genes (Table 1). One such HTR study screened 518 protein kinases in 26 primary lung neoplasms and seven cell lines, and identified 188 mutations in 141 genes (Davies et al, 2005).
Table 1.
Catalogue of main recent high-throughput cancer genomic studies and initiatives
| First author | Publication date | Genes | Tumours | Screen sizes | PMID |
|---|---|---|---|---|---|
| Bardelli | 2003 | Tyrosine kinase | Colon | 138 genes, 35 samples, a subset in 147 additional samples | 12738854 |
| Wang | 2004 | Tyrosine phosphatase | Colon | 87 genes, 18 samples, a subset in 157 additional samples | 15155950 |
| Stephens | 2005 | Kinase | Breast | 518 genes, 25 samples, a subset in 56 additional samples | 15908952 |
| Davies | 2005 | Kinase | Lung | 518 genes, 33 samples, a subset in 56 additional samples | 16140923 |
| Sjöblom | 2006 | All | Breast and colon | 13,023 genes, 22 samples, a subset in 48 additional samples | 16959974 |
| Greenman | 2007 | Kinase | 210 human cancers | 518 genes in 210 samples | 17344846 |
| Wood | 2007 | All | Breast and colon | 18,191 genes, 22 samples, a subset in 48 additional samples | 17932254 |
| Loriaux | 2008 | Tyrosine kinase | Acute myeloid leukaemia | 85 genes, 188 samples | 18252861 |
| Tomasson | 2008 | Tyrosine kinase | Acute myeloid leukaemia | 26 genes, 94 samples, a subset in 94 additional samples | 18270328 |
| Brown | 2008 | Tyrosine kinase | Chronic lymphocytic leukaemia | 70 genes, 95 samples | 18754031 |
| Jones | 2008 | All | Pancreas | 20,661 genes, 24 samples | 18772397 |
| Parsons | 2008 | All | Glioblastoma | 20,661 genes, 22 samples, a subset in 83 additional samples | 18772396 |
| CGARN | 2008 | 601 genes | Glioblastoma | 601 genes, 91 samples | 18772890 |
| Ding | 2008 | 623 genes | Lung | 623 genes, 188 samples | 18948947 |
CGARN, Cancer Genome Atlas Research Network; PMID, PubMed identifier.
Regardless of the strategy used, these studies produce an overwhelming amount of information. The results are usually provided as raw tables in the supplementary material of a given publication and the main outcomes are briefly summarized in the published text. A number of databases aim to compile this type of information, such as Catalogue of Somatic Mutations in Cancer (COSMIC), which lists >60,000 mutations (Forbes et al, 2008), and the Cancer Gene Census (CGC), which—as of October 2008—included data for 380 cancer genes (Futreal et al, 2004); other repositories also include cancer-related information (Table 2; supplementary Table S3). However, although these genomic data are of great biological value, they are, in general, not sufficiently linked to additional information on gene annotation and regulation, or on molecular interactions and pathways, or to the clinical data about tumour and tissue types. In analysing this panorama, one realizes that the available infrastructure for organizing cancer genome information is still in its infancy and certainly lags behind the capacity of the current massive experimental approaches.
Table 2.
Main cancer-specific and non-specific repositories that contain information about cancer-associated mutations
| Acronym | Full name* | Category | URL | Reference |
|---|---|---|---|---|
| COSMIC | Catalogue of Somatic Mutations in Cancers | Mutations | http://www.sanger.ac.uk/genetics/CGP/cosmic | Forbes et al, 2008 |
| CGC | Cancer Gene Census | Cancer genes | http://www.sanger.ac.uk/genetics/CGP | Futreal et al, 2004 |
| OMIM | Online Mendelian Inheritance in Man | Disease-related genes | http://www.ncbi.nlm.nih.gov/omim | Hamosh et al, 2005 |
| Ensembl | – | Polymorphisms | http://www.ensembl.org | Flicek et al, 2008 |
| dbSNP | Single Nucleotide Polymorphism Database | Polymorphisms | http://www.ncbi.nlm.nih.gov/SNP | Sherry et al, 2001 |
*For information on additional repositories, please consult Supplementary Table 3.
