Summary
Curli are adhesive fimbriae of Escherichia coli and Salmonella enterica. Expression of curli (csgA) and cellulose (bcsA) is co-activated by the transcriptional activator CsgD. In this study, we investigated the contribution of curli and cellulose to the adhesive properties of enterohemorragic (EHEC) O157:H7 and enteropathogenic E. coli (EPEC) O127:H6. While single mutations in csgA, csgD, or bcsA in EPEC and EHEC had no dramatic effect on cell adherence, double csgAbcsA mutants were significantly less adherent than the single mutants or wild-type strains to human colonic HT-29 epithelial cells or to cow colon tissue in vitro. Over-expression of csgD (carried on plasmid pCP994) in a csgD mutant, but not in the single csgA or bscA mutants, led to significant increase in adherence and biofilm formation in EPEC and EHEC, suggesting that synchronized over-production of curli and cellulose enhances bacterial adherence. In line with this finding, csgD transcription was activated significantly in the presence of cultured epithelial cells as compared to growth in tissue culture medium. Analysis of the influence of virulence and global regulators in the production of curli in EPEC identified Fis (factor for inversion stimulation) as a, heretofore unrecognized, negative transcriptional regulator of csgA expression. An EPEC E2348/69Δfis produced abundant amounts of curli whereas a double fiscsgD mutant yielded no detectable curli production. Our data suggest that curli and cellulose act in concert to favor host colonization, biofilm formation, and survival in different environments.
Keywords: Curli, gene expression/regulation, Escherichia coli, attaching and effacing, cellulose, Fis
Introduction
Non-pathogenic as well as human, avian, and animal pathogenic Escherichia coli isolates and Salmonella enterica serovars produce adhesive fimbrial structures collectively referred to as curli (Olsen et al., 1989; Collinson et al., 1991; Collinson et al., 1992; Provence and Curtiss, 1992). Morphologically, curli are thin (2–5 nm diameter), coiled, highly aggregative fibers of varying length that protrude from the bacterial surface as an amorphous matrix (Olsen et al., 1989; Bian and Normark, 1997; