Abstract
Much of the genetic predisposition to colorectal cancer (CRC) in humans is unexplained. Studying a Caucasian-dominated population in the United States, we showed that germline allele-specific expression (ASE) of the gene encoding transforming growth factor–β (TGF-β) type I receptor, TGFBR1, is a quantitative trait that occurs in 10 to 20% of CRC patients and 1 to 3% of controls. ASE results in reduced expression of the gene, is dominantly inherited, segregates in families, and occurs in sporadic CRC cases. Although subtle, the reduction in constitutive TGFBR1 expression alters SMAD-mediated TGF-β signaling. Two major TGFBR1 haplotypes are predominant among ASE cases, which suggests ancestral mutations, but causative germline changes have not been identified. Conservative estimates suggest that ASE confers a substantially increased risk of CRC (odds ratio, 8.7; 95% confidence interval, 2.6 to 29.1), but these estimates require confirmation and will probably show ethnic differences.
The annual worldwide incidence of colorectal cancer (CRC) exceeds 1 million, being the second to fourth most common cancer in industrialized countries (1). Although diet and lifestyle are thought to have a strong impact on CRC risk, genes have a key role in the predisposition to this cancer. A positive family history of CRC occurs in 20 to 30% of all probands. Highly penetrant autosomal dominant and recessive hereditary forms of CRC account for at most 5% of all CRC cases (2). Although additional high- and low-penetrance alleles have been proposed, much of the remaining predisposition to CRC remains unexplained (3).
Aberrations in the transforming growth factor–β (TGF-β) pathway are heavily involved in CRC carcinogenesis (4). Although mutations in the TGF-β type II receptor gene have been explicitly associated with CRC (5), the type I receptor gene (TGFBR1) has received less attention, although there is evidence that a common variant may be associated with cancer risk (6, 7). We hypothesized that TGFBR1 is a notable candidate for a gene that, when mutated, causes predisposition to CRC or acts as a modifier of other genes, resulting in a predisposition. Our study was undertaken to test this assumption.
Given the previously existing evidence that inherited allele-specific expression of APC acts as a mechanism of predisposition to familial adenomatous polyposis (8) and of an analogous mechanism involving DAPK1 in chronic lymphocytic leukemia (9) we searched for a similar association of TGFBR1 with CRC. We hypothesized that the putative change might be subtle; for instance, lowered rather than extinguished expression of one allele referred to here as ASE, for allele-specific expression. To test for ASE in TGFBR1, we chose three single-nucleotide polymorphisms (SNPs) (rs334348, rs334349, and rs1590) in the 3′ untranslated region (3′UTR), to which primer extension with fluorescent nucleotides (SNaPshot) (10) was applied. These three SNPs are separated by 1916 and 1778 base pairs (bp), respectively, yet they exhibit total linkage disequilibrium.
Among a total of 242 patients with microsatellite instability (MSI)–negative CRC (10), 96 (39.7%) were heterozygous for the three 3′UTR SNPs, of whom 12 showed ASE variation ratios higher than 1.5, whereas no patient showed ratios below 0.67. Forty-nine additional cases were heterozygous for one further SNP (rs7871490) located in the 3′UTR that was not in strong linkage disequilibrium with the above three markers, and 17 out of 49 (17/49) had ASE values higher than 1.5. Thus, 29 out of 138 (21%) informative CRC patients showed ASE in the TGFBR1 gene. Three additional cases had borderline values (fig. S1 and table S1).