Drivers and passengers
Like all high-throughput approaches, HTR generates noise that is difficult to distinguish from real biological signals. This noise can be technical, coming directly from sequencing technologies or from limitations in tumour-cell collection; all methods are sensitive to the presence of the normal allele, either in tumour cells or in contaminating normal cells. Gene variants that correspond to SNPs are ideally pinpointed by sequencing both tumour and normal tissues from the same patient, or by checking polymorphism databases. However, the most important source of problems is the presence of numerous mutations that are clearly detectable but do not have a direct role in cancer. In fact, only a handful of gene mutations that have been identified in HTR studies are likely to be biologically meaningful. To distinguish these mutations from the background mutation noise is a difficult task.
Mutations can be classified as ‘drivers' or ‘passengers' depending on their involvement in cancer development and progression. This metaphor was probably used for the first time in 1964, during a keynote lecture by Sir Christopher Andrewes, in which he referred to the role of viruses in either causing cancer (drivers) or being merely passengers in infected cells (Andrewes, 1964). Today, the term driver is used to denote mutations and/or genes that are positively selected and contribute to tumour development or progression, whereas the term passenger is used to designate cancer-neutral variations that are retained during the evolution of the cancerous cells.
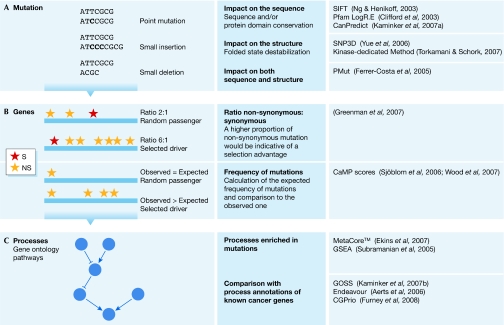
Single mutations can be responsible for the development and progression of a cancer (Fig 1A). Historically, analyses have focused on mutations that can affect protein function. These mutations are thought to be mainly non-synonymous (missense, nonsense or frameshift), in contrast to synonymous (silent) mutations. In this regard, the first oncogene identified—H-ras—was found to have a non-synonymous substitution in codon 12 that introduces an alanine in the position of a glycine, thereby blocking its GTPase function and producing a protein able to transform cells (Reddy et al, 1982). However, this corresponds to a ‘protein-centric' view of biology; one should remember that non-synonymous mutations might not always alter protein function (owing to amino-acid plasticity) and, importantly, that there is strong evidence showing that ‘silent' mutations can be biologically relevant—for example, through the modulation of splicing (Cartegni et al, 2002)—although it is difficult to assess their effects and even more difficult to predict them. Additionally, we have to keep in mind that 98% of the genome is intergenic. In this respect, it is currently impossible to interpret the consequences of mutations in non-coding DNA regions, with the exception of some favourable cases in splice sites or promoters.
Figure 1.
Driver or passenger? Multilevel strategies used to classify mutations and genes as either ‘drivers' or ‘passengers' at the level of (A) mutations, (B) genes or (C) processes. NS, non-synonymous; S, synonymous.
The current way of thinking assumes that only a small fraction of the non-synonymous mutations actually cause tumours. Historically, the identification of mutations has been followed by functional analyses to evaluate their pathogenic potential. For example, a screening of the gene encoding the tyrosine kinase FLT3 (FL cytokine receptor) identified nine non-synonymous mutations (Fröhling et al, 2007), four of which allow the growth of cultured cells independently of the presence or absence of growth factors. In general, only a small range of biological assays is used to assess pathogenicity, exploring a limited spectrum of the potential biological effects of candidate mutations and often being unable to detect small functional changes (Chin & Gray, 2008). When a direct effect on cell proliferation or the generation of apoptosis is not detected, other experiments are seldom used unless the functional annotations point directly to a crucial biological role—as is the case for proteases and kinases. It is a formidable challenge to scale up these experiments to validate the results of genome-wide HTRs, and, therefore, in silico methods are a suitable alternative. Typical computational methods are based on the assumption that somatic mutations considered as ‘drivers' would have to affect protein function markedly (Torkamani & Schork, 2007). Sequence and protein domain conservation, as well as protein structure, are used to determine the crucial positions in a given protein and to predict the causative effects of mutations (Fig 1A). The same parameters are also applied to predict the possible pathogenicity of SNPs (supplementary information online).
