Abstract
Background
Campylobacter jejuni, the commonest cause of bacterial diarrhoea worldwide, can also induce colonic inflammation. To understand how a previously identified heat stable component contributes to pro-inflammatory responses we used microarray and real-time quantitative PCR to investigate the transcriptional response to a boiled cell extract of Campylobacter jejuni NCTC 11168.
Results
RNA was extracted from the human colonocyte line HCA-7 (clone 29) after incubation for 6 hours with Campylobacter jejuni boiled cell extract and was used to probe the Affymetrix Human Genome U133A array. Genes differentially affected by Campylobacter jejuni boiled cell extract were identified using the Significance Score algorithm of the Bioconductor software suite and further analyzed using the Ingenuity Pathway Analysis program. The chemokines CCL20, CXCL3, CXCL2, Interleukin 8, CXCL1 and CXCL6 comprised 6 of the 10 most highly up-regulated genes, all with Significance Scores ≥ 10. Members of the Tumor Necrosis Factor α/Nuclear Factor-κB super-family were also significantly up-regulated and involved in the most significantly regulated signalling pathways (Death receptor, Interleukin 6, Interleukin 10, Toll like receptor, Peroxisome Proliferator Activated Receptor-γ and apoptosis). Ingenuity Pathway Analysis also identified the most affected functional gene networks such as cell movement, gene expression and cell death. In contrast, down-regulated genes were predominantly concerned with structural and metabolic functions.
Conclusion
A boiled cell extract of Campylobacter jejuni has components that can directly switch the phenotype of colonic epithelial cells from one of resting metabolism to a pro-inflammatory one, particularly characterized by increased expression of genes for leukocyte chemoattractant molecules.
Background
Campylobacter jejuni (C. jejuni) is a gram-negative micro-aerophilic bacterium responsible for the majority of human bacterial enteric infections worldwide [1,2]. C. jejuni is commonly found as a commensal organism in the intestinal tracts of a wide range of wild and domestic animals, including commercial poultry [3]. Cross-contamination from raw poultry or insufficient cooking of poultry meat are common sources of infection. Enteric infections by this pathogen are often associated with a potent localized inflammatory response. Symptoms arising from infection include watery or bloody diarrhoea with abdominal cramping and fever. In addition, C. jejuni can be invasive and is associated with septicaemia, meningitis, Guillain-Barré syndrome [4] and more recently with immuno-proliferative disease [5].
C. jejuni virulence factors for human disease include flagella based chemotaxis, adhesin-based cellular adherence, host cell invasion and the elaboration of a heat labile cytolethal distending toxin (CLDT) [2,6,7] In previous studies we have additionally shown that a heat stable C. jejuni boiled cell extract (BCE) is able to activate the transcription factor NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) [8]. This signalling molecule is responsible for inducing the expression of a number of genes involved in inflammation and cell mediated immunity [9], including chemokines capable of attracting leukocytes, resulting in inflammation. NF-κB is held inactive in the cytoplasm of a cell, whilst its nuclear localization domain is masked by inhibitory IκB proteins. If IκB is phosphorylated, leading to ubiquitin-mediated proteolysis, then NF-κB is released to transport to the nucleus of the cell, where it affects transcription of κB-responsive promoters. Therefore products that activate NF-κB can be presumed to have a strong role in triggering inflammation. Previous work has shown that live C. jejuni and a BCE can induce both NF-κB, and the synthesis and release of the chemokine interleukin-8 [8].
In order to identify a wider range of genes affected by C. jejuni products and assess the relative importance of the NF-κB response we used microarray technologies to identify genes that were both up and down-regulated in HCA-7 cells after exposure to a C. jejuni BCE [8,10]. Use of the Ingenuity Pathway Analysis (IPA) program suite enabled us to group co-regulated genes in order to identify the cellular signalling pathways activated in HCA-7 cells in response to C. jejuni BCE. The transcriptomic data were confirmed by real time quantitative PCR (RQ-PCR).
Methods
C. jejuni culture and preparation of BCE
The type strain C. jejuni National Collection of Type Cultures (NCTC) 11168 was used throughout these experiments, since it was originally isolated from a patient with diarrhoea, its genome sequence is available and it has a well-characterized pathological phenotype [11]. It was incubated on blood-agar plates (Blood Agar Base CM0271 from Oxoid, Basingstoke, UK with 5%, v/v defibrinated horse blood) under micro-aerobic conditions for 24 h. and used to inoculate Nutrient Broth no. 2 (Oxoid CM0067, 600 ml in 1000 ml flask). Inoculated flasks were shaken at 140 rpm at 42°C for 16 h. under micro-aerobic conditions. Culture purity was determined by plating samples from each overnight culture onto blood plates and incubating for 24 h., 42°C in micro-aerobic conditions. Bacteria were collected by centrifugation at 10,000 g for 15 min. The cell pellet was washed three times in Phosphate Buffered Saline (PBS), weighed and re-suspended in PBS to achieve a 10% (w/v) suspension, which was boiled for 10 min., cooled on ice for 5 min. before being centrifuged at 10, 000 g for 10 min. The supernatant was collected, passed through a 0.2 μm filter to remove residual bacteria and stored at -20°C until required.
HCA-7 cell culture and treatment with C. jejuni BCE
The human colonocyte line HCA-7 [10], clone 29, was grown to confluence in a 5% CO2 atmosphere in monolayer cultures on monolayer dishes in Dulbecco's Modified Eagle's Medium supplemented (DMEM) with 100 μg/ml penicillin, 100 μg/ml streptomycin and fetal calf serum at 10% (v/v, Fisher Scientific, Loughborough, UK) at 37°C. Twenty-four hours prior to induction by BCE, HCA-7 cells were transferred to serum-free DMEM. HCA-7 cells were then incubated for 6 h. with 25 μl BCE or PBS control in a total volume of 1 ml of DMEM. The BCE preparation was determined in parallel to induce NF-κB 300-fold using a reporter cell assay [8]. At 6 h. post induction total RNAs were extracted using RNAeasy columns (Qiagen, West Sussex, UK). Total RNA yields and purity were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies UK Limited, Stockport, UK).
cDNA synthesis
Approximately 10 μg of total RNA was reverse transcribed at 42°C for 1 h. to generate first strand DNA using 100 pmol oligo dT(24) primer containing a 5'-T7 RNA polymerase promoter sequence (5'-GCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT)24-3'), 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol (DTT), 10 mM dNTPs and 200 units SuperScript II reverse transcriptase (Invitrogen Life Technologies, Strathclyde, UK). Second strand DNA synthesis was carried out at 16°C for 2 h., using 10 units of E. coli polymerase I, 10 units of E. coli DNA ligase and 2 units of RNase H in a reaction containing 25 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 10 mM (NH4)SO4, 0.15 mM β-NAD+ and 10 mM dNTPs. 10 units of T4 DNA polymerase were added and the reaction allowed to proceed for a further 5 min. before termination with 0.5 M EDTA. Double stranded cDNA products were purified using the GeneChip Sample Cleanup Module (Affymetrix, Santa Clara, CA, USA).
cRNA synthesis
The synthetic cDNAs were in vitro transcribed using T7 RNA polymerase (ENZO BioArray High Yield RNA Transcript Labeling Kit, Affymetrix, Santa Clara, CA, USA) with biotinylated ribonucleotides to generated biotinylated complementary RNAs (cRNAs). The cRNAs were purified using the GeneChip Sample Cleanup Module before random fragmentation at 94°C for 35 min. in a buffer containing 40 mM Tris-acetate (pH 8.1), 100 mM potassium acetate and 30 mM magnesium acetate to generate molecules of approximately 35 to 200 bases long.
Array hybridization
Changes in gene transcription were analyzed by hybridization to Affymetrix Human Genome U133A array (HG-U133A) which contains probes for over 22,000 transcripts, including representation of the RefSeq database sequences and probe sets http://www.affymetrix.com/products_services/arrays/specific/hgu133.affx. The fragmented cRNAs were mixed with 0.1 mg/ml of sonicated herring sperm DNA in a hybridization buffer containing 100 mM 2-N-morpholino-ethane-sulfonic acid (MES), 1 M NaCl, 20 mM EDTA and 10% Tween 20 to make the hybridization mixture. The hybridization mixture containing the fragmented cRNA was denatured at 99°C for 5 min. and equilibrated for a further 5 min. at 45°C before centrifugation at 10,000 g for 5 min. to remove any insoluble material from the hybridization mixture. The hybridization mix was transferred to the ATH1-121501 genome array (Affymetrix, Santa Clara, CA, USA) cartridge and hybridized at 45°C for 16 h. on a rotisserie at 60 rpm.
After a 16 h. hybridization period the arrays were washed and stained in a Fluidics station (Affymetrix, Santa Clara, USA). The arrays were initially washed in a low stringency buffer A (6 × SSPE [0.9 M NaCl, 0.06 M NaH2PO4, 0.006 M EDTA], 10% Tween 20) at 25°C for 10 min. and then incubated with a high stringency buffer B (100 mM MES, 0.1 M NaCl, 10% Tween 20) at 50°C for 20 min. and stained with 10 mg/ml of streptavidin phycoerythrin (SAPE), in stain buffer containing 100 mM MES, 1 M NaCl, 0.05% Tween 20 and 2 mg/ml BSA at 25°C for 10 min. After a further wash in wash buffer A at 25°C for 20 min. they were stained with biotinylated anti-streptavidin antibody at 25°C for 10 min. After antibody staining the arrays were stained again with SAPE for signal amplification and washed with buffer A at 30°C for 30 min. The arrays were finally scanned and the intensities averaged with the Agilent GeneArray Scanner (Agilent Technology UK, West Lothian, UK).
Statistical analysis of Array data and Generation of Networks and Canonical Pathways
In order to identify genes of interest we used the S Score (Significance Score) algorithm as implemented in the Bioconductor software suite http://www.bioconductor.org[12] based on the R package http://www.r-project.org[13] that takes advantage of the fact that most genes are unchanged and calculates an S score (SD from the mean). The S score threshold of +/- 2.5 and an alpha value of P = 0.005 was used to define gene changes of interest. Data listing all genes that satisfied these criteria were analyzed by Ingenuity Pathway Analysis, Ingenuity® Systems, http://www.ingenuity.com. This generated functional networks and canonical pathways that connect the differentially expressed genes, using the IPA Knowledge base, where the interactions are supported by peer reviewed publications and which contains over 1.4 million interactions between genes, proteins, and drugs. Scores were assigned allowing ranking of the networks, using a Fisher's right tailed exact test.
Analysis of microarray data by real time quantitative PCR
To confirm microarray results, extracted HCA-7 total RNA was amplified by oligo dT(15) primers according to the Im-Prom II Kit (Promega UK, Southampton UK) methodology. Representative samples of genes from a number of the major functional groups and gene networks identified by IPA program were selected to confirm the array data using RQ-PCR analysis (Tables 1, 2 and 4) under appropriate conditions for an ABI Prism 7700. Primer and probe design utilized Primer Express software (Applied Biosystems, Warrington, UK). The primers were validated for gene specificity by agarose gel electrophoresis. Reporter dye-labelled probes were used with FAM (6-carboxyfluorescein) at the 5'-end and TAMRA (6-carboxy-tetramethyl-rhodamine) at the 3'-end. Reactions were set up in a final volume of 25 μl containing 12.5 μl of 2 × Taqman Universal PCR Mastermix (Applied Biosystems, Warrington, UK): 0.75 μl of each primer (10 pmol/μl), 0.5 μl of probe (10 pmol/μl), 2 μl of cDNA (equivalent to 5 ng total RNA/μl) and 8.5 μl of water. Samples were analyzed in triplicate and the emission released reporter dye was monitored by an ABI Prism 7700 Sequence Detector (Applied Biosystems, Warrington, UK) using the default PCR program of 2 min at 50°C and 10 min at 95°C; each cycle included denaturing at 95°C for 15 s and annealing at 60°C for 1 min. Analysis of the data was via the Sequence Detection System (SDS) software (Applied Biosystems, Warrington, UK). A no template control was included in each analysis and did not give any signal with any of the primer/probe combinations. RQ-PCR data were normalized using primers to β-actin based on the considerations outlined by Hugget et al. [14].
Table 1.
Primers and probes used in the study
| Gene | Forward Primer | Reverse Primer | Probe |
|---|---|---|---|
| β-actin | TCACCGAGCGCGGCT | TAATGTCACGCACGATTTCCC | CAGCTTCACCACCACGGCCGA |
| Interleukin-8 | ATTTTCCTAGATATTGCACGGGAG | GCAAACCCATTCAATTCCTGA | AAAATTGAGGCCAAGGGCCAAGAGAA |
| ATPase, Na+/K+ transporting, Beta1 polypeptide | GCCCAGAGGGATGACATGAT | CAGACCTTTCGCTCTCCTCG | TTTGAAGATTGTGGCGATGTGCCCA |
| Syndecan 4 | TGGGTGGTTGAGTGAGTGAATT | CCTCAACTATTCCAGCCCCAT | TTTCTCTTGCCCTGTTCCTGGTGCC |
| Retinoic acid receptor responder (tazarotene induced) 1 | ACCCTGAGGAACCTGCTGGT | TGGTTTTTTGTTTCTCAGTCTGCT | TGAGCAGAGTTCAGTGTGCATGCGCT |
| tumor necrosis factor, alpha-induced protein 3 | CTTTGAGTCAGGCTGTGGGC | TTGGATGCAATTCCTTCTTTCC | ACCACAGGGAGTAAATTGGCCTCTTTGATACA |
| nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | GGCCTCCAAACACACAGTCA | GCTGCCAGAGAGTGAGGATGA | CTCCGTGAACTCTGACTCTGTGTCATAGCTCTC |
| matrix metallo-peptidase 7 | GATCCCCCTGCATTTCAGG | CTGGCCCATCAAATGGGTAG | TCATGATTGGCTTTGCGCGAGG |
Forward primer, reverse primer and Taqman probes for RQ-PCR assays used, all listed 5' - 3' direction.
Table 2.
Up-regulated genes. Functional classes of genes shown are ordered by the S score of the most highly regulated examples in the class with S score ≥ 5.
| Function | Symbol | Name | S Score |
|---|---|---|---|
| Chemokine | CCL20 | Chemokine (C-C Motif) Ligand 20 | 13.542 |
| CXCL3 | Chemokine (C-X-C Motif) Ligand 3 | 11.866 | |
| CXCL2 | Chemokine (C-X-C Motif) Ligand 2 | 11.742 | |
| IL8 | Interleukin 8 | 11.393 | |
| CXCL1 | Chemokine (C-X-C Motif) Ligand 1 | 11.096 | |
| CXCL6 | Chemokine (C-X-C Motif) Ligand 6 | 10.79 | |
| CCL2 | Chemokine (C-C Motif) Ligand 2 | 5.294 | |
| TNF/NFkB superfamily | TNFAIP3 | Tumor Necrosis Factor, Alpha-Induced Protein 3 | 11.678 |
| IKBA | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | 10.956 | |
| TNIP1 | TNFAIP3 Interacting Protein 1 | 9.344 | |
| TNFAIP2 | Tumor Necrosis Factor, Alpha-Induced Protein 2 | 8.293 | |
| OPTN | Optineurin | 6.487 | |
| IL32 | Interleukin 32 | 6.12 | |
| NFKB1 | Nuclear Factor Kappa B (P105) | 5.355 | |
| Apoptosis/Cell death | UBD | Ubiquitin D | 11.647 |
| BIRC3 | Baculoviral IAP Repeat-Containing 3 | 11.063 | |
| CFLAR | CASP8 And FADD-Like Apoptosis Regulator | 6.224 | |
| SGK | Serum/Glucocorticoid Regulated Kinase | 5.705 | |
| ISG20 | Interferon Stimulated Exonuclease Gene 20 kda | 5.575 | |
| Extracellular Matrix | MMP7 | Matrix Metallopeptidase 7 (Matrilysin, Uterine) | 9.812 |
| SDC4 | Syndecan 4 (Amphiglycan, Ryudocan) | 8.923 | |
| LAMA3 | Laminin, Alpha 3 | 5.824 | |
| LAMC2 | Laminin, Gamma 2 | 5.32 | |
| Folate receptor | FOLR1 | Folate Receptor 1 (Adult) | 8.963 |
| Redox state | SOD2 | Superoxide Dismutase 2, Mitochondrial | 8.879 |
| TXNRD1 | Thioredoxin Reductase 1 | 6.378 | |
| Cell adhesion | ICAM1 | Intercellular Adhesion Molecule 1 | 8.879 |
| FNDC3B | Fibronectin Type III Domain Containing 3B | 5.851 | |
| Cytokines/Receptors | IFNGR1 | Interferon Gamma Receptor 1 | 8.403 |
| CSF2 | Colony Stimulating Factor 2 | 5.101 | |
| PLAT | Plasminogen Activator, Tissue | 7.464 | |
| SERPINB2 | Serpin Peptidase Inhibitor 2 | 6.319 | |
| Energy metabolism | ATP1B1 | Atpase, Na+/K+ Transporting, Beta 1 Peptide | 7.184 |
| Nuclear transcription | CEBPD | CCAAT/Enhancer Binding Protein Delta | 6.708 |
| RARRES1 | Retinoic Acid Receptor Responder | 6.179 | |
| Antibacterial | LCN2 | Lipocalin 2 | 6.6 |
| PI3 | Peptidase Inhibitor 3 (Elafin) | 5.057 | |
| Cell signalling | CDC42 | Cell Division Cycle 42 | 7.28 |
| DUSP5 | Dual Specificity Phosphatase 5 | 6.541 | |
| SGPL1 | Sphingosine-1-Phosphate Lyase 1 | 6.242 | |
| Cytoskeleton/cytokinesis | TPM1 | Tropomyosin 1 | 5.689 |
| PDLIM5 | PDZ And LIM Domain 5 | 5.169 | |
| Transcription, protein synthesis and export | SF3B1 | Splicing Factor 3b, Subunit 1, | 5.146 |
| UGCG | UDP-Glucose Ceramide Glucosyltransferase | 5.388 | |
| Cell cycle | PLK2 | Polo-Like Kinase 2 | 5.55 |
| Structural | SYNGR3 | Synaptogyrin 3 | 5.133 |
| Antigen presentation | TAP1 | Transporter 1, ATP-Binding Cassette | 5.207 |
Table 4.
Comparison of results for selected up-regulated genes determined by Affymetrix/S score and RQ-PCR.
| Gene Description | Ingenuty Name | Affymetrix Probe Set | S Score | Fold RQ-PCR | Network | Location |
|---|---|---|---|---|---|---|
| Interleukin-8 | IL8 | 211506_s_at | 11.393 | 59.4 ± 15.5 | See Figure 3 | Extra-cellular |
| ATPase, Na+/K+ transporting, Beta 1 polypeptide | ATP1B1 | 201242_s_at | 7.184 | 4.5 ± 1.8 | 10 | Plasma Membrane |
| Syndecan 4 | SDC4 | 202071_at | 8.823 | 4.0 ± 0.84 | 5 | Plasma Membrane |
| Retinoic acid receptor responder (tazarotene induced) 1 | RARRES1 | 221872_at | 6.179 | 2.4± 0.7 | 8 | Plasma Membrane |
| tumor necrosis factor, alpha-induced protein 3 | TNIP1 | 207196_s_at | 9.344 | 2.0 ± 0.2 | See Figure 3 | Nucleus |
| nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | NFKBIA | 201502_s_at | 10.956 | 4.0 ± 1.2. | See Figure 3 | Cytoplasm |
| Matrix Metallo-peptidase 7 | MMP7 | 202644_s_at | 9.812 | 2.1 ± 4.2 | 9 & See Additional file 3 | Extra-cellular |
For each gene ingenuity description, name and Affymetrix probe set, assigned network and cellular location are shown together with the S score and fold RQ-PCR change compared to β-actin control.
Chemokine and cytokine analyses
Cultured cells were prepared and induced as described above. After 6 h. incubation, the media was removed and stored at -20°C until examined using a Coulter-Alter Flow Cytometer in conjunction with a BD cytometric bead array human inflammation kit according to manufacturer's instructions (BD Biosciences, Oxford, UK). IL8 and CCL20 (MIP-3α) were specifically measured using a sandwich ELISA, by capture with a murine anti-human IL8 or CCL20 and detected using biotinylated goat anti-human IL8 using streptavidin-coupled horseradish-peroxidase, according to the manufacturer's instructions (R&D Systems, Minneapolis, MN, USA).
Results
The Bioconductor and IPA programs identified 356 genes that changed with a positive or negative S score of 2.5 or greater (maximum 13.54). Three hundred were up-regulated and 56 were down-regulated (Additional file 1).
Up-regulated genes
Table 2 shows 48 genes that were up-regulated with an S score of 5 or greater. These were grouped by class and ordered by the highest S score in each class. Chemokines dominate the most highly up-regulated genes with six of the ten highest S scores. Members of the TNFα-NF-κB super family were also highly up-regulated (Table 2). Other highly up-regulated genes were those involved in apoptosis and ubiquitination, extra-cellular matrix proteins, the folate receptor, superoxide dismutase, thioredoxin reductase, Intercellular Adhesion Molecule (ICAM) 1 and cytokines or their receptors (Colony Stimulating Factor [CSF] 2 and interferon-γ receptor 1).
Down-regulated genes
Fewer genes were down-regulated than those that were up-regulated and negative S scores were less pronounced than those for the up-regulated genes. For comparative purposes Table 3 shows down-regulated genes that were selected on the basis of a more permissive S score of -2.6 or less to yield a similar number (46). These genes were grouped by class and ordered by the highest negatively regulated (lowest value) S score in each class. The pattern of down-regulated gene classes differ markedly to those that were up-regulated. Most prominent were genes concerned with the maintenance of normal cell cycle, DNA replication and cell structure. The down-regulated group feature specific genes encoding components involved in membrane transport, mitosis, nucleotide synthesis, transcription, protein synthesis and export, membrane transport and energy metabolism.
Table 3.
Down-regulated genes Functional classes of genes shown are ordered by the S score of the most highly regulated examples in the class with S score ≤ -2.6.
| Function | Symbol | Name | S Score |
|---|---|---|---|
| Cell cycle, DNA replication and Mitosis | ID1 | Inhibitor Of DNA Binding 1 | -4.416 |
| ID3 | Inhibitor Of DNA Binding 3 | -4.304 | |
| ID2 | Inhibitor Of DNA Binding 2 | -4.054 | |
| LHX3 | LIM Homeobox 3 | -3.181 | |
| KLF1 | Kruppel-Like Factor 1 | -2.97 | |
| FOXF2 | Forkhead Box F2 | -2.684 | |
| SFN | Stratifin | -4.086 | |
| FGFBP1 | Fibroblast Growth Factor Binding Protein 1 | -3.922 | |
| SKP2 | S-Phase Kinase-Associated Protein 2 (P45) | -3.035 | |
| RPA3 | Replication Protein A3 | -2.975 | |
| RFC4 | Replication Factor C 4 | -2.845 | |
| SPBC25 | Spindle Pole Body Component 25 Homolog | -2.688 | |
| Structural | REG1A | Regenerating Islet-Derived 1 Alpha | -4.213 |
| CX36 | Connexin-36 | -3.79 | |
| COL4A5 | Collagen, Type IV, Alpha 5 | -3.69 | |
| ODF1 | Outer Dense Fiber Of Sperm Tails 1 | -3.511 | |
| CD248 | CD248 Molecule, Endosialin | -2.965 | |
| Membrane transport | SLC2A1 | Solute Carrier Family 2, Member 1 | -3.912 |
| CRIP1 | Cysteine-Rich Protein 1 (Intestinal) | -3.079 | |
| SCNN1A | Sodium Channel, Nonvoltage-Gated 1 Alpha | -2.918 | |
| HFE | Hemochromatosis Gene | -2.723 | |
| Transcription, protein synthesis and export | CHMP6 | Chromatin Modifying Protein 6 | -3.599 |
| RANBP1 | RAN Binding Protein 1 | -3.48 | |
| EHBP1 | EH Domain Binding Protein 1 | -3.106 | |
| RRM2 | Ribonucleotide Reductase M2 Polypeptide | -2.957 | |
| CTDSPL | Small Carboxy-Terminal Domain Phosphatase | -2.838 | |
| DARS2 | Aspartyl-Trna Synthetase 2 (Mitochondrial) | -2.795 | |
| POLR3K | Polymerase (RNA) Subunit K | -2.701 | |
| Nucleotide synthesis | UNG | Uracil-DNA Glycosylase | -3.553 |
| GLRX | Glutaredoxin | -3.325 | |
| DUT | dUTP Pyrophosphatase | -2.967 | |
| TYMS | Thymidylate Synthetase | -2.687 | |
| Energy metabolism | ATAD4 | ATPase Family, AAA Domain Containing 4 | -3.185 |
| COX7B | Cytochrome C Oxidase Subunit 7B | -2.893 | |
| Cytoskeleton/cytokinesis | M-RIP | Myosin Phosphatase-Rho Interacting Protein | -2.954 |
| MALL | Mal, T-Cell Differentiation Protein-Like | -2.918 | |
| ARHGAP29 | Rho Gtpase Activating Protein 29 | -2.909 | |
| ROCK2 | Rho-Associated, Coiled-Coil Containing Protein Kinase 2 | -2.701 | |
| Cytokine | TGFB2 | Transforming Growth Factor, Beta 2 | -2.909 |
| C1QTNF3 | C1q And TNF Related Protein 3 | 2.701 | |
| Protease | SPINK1 | Serine Peptidase Inhibitor, Kazal Type 1 | -2.889 |
| Cell adhesion | LGALS4 | Galectin 4 | -2.869 |
| Redox | TXNIP | Thioredoxin Interacting Protein | -2.843 |
| Cell signalling | HS1BP3 | HS1-Binding Protein 3 | -2.755 |
| Anti-inflammatory | ANXA1 | Annexin A1 (Lipocortin 1) | -2.703 |
| Matrix | LAMB1 | Laminin, Beta 1 | -2.702 |
Signalling pathways
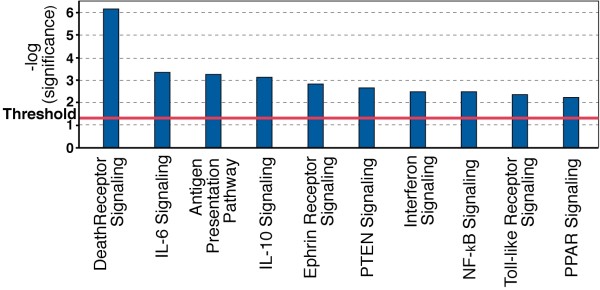
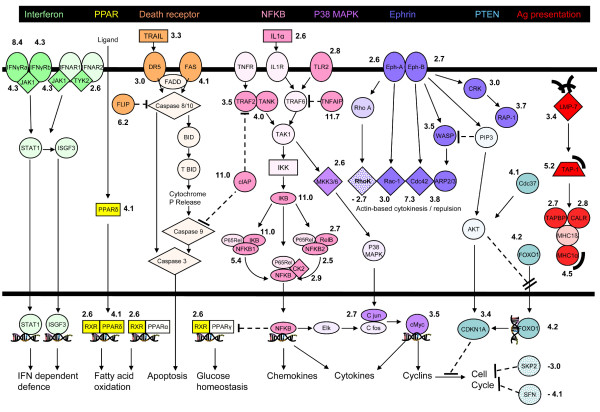
IPA identified a number of canonical signalling pathways that were most significantly affected (Figure 1). Figure 2 shows a simplified composite of all genes identified by IPA as being part of specific signalling pathways that are most significantly regulated, together with their individual S scores. Here the central mediator is the NF-κB signalling pathway that is clearly contributory in affecting the signalling through the Death Receptor, IL6, IL10, Toll-like receptor and PPAR pathways (also see Gene Networks section below and Figure 3 which also features NF-κB). In addition, several other canonical signalling pathways, some of which do not feature NF-κB, were also identified as significantly affected.
Figure 1.
Canonical Signalling Pathways identified by IPA software as significantly regulated by C. jejuni BCE. A Fisher's exact test was used to calculate a p-value (Bars) determining the probability that the association between the genes in the dataset and the canonical pathway can be explained by chance alone. Threshold refers to the cut off for p < 0.05.
Figure 2.
Regulated molecules in canonical signalling pathways identified by IPA. Individual pathways are identified by colours assigned in the black-backed heading at the top. Significantly up-regulated genes are shown in darker colour. Significantly down-regulated genes are shown stippled. Numerical values beside regulated genes show the S score. All genes identified by the IPA programme as significantly regulated have been included, together with a limited number of non-regulated genes to portray a simplified view of pathway continuity. For simplicity, molecules participating in more than one pathway are arbitrarily shown once in the most dominant pathway. Common transcriptional and other consequences of pathway activation are indicated in the Figure. Symbols are as in Figure See Figure 3 except that ---l = Inhibition (direct or indirect), ---ll = blocks translocation,) = Peptide, double helix = transcription.
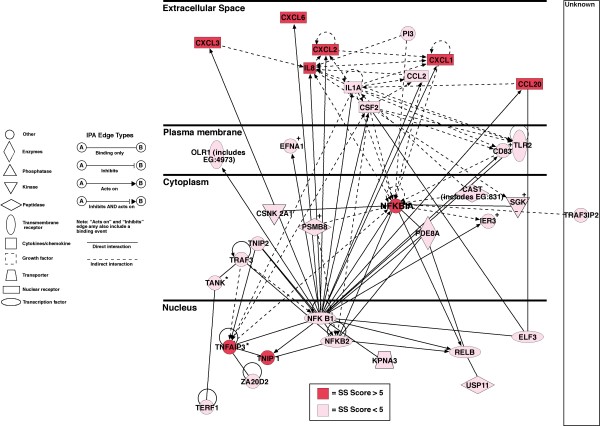
Figure 3.
IPA generated NF-κB-centred gene network. Network contains nodes (gene/gene product) and edges (indicating a relationship between the nodes) showing the cellular/subcellular location as indicated. An asterisk indicates that duplicates were identified in each dataset. Function classes of nodes indicated by shape to represent functional class, a plus sign indicates node is contained in other networks. All 35 focused genes are significantly up-regulated. Genes with an S score of ≥ 7 are shown in red and those with an S score of between 2.5–7 are shown pink. Explanation of edge types and shapes is indicated.
The antigen presentation pathway was identified through up-regulation of the Large Multifunctional Protease (LMP)-7, Transporter Associated with Antigen Processing (TAP) 1, TAP-binding protein (TAPBP), Calreticulin (CALR) and the Major Histocompatibility Complex (MHC)1-α.
Activation of the interferon-γ receptor defence signalling pathway was noted through up-regulation of both components of interferon-γ receptor, Janus kinase (JAK) 1 and Tyrosine Kinase (TYK) 2.
Activation of the ephrin signalling pathway, indicating activation of actin-based cytokinesis and repulsion. The pathway included up-regulation of ephrin receptor sub components, RHO family, GTP binding protein (Rac1), Cell Division Cycle (CDC) 42, Wiskott-Aldrich syndrome protein (WASP), actin-related protein 2 (ARP2), V-crk homologue (CRK) and Ras oncogene family member (RAP)1B with rho-associated coiled-coil containing protein kinase (ROCK) 2.
Finally, up-regulation of most components of the PI3K-phosphatase signalling pathway were noted, including phosphatase and tensin homology (PTEN) pathway indicating possible effects on the cell cycle, including Cell Division Cycle (CDC) 37, Forkhead Box (FOX)O1A and Cyclin Dependent Kinase Inhibitor (CDKN)1a (P21). SFN (Stratifin or 14-3-3σ) however, was down-regulated.
Predicted functional effects
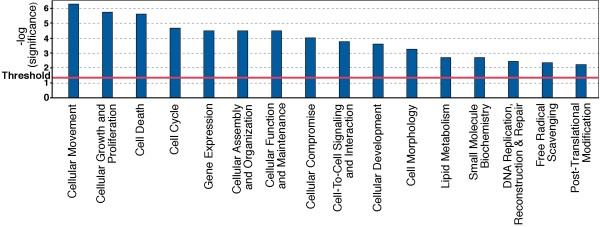
The IPA program can determine if groups of significantly changed genes have related cellular and molecular functions (Figure 4). Here IPA identified 16 functional categories that were significantly affected by the C. jejuni BCE. The most prominent functions implicated were cellular movement (reflecting changes in chemokines, adhesion receptors and molecules affecting cytokinesis), cell growth and proliferation and cell death.
Figure 4.