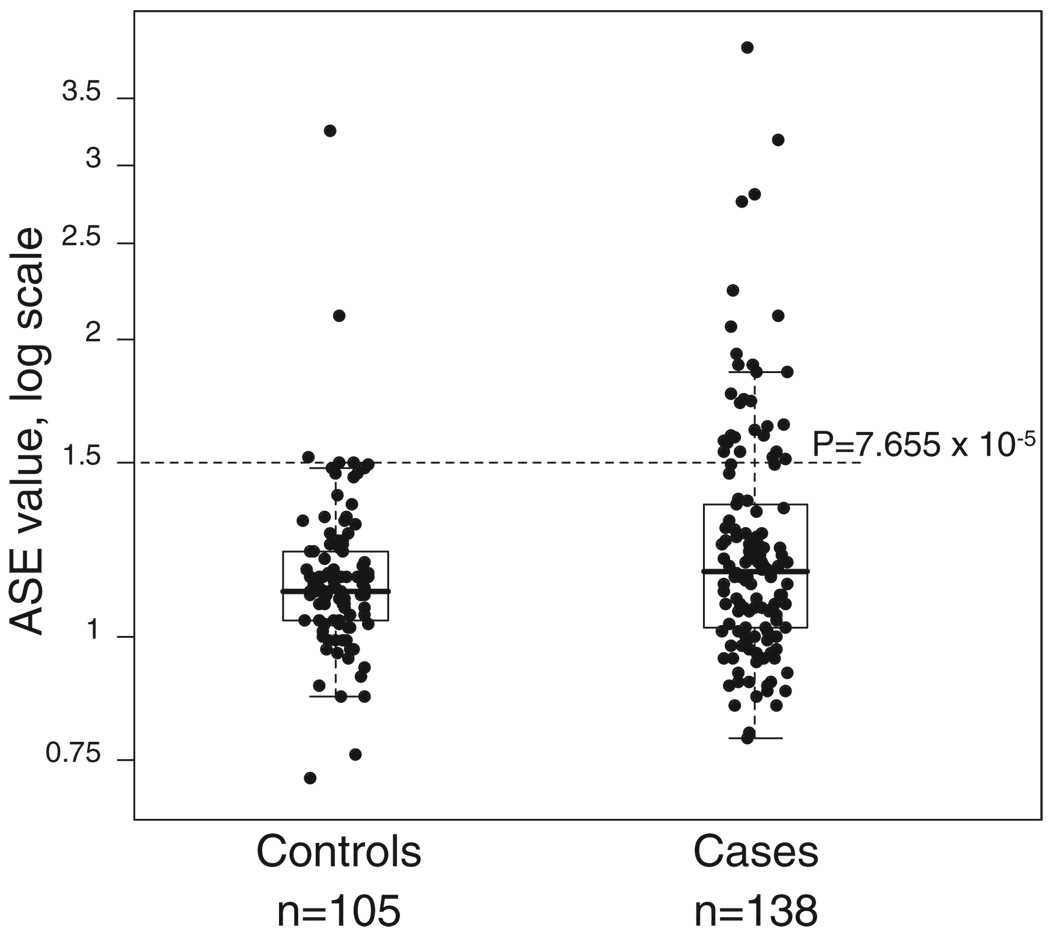
DNA samples from the blood of healthy Columbus, Ohio–area controls (195 individuals) (10) were genotyped for the four SNPs. One hundred and nine (55.9%) were heterozygous, and ASE analysis in 105 of them revealed ratios ranging between 0.72 and 3.25 (fig. S1). Only three controls showed ratios above 1.5. Our results in both the CRC patients and controls suggest that the degree of ASE is a quantitative trait (Fig. 1). Differences in the degree of ASE between patients and controls showed a P value of 0.1208 when a Wilcoxon rank sum test was applied and a P value of 0.0207 when a permutation test (100,000 permutations) was applied.
Fig. 1.
TGFBR1 ASE distribution in 138 CRC patients and 105 controls studied by SNaPshot The ASE cutoff value of 1.5 chosen to categorize the cases is indicated, together with its associated P value obtained from comparing the proportions of cases (29/138) and controls (3/105) above the indicated value.