Bioinformatic predictions based on sequence analysis—made by the SIFT (Ng & Henikoff, 2003) and PMut (Ferrer-Costa et al, 2005) programs—and experimental results have been compared for nine FLT3 mutations (Fröhling et al, 2007). The consensus bioinformatic predictions failed to identify two mutations that were experimentally shown to affect function, whereas one predicted driver mutation was not found to have a functional effect. By contrast, all four of the mutations predicted to be passengers in silico were confirmed by the functional analysis. The availability of more case studies will allow a better assessment of the predictive capacity of computational tools.
It is important to remember that there is a great difference between demonstrating that a mutation alters the function of a protein and claiming that it has a pathogenic involvement in cancer. The effects of candidate mutations in the development of cancer are probably highly context dependent and the assessment of their biological significance in the context of human cancer needs to be largely extrapolated.
At a second level of analysis, the overall frequency of mutations in a given gene can help to detect a positive selection that would support its involvement as a driver for oncogenesis (Fig 1B). The usual assumption is that positive selection is exerted mainly on non-synonymous mutations. The genetic code provides a random ratio of approximately two non-synonymous mutations for each synonymous one (2:1); higher ratios are interpreted as evidence of positive selection and competitive advantage. In reality, more complex models—borrowed from the field of molecular evolution—are applied, which can also take into account the types of mutation (transition and transversion) or the neighbouring sequence (for example, whether a C-to-T transition occurs in a CpG island; a more detailed explanation of the models used can be found in the supplementary information online). Another set of methods calculates differences between the observed and the expected frequencies of non-synonymous mutations. If a gene contains—in all sequenced tumours—more mutations than would have been expected to occur by chance, these have been positively selected during the process of tumorigenesis and, therefore, confer an advantage in this process (supplementary information online).
An obvious limitation of these approaches to the identification of cancer genes is the need to sequence many samples. Without enough observations, the less-frequently mutated genes would not meet the statistical thresholds. In fact, they would be indistinguishable from unselected passengers, although they can be revealed by functional assays (Fröhling et al, 2007). Furthermore, these statistical techniques do not provide information about the specific alleles—point mutations—involved in cancer evolution. A perhaps less obvious—albeit not less important—limitation of current studies is that they usually consider mutations individually, without modelling epistatic interactions. In a few cases, the importance of the combination of otherwise neutral (passenger) mutations has been shown (Chen et al, 2008). Epistatic effects (Moore, 2005), which are not commonly considered in cancer genome studies, might be even more important when taking genetic background into consideration, either alone or together with somatic mutations.
Among known mutations, a large proportion occurs in a few genes, such as TP53 or K-RAS (Forbes et al, 2008; Soussi et al, 2000). Hence, cancer genomes are composed of a handful of frequently mutated genes and a much larger number of infrequently mutated genes. The number of genes that are mutated in cancers, although large, possibly reflects alterations in a relatively small number of signalling pathways (Wood et al, 2007; Fig 1C). Indeed, many recent HTR studies provide an interpretation of their results in terms of alterations in ‘core pathways' (Jones et al, 2008; Parsons et al, 2008). This point is crucial—particularly from a therapeutic point of view—because designing strategies to target proteins individually is different from targeting a well-defined pathway (Check Hayden, 2008). Additionally, cancer genes might share other structural or functional properties (Furney et al, 2006), such as good evolutionary conservation or a role in essential cellular processes such as the cell cycle or DNA repair. The analyses based on previous knowledge of known pathways and functions can be useful for the interpretation of genome-wide results. However, to obtain new insights into the oncogenic process, it is important to avoid the constant re-identification of the same genes for which significant functional information is already available.
Cancer-evolution models and cancer genomics
Molecular biologists have been working for the past 20 years to determine the molecular mechanisms of cancer evolution. Modelling cancer evolution is more than an academic exercise as it has profound implications on the detection of early recurrence and in the choice of adjuvant therapy, among other aspects (Fig 2). To be useful in this context, large-scale genomic studies would have to complement these efforts, and help to improve our understanding of tumour development and progression.
Figure 2.
Modelling cancer evolution. Using the available technologies, the modelling of cancer evolution should provide insights into its development and progression.