Functional Molecular and Cellular pathways significantly affected by C. jejuni BCE. A Fisher's exact test was used to calculate a p-value (Bars) determining the probability that the association between the genes in the dataset and the canonical pathway can be explained by chance alone. Threshold refers to the cut off for p < 0.05.
Gene networks
The IPA program constructed 16 interconnected gene networks that were significantly altered as a result of treatment of HCA-7 cells with C. jejuni BCE, all with network scores of ≥ 8. The network score is the probability that a network would be assembled by chance where a level of > 3 is statistically significant, at p < 0.001. In the four most significantly regulated all 35 focus genes of the network were affected, all giving an identical score of 52 (P < 1E-52).
The first network (Figure 3) contains genes concerned with cellular movement, particularly chemotaxis. NF-κB occupies a central position in the network and includes a number of genes which are known to up-regulate including a number of chemokines.
The second network (Additional file 2) likewise contains genes associated with cellular movement, including cytokinesis and inflammatory responses. Up-regulated genes include Ephrin Receptor B2 (EPHB2), PTGS2 (COX-2), ICAM1, both components of interferon-γ receptor, IL23A, IL27RA, JAK1, JUNB proto oncogene, Mitogen Activated Protein Kinase Kinase Kinase Kinase (MAP4K4), TYK2, Mothers Against DPP homologues (SMAD) 3, with 2 genes shown to be significantly down-regulated (SH2B and Transforming Growth Factor [TGF] β2).
MYC occupies a central position in the third network (Additional file 3), which contains genes concerned with the regulation of the cell cycle. Up-regulated genes include MYC as well as FAS, folate receptor (FOLR1), HLA molecules E, F and G, laminins β3, α3 (LAM-B3, A3) and γ2 (LAMC2), Matrix Metallo Proteinase (MMP)7, and SOD2. Down-regulated were Laminin β1 (LAMB1), RAN Binding Protein 1 (RANBP1) Thioredoxin Interacting Protein (TXNIP) and Thymidylate Synthetase (TYMS).
Finally, a network (Additional file 4) contains genes affecting cell death and gene expression. The network contains 25 genes that were up-regulated, including Activating Transcription Factor (ATF) 3, cellular Inhibitor of Apoptosis Proteins (cIAP) 1 and 2 (BIRC 2 and 3), cyclin dependent kinase (CDK) 7, cyclin dependant kinase inhibitor (CDKN) 1A, GATA binding protein (GATA) 6, TNFα-Induced Protein (TNFAIP) 2, the TNF-Related Apoptosis-Inducing Ligand (TRAIL or TNFSF10), its receptor TRAILR2 (TNFRSF10B or Death receptor [DR] 5) and TNF Receptor Associated Factor (TRAF) 2. Whilst CDKN1A is up-regulated, CDKN3 is down-regulated, as are the Inhibitors of DNA Binding (ID)1,2 and 3, Mini-Chromosome Maintenance homologue (MCM) 6, RCF4, rho-associated, coiled-coil containing protein kinase (ROCK) 2 and S-Phase Kinase-Associated Protein (SKP) 2.
Validation of Microarray data
Changes in gene expression identified by microarray were confirmed by RQ-PCR (Table 4). However, hierarchical differences are apparent between the RQ-PCR values normalized against β-actin compared with the S score associated with the significantly regulated genes as indicated by differential hybridization of the cRNA preparations to the microarray.
Chemokine and cytokine responses
To further validate the gene transcriptional changes using microarray and RQ-PCR methods, we measured the levels of secretory immunomodulatory proteins in parallel cell supernatants of HCA-7 cells pre- and post-induction with C. jejuni BCE. Table 5 presents the chemokine and cytokine levels of pro- and anti-inflammatory secretory proteins. Consistent with the microarray observations the pro-inflammatory chemokine CCL20 showed a 12.6-fold increase in levels 6 h. post treatment. IL8 levels were also found to increase, but far more dramatically than CCL20 with a 460-fold induction. HCA-7 colonocytes are particularly IL8 responsive with post-induction levels of 18.4 ng/ml, an observation that is consistent with previous reports with this cell line [8]. The pro-inflammatory cytokine IL1β showed a weak response consistent with the transcriptional response recorded in the microarray study. Pro-inflammatory cytokine IL6 showed a 5-fold increase, whereas the anti-inflammatory cytokine IL10 remained static. The transcriptional response of the genes encoding IL6 and IL10 did not show marked transcriptional changes but the pathways associated with these immunomodulatory proteins were recognized by IPA and are responsive to NF-κB.
Table 5.
Cytokine and chemokine levels (pg/ml) pre- and post-induction of HCA-7 cells with C. jejuni BCE for 6 h.
| Pre-Induction | Post-Induction | Fold-Induction | |
|---|---|---|---|
| IL10 | 12 (± 2) | 15 (± 3) | 1.25 |
| IL6 | 30 (± 3) | 150 (± 5) | 5 |
| IL1β | 20 (± 4) | 30 (± 6) | 1.5 |
| IL8 | 40 (± 16) | 18,400 (± 400) | 460 |
| CCL20 | 30 (± 6) | 380 (± 40) | 12.6 |
Discussion
Understanding the pathogenesis of C. jejuni enteric disease is important both because C. jejuni is a major cause of diarrhoeal illness worldwide and because it may serve as a model for ulcerative colitis, the pathology of which it closely resembles [15]. Previous work has shown that direct interaction between C. jejuni and epithelial cells is capable of inducing pro-inflammatory and pro-secretory processes [8,16]. These are associated with cellular invasion [17] and secretion of IL8 by CLDT dependent and independent mechanisms [16,18]. Direct use of a BCE has allowed us to use a reductionist approach to investigate effects of C. jejuni that are not dominated by these linked processes of cellular invasion by live bacteria and by toxin based cell lysis. BCE has been determined to contain polysaccharide and protein components of the cell. As demonstrated previously the NF-κB inducing activity of C. jejuni BCE is relatively insensitive to digestion by protease K [8]. However the protein content has been determined using tryptic digests of SDS-polyacryamide extracted protein bands using MALDI-TOF mass spectrometry as flagellin (Cj1339c), trigger factor (Cj0193c), lipoprotein (Cj0983), major outer membrane protein (Cj0599), cytochrome-c peroxidase (Cj0358), bacterioferritin (Cj1534c), cell binding factor PEB4A (Cj0496), hypothetical protein (Cj0706), periplasmic protein (Cj0772c), fibronectin binding protein (Cj1478c), non-heme iron protein (Cj0012c), periplasmic protein (Cj1380), periplasmic protein (Cj0420), periplasmic protein (Cj0998c), DNA-binding protein HU (Cj0913c), periplasmic cytochrome C (Cj1153) and thioredoxin (Cj0147c) [11]. The polysaccharide component features α-glucan oligomers. The C. jejuni extract is notably devoid of the dominating heat-labile effects of the CLDT. C. jejuni BCE, like infection with live C. jejuni, has been shown to be a potent inducer of NF-κB using either luciferase based reporter assays, western blots with antibodies against IκB or electrophoretic mobility shift assays in epithelial cells [8] but, unlike treatment with live C. jejuni, this does not lead to host cell lysis. These observations are consistent with the hypothesis that a heat stable component plays a significant role in the pro-inflammatory response upon exposure to C. jejuni.
We hypothesize that NF-κB modulation is central to the response of enterocytes to C. jejuni BCE; to study this we determined the global changes in gene expression induced by C. jejuni BCE treatment of the well-differentiated human colonocyte line HCA-7, clone 29. In order to ensure the relevance of our results we have adopted stringent criteria for the identification of significantly affected genes and used the IPA program to determine the functional links between these gene products, identify the signalling pathways and networks to which they belong. These changes were validated by showing similar affects on mRNA levels when genes of interest were investigated by real-time quantitative PCR.