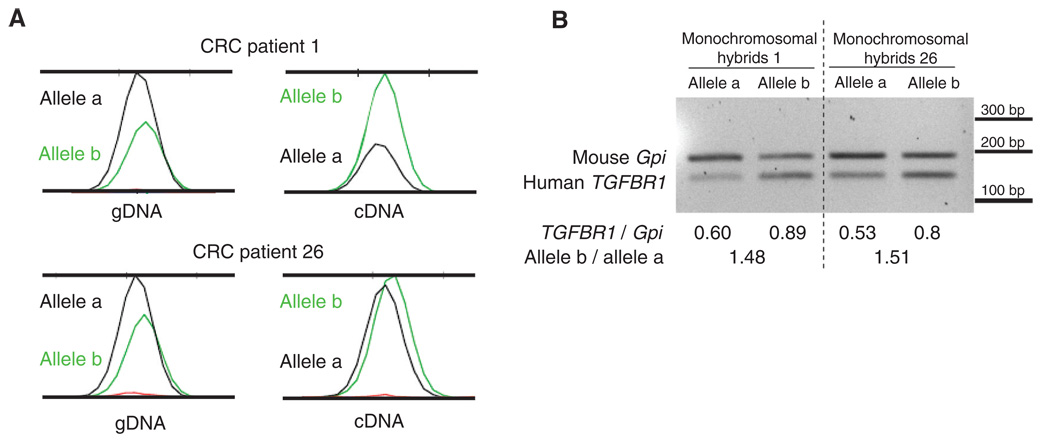
At this stage, it is not possible to determine whether the degree of predisposition to CRC is proportional to the degree of ASE or whether there is a threshold value that separates “abnormal” values that predispose to CRC from “normal” values that do not. A ratio of 1 means that both alleles are equally expressed, whereas a ratio of 1.5 means a 33% difference, as does a ratio of 0.67. To define a cutoff point, we applied receiver operating characteristic (ROC) analysis, which estimates the sensitivity and specificity of cutoff points. As shown in table S2, the value of 1.5 maximizes both characteristics, providing the highest Youden’s index. When a cutoff of 1.5 was used, the P value comparing cases and controls was 7.655 × 10−5. Although there is no overall need to define a firm cutoff point, we used the value of 1.5 to categorize CRC cases and controls into ASE and non-ASE. In order to determine whether the observed ratios falling outside this range represent an increase or decrease in the transcript of one allele, a reverse transcription polymerase chain reaction (RT-PCR) experiment was performed, taking advantage of hybrid clones monoallelic for chromosome 9 created from two individuals with ASE (patients 1 and 26, table S1). Each of the four hybrid clones contained either the maternal or paternal copy of chromosome 9, plus the mouse genome (10). As shown in Fig. 2A, ASE determination in the diploid samples indicated that the expression of one allele (a) was reduced as compared to that of the other allele (b). In the four monoallelic hybrid clones, the densitometric values of the RT-PCR of human TGFBR1 were compared with the corresponding values for mouse Gpi (10). One allele (a) showed reduced expression in both patients. These experiments support the notion of lowered expression of one allele, and in both patients the same allele was affected (Fig. 2, A and B).
Fig. 2.
ASE determination in two ASE CRC probands. (A) ASE detection in blood DNA by SNaPshot. The ASE ratio was calculated by normalizing the ratio between the peak areas of the two alleles in cDNA with the same parameters in genomic DNA (gDNA). In both examples, the transcript from the a allele is reduced with respect to that from the b allele. (B) Semiquantitative RT-PCR of the cDNA from monochromosomal hybrids of the same two patients. Human TGFBR1 expression (amplicon size 135 bp) was assessed and mouse Gpi was used as a control (176 bp). The values shown below the gel represent the ratios of the densitometric values of human TGFBR1 versus mouse Gpi, showing reduced expression of human TGFBR1 in the hybrids that contain the a allele.
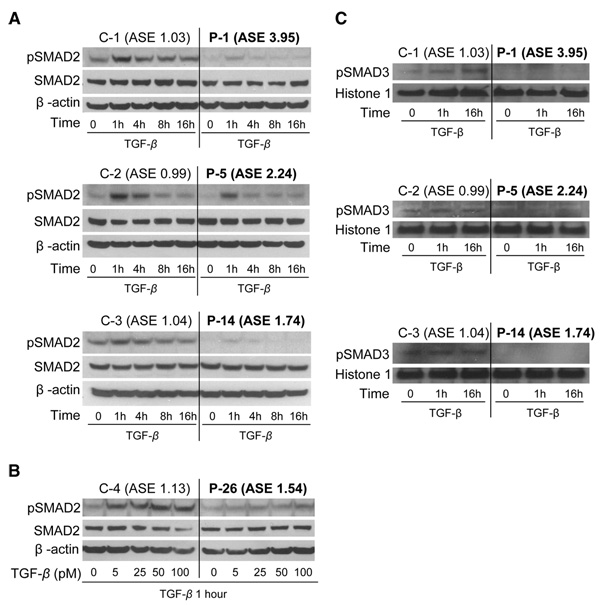
To assess the effect of ASE on TGF-β signaling, lymphoblastoid cell lines from four ASE patients and four non-ASE healthy controls were exposed to TGF-β (10), which binds TGFBR2 and leads to the formation of the TGFBR2/TGFBR1/TGF-β heteromeric complex. We observed differences in levels of phosphorylated SMAD2 (pSMAD2), an important downstream effector and surrogate marker of TGF-β signaling (11, 12). There were constitutive differences in pSMAD2 expression between ASE patients and non-ASE controls in the absence of exogenously added TGF-β (time 0; Fig. 3A). Differences in pSMAD2 levels became more pronounced upon exposure to TGF-β. These differences were observed at low TGF-β concentrations (<5 pM) (Fig. 3B) and occurred in four out of four ASE cases as compared to non-ASE controls.