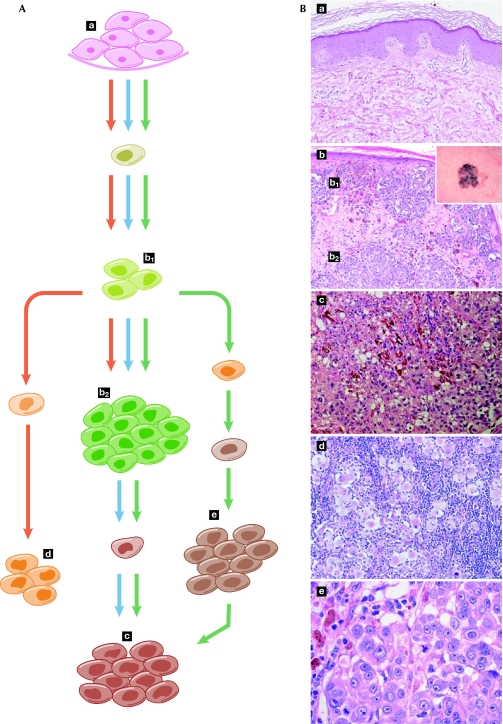
Historically, cancer research has been dominated by the ‘clonal evolution' model of tumour development and progression (Fig 3, blue arrows). This model postulates that tumour cells acquire specific genetic changes, leading to clonal expansion. These changes are selected in competition with other tumour cells through a Darwinian process, and those that confer a selective advantage become fixed, thereby allowing a phylogenetic tracing of the history of the evolving cell populations. In this model, it is generally considered that benign lesions are precursors of malignant tumours, genomically stable tumours precede genomically unstable ones and metastases are the ultimate step in tumour evolution. More recently, it has become evident that some experimental results do not fit this model. For example, some metastases of breast cancers bear little genetic resemblance to the primary tumour (Schmidt-Kittler et al, 2003). Moreover, a recent study showed that a small proportion of normal mouse mammary epithelial cells injected intravenously can survive at distant sites and eventually develop into tumours (Podsypanina et al, 2008). These observations have led to the proposal of the ‘parallel evolution' model (Gray, 2003; Yokota & Kohno, 2004), in which cells that generate metastases are separated relatively early from the primary tumour and evolve independently (Fig 3, red arrows). This model is reinforced by data from gene-expression profiles that are predictive of metastases in certain primary breast tumours (Bernards & Weinberg, 2002).
Figure 3.
Models of cancer evolution. (A) The ‘clonal selection model' (blue arrows) is the prevailing view to explain the successive steps of mutation and selection from normal tissue to primary tumour and metastasis. However, metastasis-generating cells can emerge relatively early in the tumorigenic process and ‘seed' distant tissues, thereby evolving in parallel with the primary tumour and delineating the ‘parallel evolution' model (red arrows). Finally, these two models can occur simultaneously and metastatic deposits can act as sites from which additional metastases can be generated, therefore leading to an integrated model of cancer evolution (green arrows). (B) Microphotographs provide a histological snapshot of normal skin tissue (a), primary tumour (superficial b1 and deep b2, macroscopic appearance inset in b), subcutaneous metastasis (c), metastasis in the lymph node (d) and metastasis in the lung (e), and are shown in correspondence with the cancer-evolution models. This melanoma—which originates from the transformation of pigmented skin cells—provides a visual example of the modelling paradigms, illustrating the gap between ideal models and actual observations.
The differences between both models have important consequences for the interpretation and clinical use of the knowledge about cancer-associated mutations. In the framework of the ‘clonal evolution' model, the significance of a mutation detected in a metastasis—in the absence of information about its presence in the primary tumour or in pre-neoplastic lesions—is unclear. In the context of the ‘parallel evolution' model, targeting this mutation for therapy would have no effect on the growth of the primary tumour, which could evolve to metastasis through other mutations. Given the complexity of cancer and the diversity of its phenotypic presentation, it is unlikely that a single paradigm will universally account for cancer development and progression; the different models might be complementary rather than exclusive, at least when considering cancer globally rather than at the individual level. It seems possible that these two models might explain different, albeit concurrent, biological processes (Fig 3, green arrows). This integration is also reinforced by evidence that metastases can act as repositories from which additional systemic tumour-cell seedings can take place (Nguyen & Massagué, 2007).