Consistent with the initial hypothesis that NF-κB plays a major role in the response of HCA-7 cells to C. jejuni BCE, and features in 8 of the 11 designated signalling pathways identified by IPA as up-regulated. Moreover, all genes in the NF-κB associated network (Figure 3) were up-regulated by C. jejuni BCE. The dominant component of this response concerned up regulation of chemokines that would act to induce the influx of acute inflammatory cells that characterize Campylobacter colitis. Our data are remarkably similar to transcriptomic data reported by Hinata et al., who activated NF-κB by transfecting clones expressing subunits of NF-κB to show up-regulation of the chemokines CXCL3 (GRO3) IL8, CXCL6, CXCL2 (GRO2), CXCL20 (SCYA20), CXCL1 (GRO1), CCL2 (CXYA2) as well as IL1α and CSF2, all of which were also significantly up-regulated in our study [19]. The NFKB1, NFKB2 and RELB components of NF-κB are also similarly up-regulated in our study. Other changes that are likely to be of functional importance and are the up-regulation of COX2 (PTGS2), TNIP2, MYC, SOD2, ELF3 and ICAM1 (Additional file 1), where all of these processes are also downstream targets of NF-κB [20] and mediators of feedback inhibition of NF-κB activation such as NFKBIA (IκB) [9], TNIP1 [21] and TNIP2 (Figure 3) [22]. A central role for NF-κB is also supported by data using the monocytic cell line THP-1 [23]. Studies in which Caco-2 cells were incubated with live bacteria resulted in expression of many genes similar to those reported here, including chemokines, but additionally, the NF-κB inhibitor NFKBIZ [24]. This difference may reflect the ability of live bacteria to invade cells and/or elaborate a CLDT with DNase activity [6].
The pattern of significantly down-regulated genes (Table 3) is remarkably different with a reduction in expression in constitutively expressed genes concerned with nucleotide synthesis, transcription, DNA replication, mitosis, structural protein synthesis, membrane transport and energy metabolism. These changes likely reflect the reprioritization of cellular metabolism in response to pro-inflammatory products.
Whether the changes caused by the C. jejuni BCE would lead to increased or reduced apoptosis is difficult to predict, especially as HCA-7 lack a functional TP53 protein, although these cells are capable of apoptosis given the appropriate signal [25]. Invasive C. jejuni infection can cause cell death in HCA-7 cells [16], although we did not see this with the addition of BCE [8]. Increased expression of members of the death receptor pathway, the TNFα superfamily and their receptors, but also of TNFα agonists may imply regulated activation of pro-apoptotic activity [26-30]. Up-regulation of TRAIL, DR5, and FAS ligand acting via FADD, the universal adaptor protein known domain-containing members of the TNF receptor superfamily, would successively activate caspases 8, 10 and 3 as well as possible G1-S cell cycle progression [27]. However, the antagonists TNFAIP3, FLIP and cIAP, which respectively inhibit apoptosis via TRAF6, caspases 8, 9, 10 and TRAF-2 directly or indirectly are also prominent amongst the up-regulated genes [29-32].
Moreover, several other key proteins for the cell cycle and apoptosis are affected. Thus CDKN1A (P21, WAF, WAF1 or CIP1) which plays a pivotal role in inhibiting cell cycle progression at several points in response to DNA damage [33], is up-regulated, as are FOXO1A and SMAD 2 (Additional file 1) and 3 (Additional file 2), which act together to increase CDKN1A activity [34,35]. Conversely, other genes that inhibit cell cycle progression are down-regulated. These include SKP2, the F-box receptor that interacts with p19 and the CDK2/cyclin A to prevent entry into G1 [36] and SFN (stratifin or 14-3-3σ) a key target of the tumour suppressor gene TP53 which acts to cause G2 arrest [37].
Five other changes of potential functional importance are of note. Firstly, a number of potentially antibacterial agents are highly induced, including LCN2 (lipocalin-2) [38,39] and PI3 (peptidase inhibitor 3, aka ELAFIN) [40], whilst MMP7 is thought to activate defensins [41]. Secondly, five key molecules involved in antigen processing and presentation (Figure 1, 2) [42] were also up-regulated and could play a role in the development of immune responses to C. jejuni. Thirdly, alterations in matrix metalloproteinases and leukocyte receptors would influence the inflammatory response, with MMP9 acting to facilitate neutrophil transfer by activating interleukin-8 [43] and MMP7 acting to localize them to sites of tissue damage [44]. Fourthly, the ephrin pathway (Figure 2), including Ephrin A2 and B2 receptors (EPHA2, EPHB2) and Ephrin A1 (EFNA1, Figure 3), rho kinase (ROCK2), Rac, ARP2/3, CDC42 and WASP appeared to be strongly up-regulated. This pathway is concerned with activation of cytokinetic changes that may potentially play a role in rapid restitution [45,46]. Finally, up-regulation of the folate receptor (FOLR1) may reflect preparation for reparative nucleotide synthesis dependent upon one-carbon transfer activity [47].
Conclusion
The data we have generated using a BCE of C. jejuni represents a reductionist approach to determine some of the cellular responses associated with C. jejuni infection. However, because C. jejuni BCE represents a robust NF-κB inducing activity that is not only heat-stable but resistant to protease and acidic pH (pH 3) [8], these may indeed be of clinical significance if these products are shed upon C. jejuni infection or co-delivered through the diet. C. jejuni has been detected in many commercially available chicken portions [2] and clinical cases of Campylobacter enterocolitis are frequently associated with ingestion of partially cooked poultry meat [48].
Changes in host gene expression following C. jejuni BCE interestingly reflects some of the changes that are known to occur in inflammatory bowel diseases (IBD) such as ulcerative colitis, for which C. jejuni colitis can be considered a model, and may therefore indicate other potential targets for investigation of epithelial-derived mediators of inflammation in ulcerative colitis/IBD. Up-regulation of NF-κB is well recognized and considered a possible target of mesalazine [49,50]. Genes up-regulated by C. jejuni that have been associated with active ulcerative colitis/IBD include chemokines [51], such as IL8 and CCL20 (macrophage inflammatory protein 3α) [52-54] cytokines, including TNFα [55], eicosanoids [53] and elafin [56]. IL23, IL32 [57-59] and receptors such as interferon-γ receptor, and TLR2 [60] have all been demonstrated to be altered here (Table 2, Additional file 1). Activation of pro-apoptotic pathways involving the TNF superfamily and death domain signalling pathway have been reported to be up-regulated in colonic enterocytes isolated from patients with ulcerative colitis, from which C-IAP2 (BIRC3) has been proposed as a disease marker [61], whilst the leukocytes serine anti-proteinase elafin has recently been identified as a candidate biomarker for ulcerative colitis but with attenuated induction in Crohn's disease [56]. Thus, the data we report here include a number of pathways and mediators that may be realistic anti-inflammatory therapeutic targets to prevent or reduce the activity of C. jejuni colitis or ulcerative colitis. These targets include mechanisms for chemoattraction of inflammatory cells, cellular processes associated with repair and the processes associated with apoptosis, as well as NF-κB itself, the utilization of which can be investigated by intervention studies in model systems and humans.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KM conceived the study, designed, co-ordinated, interpreted the experiments, and co-wrote the manuscript. IC determined the characteristics of the BCE, contributed to experimental design, interpretation of data, and to the writing of the manuscript. ML drafted the original manuscript, performed some of the cytokine analysis and contributed to analysis of data. PC performed the analysis of transcriptomics by Bioconducter and IPA. JS performed the BCE induction experiment. ED performed RQ-PCR analysis. FM and PC analysed components of BCE. CH Co-wrote the manuscript and interpreted the data. All authors read contributed to and approved the final manuscript.
Supplementary Material
Complete listed of significantly regulated genes induced by C. Jejuni BCE. File contains all genes identified by the Bioconductor and IPA programmes as significantly regulated (S score ≤ -2.5 or ≥ 2.5). All genes are shown together with their synonym, description, Genbank name, S score, network allocation, location, family, Entrez ID for Human, Mouse and Rat, and NCBI Entrez Gene web-link. Network 1 is displayed in additional file 3, network 2 is displayed in additional file 4, network 3 is displayed in figure 3, and network 4 is displayed in additional file 2.
IPA generated cell movement associated gene network. All 35 focus genes in this pathway are significantly up or down-regulated. Labeling of Network is similar to that of figure 3. Genes with an S score of ≥ 7 are shown in red and those with an S score between 2.5–7 are shown pink. Down-regulated genes with an S score between -2.5 and -7 are shown green.
IPA generated MYC associated gene network. All 35 focus genes in this pathway are significantly up or down-regulated. Labeling of Network is similar to that of figure 3. Genes with an S score of ≥ 7 are shown in red and those with an S score between 2.5–7 are shown pink. Down-regulated genes with an S score between -2.5 and -7 are shown green.
IPA generated cell death associated gene network. All 35 focus genes in this pathway are significantly up or down-regulated. Labeling of Network is similar to that of figure 3. Genes with an S score of ≥ 7 are shown in red and those with an S score between 2.5–7 are shown pink. Down-regulated genes with an S score between -2.5 and -7 are shown green.
Contributor Information
Kenneth H Mellits, Email: ken.mellits@nottingham.ac.uk.
Ian F Connerton, Email: ian.connerton@nottingham.ac.uk.
Michael F Loughlin, Email: michael.loughlin@ntu.ac.uk.
Peter Clarke, Email: resurgo@gmail.com.
Julie Smith, Email: j.smith181@btinternet.com.
Eleanor Dillon, Email: svzerd@nottingham.ac.uk.
Phillippa L Connerton, Email: Phillippa.connerton@nottingham.ac.uk.
Francis Mulholland, Email: francis.mulholland@bbsrc.ac.uk.
Christopher J Hawkey, Email: cj.hawkey@nottingham.ac.uk.
Acknowledgements
We acknowledge the assistance if Drs. Janet Higgins and John Okyere, at NASC array service, University of Nottingham and Mr. Lyndon Cochrane for help with illustrations. IFC and KHM were the recipients of a grant award from the Biotechnology and Biological Research Council, UK
References
- Snelling WJ, Matsuda M, Moore JE, Dooley JS. Campylobacter jejuni. Lett Appl Microbiol. 2005;41(4):297–302. doi: 10.1111/j.1472-765X.2005.01788.x. [DOI] [PubMed] [Google Scholar]
- Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5(9):665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- Jorgensen F, Bailey R, Williams S, Henderson P, Wareing DR, Bolton FJ, Frost JA, Ward L, Humphrey TJ. Prevalence and numbers of Salmonella and Campylobacter spp. on raw, whole chickens in relation to sampling methods. Int J Food Microbiol. 2002;76(1–2):151–164. doi: 10.1016/S0168-1605(02)00027-2. [DOI] [PubMed] [Google Scholar]
- Hughes RA, Cornblath DR. Guillain-Barre syndrome. Lancet. 2005;366(9497):1653–1666. doi: 10.1016/S0140-6736(05)67665-9. [DOI] [PubMed] [Google Scholar]
- Lecuit M, Abachin E, Martin A, Poyart C, Pochart P, Suarez F, Bengoufa D, Feuillard J, Lavergne A, Gordon JI. Immunoproliferative small intestinal disease associated with Campylobacter jejuni. N Engl J Med. 2004;350(3):239–248. doi: 10.1056/NEJMoa031887. [DOI] [PubMed] [Google Scholar]
- Guerry P. Campylobacter flagella: not just for motility. Trends Microbiol. 2007;15(10):456–461. doi: 10.1016/j.tim.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Smith JL, Bayles DO. The contribution of cytolethal distending toxin to bacterial pathogenesis. Crit Rev Microbiol. 2006;32(4):227–248. doi: 10.1080/10408410601023557. [DOI] [PubMed] [Google Scholar]
- Mellits KH, Mullen J, Wand M, Armbruster G, Patel A, Connerton PL, Skelly M, Connerton IF. Activation of the transcription factor NF-kappaB by Campylobacter jejuni. Microbiology. 2002;148(Pt 9):2753–2763. doi: 10.1099/00221287-148-9-2753. [DOI] [PubMed] [Google Scholar]
- Brasier AR. The NF-kappaB regulatory network. Cardiovasc Toxicol. 2006;6(2):111–130. doi: 10.1385/CT:6:2:111. [DOI] [PubMed] [Google Scholar]
- Kirkland SC. Dome formation by a human colonic adenocarcinoma cell line (HCA-7) Cancer Res. 1985;45(8):3790–3795. [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403(6770):665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Kennedy RE, Kerns RT, Kong X, Archer KJ, Miles MF. SScore: an R package for detecting differential gene expression without gene expression summaries. Bioinformatics. 2006;22(10):1272–1274. doi: 10.1093/bioinformatics/btl108. [DOI] [PubMed] [Google Scholar]
- Zhang J, Carey V, Gentleman R. An extensible application for assembling annotation for genomic data. Bioinformatics. 2003;19(1):155–156. doi: 10.1093/bioinformatics/19.1.155. [DOI] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6(4):279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Colgan T, Lambert JR, Newman A, Luk SC. Campylobacter jejuni enterocolitis. A clinicopathologic study. Arch Pathol Lab Med. 1980;104(11):571–574. [PubMed] [Google Scholar]
- Beltinger J, Brough J, Skelly MM, Thornley J, Spiller RC, Stack WA, Hawkey CJ. Disruption of colonic barrier function and induction of mediator release by strains of Campylobacter jejuni that invade epithelial cells. 2008. pp. 7345–52. [DOI] [PMC free article] [PubMed]
- Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. Identification of proteins required for the internalization of Campylobacter jejuni into cultured mammalian cells. Adv Exp Med Biol. 1999;473:215–224. doi: 10.1007/978-1-4615-4143-1_22. [DOI] [PubMed] [Google Scholar]
- Hickey TE, McVeigh AL, Scott DA, Michielutti RE, Bixby A, Carroll SA, Bourgeois AL, Guerry P. Campylobacter jejuni cytolethal distending toxin mediates release of interleukin-8 from intestinal epithelial cells. Infect Immun. 2000;68(12):6535–6541. doi: 10.1128/IAI.68.12.6535-6541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinata K, Gervin AM, Jennifer Zhang Y, Khavari PA. Divergent gene regulation and growth effects by NF-kappa B in epithelial and mesenchymal cells of human skin. Oncogene. 2003;22(13):1955–1964. doi: 10.1038/sj.onc.1206198. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Gaynor RB. IkappaB kinases: key regulators of the NF-kappaB pathway. Trends Biochem Sci. 2004;29(2):72–79. doi: 10.1016/j.tibs.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Heyninck K, Kreike MM, Beyaert R. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. FEBS Lett. 2003;536(1–3):135–140. doi: 10.1016/S0014-5793(03)00041-3. [DOI] [PubMed] [Google Scholar]
- Van Huffel S, Delaei F, Heyninck K, De Valck D, Beyaert R. Identification of a novel A20-binding inhibitor of nuclear factor-kappa B activation termed ABIN-2. J Biol Chem. 2001;276(32):30216–30223. doi: 10.1074/jbc.M100048200. [DOI] [PubMed] [Google Scholar]
- Jones MA, Totemeyer S, Maskell DJ, Bryant CE, Barrow PA. Induction of proinflammatory responses in the human monocytic cell line THP-1 by Campylobacter jejuni. Infect Immun. 2003;71(5):2626–2633. doi: 10.1128/IAI.71.5.2626-2633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinella ES, Eversley CD, Carroll IM, Andrus JM, Threadgill DW, Threadgill DS. Human epithelial-specific response to pathogenic Campylobacter jejuni. FEMS Microbiol Lett. 2006;262(2):236–243. doi: 10.1111/j.1574-6968.2006.00396.x. [DOI] [PubMed] [Google Scholar]
- Huang X, Guo B. Adenomatous polyposis coli determines sensitivity to histone deacetylase inhibitor-induced apoptosis in colon cancer cells. Cancer Res. 2006;66(18):9245–9251. doi: 10.1158/0008-5472.CAN-06-0887. [DOI] [PubMed] [Google Scholar]
- Yan N, Shi Y. Mechanisms of apoptosis through structural biology. Annu Rev Cell Dev Biol. 2005;21:35–56. doi: 10.1146/annurev.cellbio.21.012704.131040. [DOI] [PubMed] [Google Scholar]
- Werner MH, Wu C, Walsh CM. Emerging roles for the death adaptor FADD in death receptor avidity and cell cycle regulation. Cell Cycle. 2006;5(20):2332–2338. doi: 10.4161/cc.5.20.3385. [DOI] [PubMed] [Google Scholar]
- Wajant H, Scheurich P. Tumor necrosis factor receptor-associated factor (TRAF) 2 and its role in TNF signaling. Int J Biochem Cell Biol. 2001;33(1):19–32. doi: 10.1016/S1357-2725(00)00064-9. [DOI] [PubMed] [Google Scholar]
- Beyaert R, Heyninck K, Van Huffel S. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis. Biochem Pharmacol. 2000;60(8):1143–1151. doi: 10.1016/S0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda JE, MacKenzie A. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379(6563):349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- Kataoka T. The caspase-8 modulator c-FLIP. Crit Rev Immunol. 2005;25(1):31–58. doi: 10.1615/CritRevImmunol.v25.i1.30. [DOI] [PubMed] [Google Scholar]
- Li X, Yang Y, Ashwell JD. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature. 2002;416(6878):345–347. doi: 10.1038/416345a. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Arden KC. FoxO: linking new signaling pathways. Mol Cell. 2004;14(4):416–418. doi: 10.1016/S1097-2765(04)00213-8. [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117(2):211–223. doi: 10.1016/S0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- Pagano M. Control of DNA synthesis and mitosis by the Skp2-p27-Cdk1/2 axis. Mol Cell. 2004;14(4):414–416. doi: 10.1016/S1097-2765(04)00268-0. [DOI] [PubMed] [Google Scholar]
- Wilker EW, van Vugt MA, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM. 14-3-3sigma controls mitotic translation to facilitate cytokinesis. Nature. 2007;446(7133):329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432(7019):917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- Borregaard N, Cowland JB. Neutrophil gelatinase-associated lipocalin, a siderophore-binding eukaryotic protein. Biometals. 2006;19(2):211–215. doi: 10.1007/s10534-005-3251-7. [DOI] [PubMed] [Google Scholar]
- Sallenave JM. The role of secretory leukocyte proteinase inhibitor and elafin (elastase-specific inhibitor/skin-derived antileukoprotease) as alarm antiproteinases in inflammatory lung disease. Respir Res. 2000;1(2):87–92. doi: 10.1186/rr18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286(5437):113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- Kloetzel PM. Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat Immunol. 2004;5(7):661–669. doi: 10.1038/ni1090. [DOI] [PubMed] [Google Scholar]
- Steen PE Van den, Proost P, Wuyts A, Van Damme J, Opdenakker G. Neutrophil gelatinase B potentiates interleukin-8 tenfold by aminoterminal processing, whereas it degrades CTAP-III, PF-4, and GRO-alpha and leaves RANTES and MCP-2 intact. Blood. 2000;96(8):2673–2681. [PubMed] [Google Scholar]
- Li Q, Park PW, Wilson CL, Parks WC. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell. 2002;111(5):635–646. doi: 10.1016/S0092-8674(02)01079-6. [DOI] [PubMed] [Google Scholar]
- Kullander K, Klein R. Mechanisms and functions of Eph and ephrin signalling. Nat Rev Mol Cell Biol. 2002;3(7):475–486. doi: 10.1038/nrm856. [DOI] [PubMed] [Google Scholar]
- Anton IM, Jones GE, Wandosell F, Geha R, Ramesh N. WASP-interacting protein (WIP): working in polymerisation and much more. Trends Cell Biol. 2007;17(11):555–562. doi: 10.1016/j.tcb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Stanger O. Physiology of folic acid in health and disease. Curr Drug Metab. 2002;3(2):211–223. doi: 10.2174/1389200024605163. [DOI] [PubMed] [Google Scholar]
- Friedman CR, Neimann J, Wegener HC, Tauxe RV. In: Campylobacter. 2. Nachamkin I, Blaser MJ, editor. Washington, DC ASM Press; 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations; pp. 121–138. [Google Scholar]
- Zhang SZ, Zhao XH, Zhang DC. Cellular and molecular immunopathogenesis of ulcerative colitis. Cell Mol Immunol. 2006;3(1):35–40. [PubMed] [Google Scholar]
- Bantel H, Berg C, Vieth M, Stolte M, Kruis W, Schulze-Osthoff K. Mesalazine inhibits activation of transcription factor NF-kappaB in inflamed mucosa of patients with ulcerative colitis. Am J Gastroenterol. 2000;95(12):3452–3457. doi: 10.1111/j.1572-0241.2000.03360.x. [DOI] [PubMed] [Google Scholar]
- Papadakis KA. Chemokines in inflammatory bowel disease. Curr Allergy Asthma Rep. 2004;4(1):83–89. doi: 10.1007/s11882-004-0048-7. [DOI] [PubMed] [Google Scholar]
- Mahida YR, Ceska M, Effenberger F, Kurlak L, Lindley I, Hawkey CJ. Enhanced synthesis of neutrophil-activating peptide-I/interleukin-8 in active ulcerative colitis. Clinical Science. 1992;82(3):273–275. doi: 10.1042/cs0820273. [DOI] [PubMed] [Google Scholar]
- Cole AT, Pilkington BJ, McLaughlan J, Smith C, Balsitis M, Hawkey CJ. Mucosal factors inducing neutrophil movement in ulcerative colitis: the role of interleukin 8 and leukotriene B4. Gut. 1996;39(2):248–254. doi: 10.1136/gut.39.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Yamakawa M, Hiroaki T, Kawata S, Kimura O. Correlation of dendritic cell infiltration with active crypt inflammation in ulcerative colitis. Clin Immunol. 2007;122(3):288–297. doi: 10.1016/j.clim.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, Bamba T, Fujiyama Y. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52(1):65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach CF, Eriksson A, Jennische E, Lange S, Gunnerek C, Lonnroth I. Detection of elafin as a candidate biomarker for ulcerative colitis by whole-genome microarray screening. Inflamm Bowel Dis. 2006;12(9):837–842. doi: 10.1097/01.mib.0000232469.23574.11. [DOI] [PubMed] [Google Scholar]
- McGovern D, Powrie F. The IL23 axis plays a key role in the pathogenesis of IBD. Gut. 2007;56(10):1333–1336. doi: 10.1136/gut.2006.115402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos PL, Szamosi T, Szilvasi A, Molnar E, Lakatos L, Kovacs A, Molnar T, Altorjay I, Papp M, Tulassay Z. ATG16L1 and IL23 receptor (IL23R) genes are associated with disease susceptibility in Hungarian CD patients. 2008. pp. 867–73. [DOI] [PubMed]
- Shioya M, Nishida A, Yagi Y, Ogawa A, Tsujikawa T, Kim-Mitsuyama S, Takayanagi A, Shimizu N, Fujiyama Y, Andoh A. Epithelial overexpression of interleukin-32alpha in inflammatory bowel disease. Clin Exp Immunol. 2007;149(3):480–486. doi: 10.1111/j.1365-2249.2007.03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Bores L, Fonseca GC, Villeda MA, Yamamoto-Furusho JK. Novel genetic markers in inflammatory bowel disease. World J Gastroenterol. 2007;13(42):5560–5570. doi: 10.3748/wjg.v13.i42.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidelin JB, Nielsen OH. Expression profiling of apoptosis-related genes in enterocytes isolated from patients with ulcerative colitis. Apmis. 2006;114(7–8):508–517. doi: 10.1111/j.1600-0463.2006.apm_116.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete listed of significantly regulated genes induced by C. Jejuni BCE. File contains all genes identified by the Bioconductor and IPA programmes as significantly regulated (S score ≤ -2.5 or ≥ 2.5). All genes are shown together with their synonym, description, Genbank name, S score, network allocation, location, family, Entrez ID for Human, Mouse and Rat, and NCBI Entrez Gene web-link. Network 1 is displayed in additional file 3, network 2 is displayed in additional file 4, network 3 is displayed in figure 3, and network 4 is displayed in additional file 2.
IPA generated cell movement associated gene network. All 35 focus genes in this pathway are significantly up or down-regulated. Labeling of Network is similar to that of figure 3. Genes with an S score of ≥ 7 are shown in red and those with an S score between 2.5–7 are shown pink. Down-regulated genes with an S score between -2.5 and -7 are shown green.
IPA generated MYC associated gene network. All 35 focus genes in this pathway are significantly up or down-regulated. Labeling of Network is similar to that of figure 3. Genes with an S score of ≥ 7 are shown in red and those with an S score between 2.5–7 are shown pink. Down-regulated genes with an S score between -2.5 and -7 are shown green.
IPA generated cell death associated gene network. All 35 focus genes in this pathway are significantly up or down-regulated. Labeling of Network is similar to that of figure 3. Genes with an S score of ≥ 7 are shown in red and those with an S score between 2.5–7 are shown pink. Down-regulated genes with an S score between -2.5 and -7 are shown green.