Fig. 3.
Analysis of SMAD-mediated TGF-β signaling in lymphoblastoid cell lines from ASE CRC patients and non-ASE healthy controls. (A) SMAD2 and phosphorylated SMAD2 (pSMAD2) expression were assessed by Western blotting in lymphoblastoid cell lines from ASE patients (P-1, P-5, and P-14) and non-ASE controls (C-1, C-2, and C-3), after exposure to TGF-β (100 pM) at various time points from 0 to 16 hours (h) and using β-actin as a loading control. In all three ASE cases, less constitutive pSMAD2 was observed than in non-ASE controls. The differences in pSMAD2 expression between ASE and non-ASE cell lines were further enhanced after exposure to TGF-β. (B) SMAD2 and p-SMAD2 expression 1 hour after exposure to different TGF-β concentrations. The effect shown in (A) also occurs at low concentrations of TGF-β (5 pM). (C) pSMAD3 detection in nuclear extracts from three ASE patients and three non-ASE controls after exposure to TGF-β1. The three non-ASE lymphoblastoid cell lines had pSMAD3 expression in the nucleus, whereas nuclear pSMAD3 expression was undetectable in two ASE cases (P-1 and P-14) and barely detectable in one case (P-5).
It has been shown that phosphorylation of SMAD3 is an essential step in signal transduction by TGF-β for the inhibition of cell proliferation (13). Furthermore, Smad3-deficient mice are prone to developing colon cancer (14, 15). To assess the impact of TGFBR1 ASE on the phosphorylation of SMAD3, we used an antibody targeting the Ser423/425 site in SMAD3 (10, 16). Constitutive levels of pSMAD3 were detectable in the lymphoblastoid cell lines of three non-ASE controls, whereas pSMAD3 was barely detectable in one ASE case (Fig. 3C). Exposure to TGF-β did not result in any detectable increase in pSMAD3 in the lymphoblastoid cell lines of the ASE patients. The pSMAD2 and pSMAD3 results indicate that patients with ASE exhibit decreased SMAD-mediated signaling when compared with non-ASE controls.
A GCG trinucleotide variable number of tandem repeat polymorphism occurs in exon 1 of TGFBR1. The most common allele contains nine repeats leading to a stretch of nine alanines (9A) in the signal peptide of the receptor protein. The second most common allele has six repeats (6A) and occurs in approximately 14% of all individuals in most Caucasian populations (6). The 6A allele has been associated with a low-level but statistically significant predisposition to several forms of cancer (17–20). Recent studies suggest that the association of 6A with colon cancer is either weak [odds ratio (OR) 1.2, 95% confidence interval (CI) 1.01 to 1.43] (17) or borderline significant (OR 1.13, CI 0.98 to 1.30) (21). We typed this polymorphism in all 242 CRC cases studied by us and found 9A/9A in 197, 9A/6A in 40, 6A/6A in 4, and 1 failed (table S3). There were clearly more 9A/6A heterozygotes among the patients with ASE (14/29) than in those without ASE (22/108) (P = 0.0052, chi-square test). We tentatively concluded from these data that the 6A allele is probably in linkage disequilibrium with one of the putative mutations that causes ASE, but 6A is not in itself causative of ASE.