HTR studies are usually performed using DNA from cell lines, xenografts or large and advanced tumours. This bias in sample selection, owing to the fact that earlier stage tumours are underrepresented, is to some extent also present in low-scale studies. During the advanced stages of tumorigenesis, all the mutations necessary for cancer development and progression are already present; as is commonly assumed, “such tumours contain all the mutations found in the early stage tumours, but the converse is not true” (Wood et al, 2007). The information derived from HTR studies is therefore intrinsically far from providing information on other stages of cancer evolution, and hence does not contribute to our understanding of the development and evolution of cancer. Furthermore, the identification of causative cancer genes and mutations—based on the methods adapted from evolutionary biology described above—tends to be too general to give specific information at the level of resolution required by the current cancer-evolution models. Hence, we continually have to revisit our understanding of the contribution of genetic variants based only on the study of snapshots in tumour evolution, which do not provide sufficient insight to elucidate the true relevance of these genes in the tumorigenic process.
In order to attain more insight into the contribution of the different models to cancer development, and to validate more precisely the significance of the genetic and genomic changes found in advanced tumours, one would need to obtain information about specific genes and mutations at different stages of tumour evolution. Knowing the time of appearance of a given mutation would allow for a better estimate of its contribution to the fitness of cancer cells, which is essential to distinguish between the various evolution models. This involves several difficulties and only a few metastasis-specific alterations have been identified (Nguyen & Massagué, 2007). First, it is conceivable that individual genetic alterations, or a given genetic programme, could render a stage-specific advantage to tumours and be either neutral or deleterious at later stages, as is the case for the epithelial–mesenchymal transition programme, which is activated in the invasive front of tumours but might be repressed in metastases (Thiery & Sleeman, 2006). Additionally, changes in the tumour microenvironment—either locally or at metastatic sites—might impose different selection pressures on genetic changes and thereby modulate the influence of the individual mutations. Furthermore, the effects of genetic alterations might be incremental rather than qualitative, thereby allowing for epistatic interactions, which are often not considered when modelling molecular pathogenesis. Improvements in technology and more focused research on early-stage tumours are needed to fill these gaps. For such applications, the lack of sensitivity for detecting a given mutation in a low proportion of alleles is a major technical concern when standard sequencing technology is used. However, this limitation might be overcome with ultra-sequencing technologies (Gupta, 2008). These technologies—which are already able to detect rare subclones with a sensitivity as low as 1 in 5,000 copies—would be relevant to track the subpopulations of cells that are responsible for initiating the genetic lesions, for drug resistance or for metastasis (Campbell et al, 2008).
Integrative approaches would be a solution to overcome the limitations specific to both genomic and functional methods. The findings obtained using diverse high-throughput genomic techniques—such as gene mutation, copy-number variation, expression analyses and epigenetic changes—have recently been combined for glioblastoma and pancreatic ductal adenocarcinoma (Cancer Genome Atlas Research Network, 2008; Jones et al, 2008; Parsons et al, 2008). The gathering of independent evidence supported the causative implication of genes in tumours, for example, by showing that a subset of the genes recurrently found in copy-number-alteration regions has an expression pattern that correlates with copy number (Cancer Genome Atlas Research Network, 2008). In practice, the interpretation of heterogeneous high-throughput information is still a formidable challenge, and multidimensional analyses of data coming from high-throughput studies still face the problems of data standardization, database annotations and normalization of phenotypic descriptions.
The combination of all these efforts should have an impact on the development of improved strategies for early detection, improved tumour subclassification, a more rational selection of therapy and more accurate prognostication, all of which represent important aspects of patient management.
Conclusion
Cancer genome studies—including the inevitably associated computational analyses—have the potential to predict which genes and mutations contribute to tumour development (known as driver genes or mutations) on a large scale. However, despite the enormous capacity of the experimental resequencing methodologies and the expected improvements therein, limitations still exist. Indeed, the reliable detection of less-frequent mutations is still arduous, and it is difficult to obtain a sufficiently systematic mutation analysis that will allow conclusions to be drawn about the prevalence and distribution of mutations according to tumour stage. Furthermore, mutation analysis can by itself provide only statistical information on potential associations with cancer and not direct causative information, and it is a major challenge in molecular terms to go from genomic information to data interpretation. For example, the classification of mutations as drivers or passengers depends on the analysis of the possible functional consequences of these mutations, which is a technology that is not free from limitations and, in addition, does not provide a complete picture of the actual implication of the mutations in the development of cancer. In other words, the future challenge will be to support—or to refute—the current cancer models with high-throughput experimental methods within a reasonable time scale at an affordable cost. This would involve both the descriptive large-scale genomic analysis of pre-neoplastic lesions and early cancers, and the functional analysis of genetic variants: a combined effort that is crucial to translate genomic knowledge into molecular pathophysiology and patient management.
We must note that many of the difficulties in the application of high-throughput variation approaches are similar to those found in the study of other complex diseases. Cancer is particularly challenging—and therefore attractive—as this is the field in which the largest amount of molecular information is available, the diversity of phenotypes and pathologies is more notable, and the complex evolution of disease at the cellular and/or tissue level has been most directly addressed. These are all good reasons to believe that the symbiosis of high-throughput technologies, molecular and cellular mechanistic models, and new experimental systems and models will be effective first in cancer research.
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Sidebar A | In need of answers.
How can we define a cancer gene and how many cancer genes exist?
How can the functional effects of mutations in cancer cells be predicted?
How can cancer genes and their associated functional roles be precisely assessed by detailed biological research?
How can the gene variants that are causally involved in tumour development or progression be identified among all the gene variants in a tumour?
How can the gap between the genetic variants observed in cancers and the current models of cancer evolution be bridged?
How can the experimental analysis of gene variants involved in cancer be accelerated?
Supplementary Material
Francisco X. Real, José M. G. Izarzugaza, Alfonso Valencia & Anaïs Baudot
Acknowledgments
We thank M. Soengas and A. Toll for providing microphotographs. This work is supported by Instituto de Salud Carlos III (ISCIII) grant COMBIOMED (RD07/0067/0014), BIO2007-66855, the Spanish Ministry of Education and Science, and European Union grant LSHG-CT-2003-503265 (BioSapiens). Work in the F.X.R. laboratory is supported by grant SAF2007-60860 and CONSOLIDER INGENIO BioCancer from Ministerio de Ciencia e Innovación. A.B. is supported by the Juan de la Cierva fellowship.
References
- Aerts S et al. (2006) Gene prioritization through genomic data fusion. Nat Biotechnol 24: 537–544 [DOI] [PubMed] [Google Scholar]
- Andrewes C (1964) Tumour-viruses and virus-tumours. BMJ 1: 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardelli A et al. (2003) Mutational analysis of the tyrosine kinome in colorectal cancers. Science 300: 949. [DOI] [PubMed] [Google Scholar]
- Bernards R, Weinberg RA (2002) A progression puzzle. Nature 418: 823. [DOI] [PubMed] [Google Scholar]
- Brown JR, Levine RL, Thompson C, Basile G, Gilliland DG, Freedman AS (2008) Systematic genomic screen for tyrosine kinase mutations in CLL. Leukemia 22: 1966–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ et al. (2008) Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3: 285–298 [DOI] [PubMed] [Google Scholar]
- Check Hayden E (2008) Cancer complexity slows quest for cure. Nature 455: 148. [DOI] [PubMed] [Google Scholar]
- Chen Z, Feng J, Saldivar J, Gu D, Bockholt A, Sommer SS (2008) EGFR somatic doublets in lung cancer are frequent and generally arise from a pair of driver mutations uncommonly seen as singlet mutations: one-third of doublets occur at five pairs of amino acids. Oncogene 27: 4336–4343 [DOI] [PubMed] [Google Scholar]
- Chin L, Gray JW (2008) Translating insights from the cancer genome into clinical practice. Nature 452: 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng WJ et al. (2007) Limits to the Human Cancer Genome Project? Science 315: 762; author reply 764–765 [DOI] [PubMed] [Google Scholar]
- Clifford RJ, Edmonson MN, Nguyen C, Buetow KH (2004) Large-scale analysis of non-synonymous coding region single nucleotide polymorphisms. Bioinformatics 20: 1006–1014 [DOI] [PubMed] [Google Scholar]
- Collins FS, Brooks LD, Chakravarti A (1998) A DNA polymorphism discovery resource for research on human genetic variation. Genome Res 8: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Davies H et al. (2005) Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res 65: 7591–7595 [DOI] [PubMed] [Google Scholar]
- Ding L et al. (2008) Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P (2007) Chromosomal chaos and cancer. Sci Am 296: 52–59 [DOI] [PubMed] [Google Scholar]
- Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T (2007) Pathway mapping tools for analysis of high content data. Methods Mol Biol 356: 319–350 [DOI] [PubMed] [Google Scholar]
- Ferrer-Costa C, Gelpí JL, Zamakola L, Parraga I, de la Cruz X, Orozco M (2005) PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics 21: 3176–3178 [DOI] [PubMed] [Google Scholar]
- Flicek P et al. (2008) Ensembl 2008. Nucleic Acids Res 36: D707–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR (2008) The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum Genet 10: 10.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhling S et al. (2007) Identification of driver and passenger mutations of FLT3 by high-throughput DNA sequence analysis and functional assessment of candidate alleles. Cancer Cell 12: 501–513 [DOI] [PubMed] [Google Scholar]
- Furney SJ, Higgins DG, Ouzounis CA, López-Bigas N (2006) Structural and functional properties of genes involved in human cancer. BMC Genomics 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney SJ, Calvo B, Larrañaga P, Lozano JA, Lopez-Bigas N (2008) Prioritization of candidate cancer genes—an aid to oncogenomic studies. Nucleic Acids Res 36: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR (2004) A census of human cancer genes. Nat Rev Cancer 4: 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JW (2003) Evidence emerges for early metastasis and parallel evolution of primary and metastatic tumors. Cancer Cell 4: 4–6 [DOI] [PubMed] [Google Scholar]
- Greenman C et al. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PK (2008) Single-molecule DNA sequencing technologies for future genomics research. Trends Biotechnol 26: 602–611 [DOI] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA (2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 33: D514–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S et al. (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, Zhang Y, Watanabe C, Zhang Z (2007a) CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res 35: W595–W598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS et al. (2007b) Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res 67: 465–473 [DOI] [PubMed] [Google Scholar]
- Loeb LA, Bielas JH (2007) Limits to the Human Cancer Genome Project? Science 315: 762; author reply 764–765 [PubMed] [Google Scholar]
- Loeb LA, Bielas JH, Beckman RA (2008) Cancers exhibit a mutator phenotype: clinical implications. Cancer Res 68: 3551–3557 [DOI] [PubMed] [Google Scholar]
- Loriaux MM et al. (2008) High-throughput sequence analysis of the tyrosine kinome in acute myeloid leukemia. Blood 111: 4788–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER (2008) The impact of next-generation sequencing technology on genetics. Trends Genet 24: 133–141 [DOI] [PubMed] [Google Scholar]
- Moore JH (2005) A global view of epistasis. Nat Genet 37: 13–14 [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31: 3812–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Massagué J (2007) Genetic determinants of cancer metastasis. Nat Rev Genet 8: 341–352 [DOI] [PubMed] [Google Scholar]
- Parsons DW et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsypanina K, Du YN, Jechlinger M, Beverly LJ, Hambardzumyan D, Varmus H (2008) Seeding and propagation of untransformed mouse mammary cells in the lung. Science 321: 1841–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy EP, Reynolds RK, Santos E, Barbacid M (1982) A point mutation is responsible for the acquisition of transforming properties by the T24 human bladder carcinoma oncogene. Nature 300: 149–152 [DOI] [PubMed] [Google Scholar]
- Schmidt-Kittler O et al. (2003) From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci USA 100: 7737–7742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29: 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom T et al. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [DOI] [PubMed] [Google Scholar]
- Soussi T, Dehouche K, Béroud C (2000) p53 website and analysis of p53 gene mutations in human cancer: forging a link between epidemiology and carcinogenesis. Hum Mutat 15: 105–113 [DOI] [PubMed] [Google Scholar]
- Stephens P et al. (2005) A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet 37: 590–592 [DOI] [PubMed] [Google Scholar]
- Strauss BS (2007) Limits to the Human Cancer Genome Project? Science 315: 762–764; author reply 764–765 [PubMed] [Google Scholar]
- Subramanian A et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium (2003) The International HapMap Project. Nature 426: 789–796 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP (2006) Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol 7: 131–142 [DOI] [PubMed] [Google Scholar]
- Tomasson MH et al. (2008) Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood 111: 4797–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Schork NJ (2007) Accurate prediction of deleterious protein kinase polymorphisms. Bioinformatics 23: 2918–2925 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW (2004) Cancer genes and the pathways they control. Nat Med 10: 789–799 [DOI] [PubMed] [Google Scholar]
- Wang Z et al. (2004) Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 304: 1164–1166 [DOI] [PubMed] [Google Scholar]
- Wood LD et al. (2007) The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113 [DOI] [PubMed] [Google Scholar]
- Yokota J, Kohno T (2004) Molecular footprints of human lung cancer progression. Cancer Sci 95: 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Melamud E, Moult J (2006) SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics 7: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L et al. (2008) An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.