All 29 patients showing ASE and three patients with borderline ASE values (1.49, 1.49, and 1.46) (n = 32 patients) were studied for genetic changes occurring in the germ line. By sequencing of all nine exons, 2 kb upstream of exon 1, and the entire 3′UTR (10), a single sequence change in the coding exons was identified in patient 30, consisting of a coding DNA 1204 T→A (c.1204T>A) missense change in exon 7 that changes a tyrosine to asparagine (p.Tyr401Asn). Its pathogenicity is currently being assessed. Several changes, all previously reported as polymorphic, were identified in the 3′UTR and promoter regions. In three patients, a deletion (del) of two bases (c.1-1782_1783delCA) at 1783 bp upstream of exon 1 was identified in a repetitive sequence of short interspersed nuclear elements. Multiplex ligation-dependent probe amplification (10) did not suggest any large rearrangements, deletions, or duplications of exons. In a study of promoter methylation, none of the comparisons of germline methylation status between ASE and non-ASE cases and ASE cases versus controls were signigicant (supporting online material text and table S4). Thus, germline promoter methylation is unlikely to play a role in ASE.
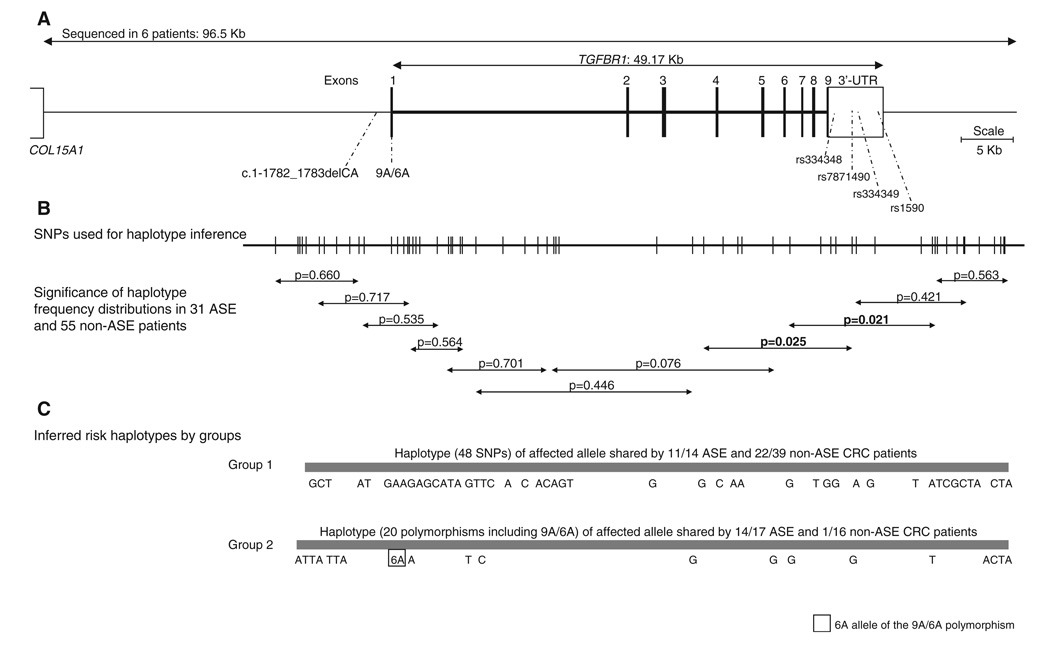
We hypothesized that changes occurring in noncoding regions of the gene could be responsible for the reduction in expression. To fully study this possibility, overlapping fragments of 1.7 to 10 kb were amplified by long-range PCR, cloned, and sequenced. In all, approximately 96.5 kb covering the whole gene and 3′UTR (49 kb), 35 kb upstream of exon 1 (up to the next gene COL15A1), and 12.5 kb downstream of the 3′UTR (Fig. 4) were fully sequenced in the four monochromosomal hybrids (patients 1 and 26) and in diploid DNA from four other ASE patients (patients 5, 11, 14, and 21) (10). Our sequencing strategy allowed us to determine the phase of every change within each amplicon and over larger regions when at least one change occurred in the overlapping fragments. In all, 25 and 104 changes were identified in the down-regulated alleles of patients 1 and 26, respectively, whereas 31 and 6 changes were detected in their wild-type counterparts. Diploid DNA from the four patients harbored 61, 37, 33, and 135 changes, respectively.
Fig. 4.
(A) Diagram of the TGFBR1 genomic region. The uppermost line depicts the 96.5-kb region sequenced in six ASE patients (four monochromosomal hybrids and four diploid DNAs). Shown are the locations of the 2-bp CA deletion upstream of exon 1, the 9A/6A polymorphism in exon 1, and the four SNPs in the 3′UTR used for ASE determinations. (B) Locations of the 60 SNPs used for haplotype inference in ASE (n = 31) and non-ASE (n = 55) CRC patients. The arrowed shorter lines each depict a 10-SNP overlapping window. P values indicate the significance of differences in haplotype distribution between ASE and non-ASE individuals. (C) Two major haplotypes identified in ASE patients are shown.
Excluding changes known to be present in the wild-type alleles, 140 changes were identified in the down-regulated alleles. Only the c.1-1782_1783delCA change stood out as a candidate mutation. It occurred in 3/29 (10.3%) ASE patients, in 0/3 ASE controls, in 1/51 (2%) non-ASE CRC patients, and 1/81 (1.2%) non-ASE controls. In summary, these investigations did not uncover the genetic changes causing ASE.
Genotyping of most changes identified by sequencing was carried out in all available ASE CRC patients, including borderline cases (n = 31), and in 55 non-ASE CRC patients. Construction of haplotypes from the available genotype and haplotype data was performed with PHASE v.2.1.1 (10). In all, 60 polymorphisms covering 73.5 kb (from 12 kb upstream of exon 1 to 12.5 kb downstream of the 3′UTR) were used for haplotype inference (table S5). For all ASE and non-ASE patients, the program was run with 1000 permutations with overlapping 10-SNP sliding windows. Haplotype frequency distributions in ASE and non-ASE populations showed significant differences in a genomic region covering the area between the 3′ end of intron 3 to ~5 kb downstream of the 3′ end of the UTR (Fig. 4).
The group of patients carrying the minor allele for the three 3′UTR SNPs in linkage disequilibrium (group 1) was very different from the other group derived from the study of SNP rs7871490 (group 2). Haplotype analysis was performed separately in the two groups, using 50 and 21 SNPs, respectively. In group 1 (n = 53), one major haplotype for the affected alleles was present in 11/14 (78.6%) of ASE but also in 22/39 (56.4%) non-ASE patients (Fig. 4). For group 2 (n = 33), another major haplotype for the affected allele was present in 14/17 (82.4%) of ASE and in 1/16 (6.3%) of non-ASE patients (Fig. 4). Fisher’s exact test to compare haplotype proportions showed P values of 0.2031 and 1.260 × 10−5 for groups 1 and 2, respectively. The 6A allele of the 9A/6A polymorphism occurred in the ASE haplotype in all 14 cases of group 2, but not in group 1, where all ASE cases except one were homozygous for the 9A allele.
In search of somatic changes in line with Knudson’s two-hit hypothesis, loss of heterozygosity (LOH) analyses as well as a search for somatic mutations in the coding sequences of the gene were performed in DNA from the tumors of 26 ASE patients. Using the described threshold (10), 6 cases out of 26 showed LOH. In three of these six cases, the wild-type allele, the one with normal expression in blood, was lost or reduced, whereas in the other three cases, the allele showing germline ASE was lost. Exon-by-exon sequencing of the entire gene in tumors from 26 ASE patients revealed somatic changes in three tumors that were not found in blood DNA. The mutations were: c.634G>A (p.Gly212Asp) in one tumor and c.682_685delAAG (p.Glu228del) in two tumors. These mutations occurred in exon 4, which encodes the kinase domain of the protein. LOH analyses and exon 4 sequencing in 49 tumors of CRC patients without ASE showed that none of these tumors had evidence of somatically acquired mutations, and five showed LOH (table S3). Fisher’s exact test comparing proportions of LOH and mutations between ASE and non-ASE cases showed P values of 0.1708 and 0.0355, respectively. The occurrence of somatic mutations in ASE cases but not in controls supports the role of TGFBR1 as a tumor suppressor gene. On the other hand, the fact that LOH affected the ASE allele as often as the wild-type allele could indicate random losses.
The cohort of MSI-negative CRC patients had been deliberately enriched in familial cases (10). In the cohort of 138 patients with available ASE values, 59 out of 136 (43.4%) were familial according to the criteria indicated above, and family information was not available in two cases. Among the cases showing ASE, 53.6% were familial (table S3). The proportion of ASE was higher among familial than nonfamilial cases: 15/59 (25.4%) familial cases versus 13/77 nonfamilial cases (16.9%). A chi-square test to compare proportions showed that this difference was not statistically significant (P = 0.314).
The above data suggest that ASE contributes somewhat more to familial than to sporadic CRC but do not allow its inheritance to be assessed. If ASE is regularly inherited as a dominant trait, the expectation is that 50% of first-degree relatives (FDRs) also have ASE. Data from four families that are informative in this regard are shown in fig. S2. In all, among 11 FDRs, ASE was greater than 1.5 in 4, borderline in 2 (ASE values 1.40 and 1.44), and low in 5. There was no instance of ASE being incompatible with Mendelian dominant inheritance. In all four families, co-segregation of ASE with the inferred risk haplotype, representing the down-regulated allele, occurred. The highest Kong and Cox nonparametric LOD score was 1.25, with a P value of 0.008 (nonparametric z score = 4.12; P value = 0.00002). Among the four to six ASE-positive FDRs, two had CRC, one had endometrial cancer and a tubular colonic adenoma, one had prostate cancer, and another had multiple polyps in the colon and rectum (table S6). Although fragmentary, these data suggest dominant inheritance of ASE with incomplete penetrance of CRC in ASE carriers.
There is indirect evidence to support the notion that ASE of TGFBR1 contributes to CRC development. The TGF-β pathway is strongly involved in the carcinogenesis of colon and other cancers, and its signaling is dependent on the integrity of both of its receptors (TGFBR1 and TGFBR2) (22, 23). In a comprehensive study of CRC tumors, somatic mutations occurred with high frequency in 69/13,023 genes. Among these 69 genes were TGFBR2, SMAD4, SMAD2, and SMAD3, attesting to the importance of the TGF-β pathway in CRC (24). There is rapidly increasing evidence that subtle variations in gene expression play central roles not only in development in various organisms but also in human disease (8, 9, 25). Linkage analysis of a cohort of sibling pairs concordant or discordant for colorectal carcinoma or adenoma highlighted a region in chromosome 9q22-31 (26). Subsequently, borderline significant linkage to the same region was observed in families segregating colorectal cancer or adenoma without microsatellite instability (27, 28). This evidence is compatible with, but in no way proves, a role for TGFBR1.
We were unable to determine what mechanism causes ASE. The haplotype data support the implication of ancestral mutations for most ASE patients. Moreover, the elusive genomic change causing ASE is likely to occur in cis, but the data do not exclude the possibility that ASE arises as a result of trans-acting genes that preferentially affect the risk haplotypes. Such genes could well be RNA genes as predicted earlier (29). Very recently, the existence of extensive quantitative trait loci for gene expression was documented in two large studies (30, 31).
How common is ASE of TGFBR1? Using our definition, it occurred in 29/138 tested CRC patients (21%) and in 3/105 tested controls (3%). In the extreme, if none of the non-informative CRC cases had ASE, the frequency would be 29/242 (12%), and for the controls, 3/195 (1.5%). Because not all individuals are informative (heterozygous for a transcribed SNP), the true frequency in cases and controls cannot be precisely assessed at present. Using the above alternative numbers, we can calculate the OR of CRC in carriers of ASE. In the first scenario, the OR is 9.0 (CI 2.7 to 30.6), and in the conservative one, OR is 8.7 (CI 2.6 to 29.1).
What proportion of all CRC is attributable to ASE of TGFBR1? From the available data of the present case-control study, we estimated the population attributable risk (PAR). If ASE occurs in 21% of cases and 3% of controls, the estimated PAR is 18.7% (CI 10.8 to 25.8). If ASE occurs in 12% of cases and 1.5% of controls, the estimated PAR is 10.6% (CI 6.0 to 14.9). These numbers are estimates, representing the Caucasian-dominated population of central Ohio, and are heavily dependent on the relevant allele frequencies, which may show strong inter-ethnic variation. We nevertheless conclude that ASE of TGFBR1 is a major contributor to the genetic predisposition to CRC.
Supplementary Material
www.sciencemag.org/cgi/content/full/1159397/DC1
Materials and Methods
SOM Text
Figs. S1 and S2
Tables S1 to S6
References
Footnotes
Information about obtaining reprints of this article or about obtaining permission to reproduce this article in whole or in part can be found at: http://www.sciencemag.org/about/permissions.dtl
References and Notes
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. CA Cancer J. Clin. 2005;55:74. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.de la Chapelle A. Nat. Rev. Cancer. 2004;4:769. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 3.Lindor NM, et al. JAMA. 2005;293:1979. [Google Scholar]
- 4.Siegel PM, Massague J. Nat. Rev. Cancer. 2003;3:807. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 5.Markowitz S, et al. Science. 1995;268:1336. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 6.Pasche B, et al. Cancer Res. 1998;58:2727. [PubMed] [Google Scholar]
- 7.Xu Y, Pasche B. Hum. Mol. Genet. 2007;16:R14. doi: 10.1093/hmg/ddl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan H, et al. Nat. Genet. 2002;30:25. doi: 10.1038/ng799. [DOI] [PubMed] [Google Scholar]
- 9.Raval A, et al. Cell. 2007;129:879. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Materials and methods are available as supporting material on Science Online.
- 11.Massague J, Gomis RR. FEBS Lett. 2006;580:2811. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 12.Massague J. Mol. Cell. 2008;29:149. doi: 10.1016/j.molcel.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, et al. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10669. [Google Scholar]
- 14.Zhu Y, Richardson JA, Parada LF, Graff JM. Cell. 1998;94:703. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]
- 15.Sodir NM, et al. Cancer Res. 2006;66:8430. doi: 10.1158/0008-5472.CAN-06-1437. [DOI] [PubMed] [Google Scholar]
- 16.Sekimoto G, et al. Cancer Res. 2007;67:5090. doi: 10.1158/0008-5472.CAN-06-4629. [DOI] [PubMed] [Google Scholar]
- 17.Pasche B, et al. J. Clin. Oncol. 2004;22:756. doi: 10.1200/JCO.2004.99.271. [DOI] [PubMed] [Google Scholar]
- 18.Pasche B, et al. Cancer Res. 1999;59:5678. [PubMed] [Google Scholar]
- 19.Pasche B, et al. JAMA. 2005;294:1634. doi: 10.1001/jama.294.13.1634. [DOI] [PubMed] [Google Scholar]
- 20.Bian Y, et al. J. Clin. Oncol. 2005;23:3074. doi: 10.1200/JCO.2005.00.281. [DOI] [PubMed] [Google Scholar]
- 21.Skoglund J, et al. Clin. Cancer Res. 2007;13:3748. doi: 10.1158/1078-0432.CCR-06-2865. [DOI] [PubMed] [Google Scholar]
- 22.Bierie B, Moses HL. Cytokine Growth Factor Rev. 2006;17:29. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Massague J, Blain SW, Lo RS. Cell. 2000;103:295. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 24.Sjoblom T, et al. Science. 2006;314:268. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 25.Yan H, Zhou W. Curr. Opin. Oncol. 2004;16:39. doi: 10.1097/00001622-200401000-00008. [DOI] [PubMed] [Google Scholar]
- 26.Wiesner GL, et al. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12961. doi: 10.1073/pnas.2132286100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kemp ZE, et al. Cancer Res. 2006;66:5003. doi: 10.1158/0008-5472.CAN-05-4074. [DOI] [PubMed] [Google Scholar]
- 28.Skoglund J, et al. J. Med. Genet. 2006;43:e7. doi: 10.1136/jmg.2005.033928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morley M, et al. Nature. 2004;430:743. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goring HH, et al. Nat. Genet. 2007;39:1208. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 31.Stranger BE, et al. Nat. Genet. 2007;39:1217. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.We thank H. He, K. Sotamaa, and W. Frankel for help. This work was supported by NIH grants CA67941, CA16058, CA112520, and CA108741; grants from the Walter S. Mander Foundation, Chicago, IL; and the Jeannik M. Littlefield–American Association for Cancer Research Grant in Metastatic Colon Cancer Research. L.V. was supported by a fellowship from the Fundación Ramón Areces.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
www.sciencemag.org/cgi/content/full/1159397/DC1
Materials and Methods
SOM Text
Figs. S1 and S2
Tables S1 to S6
References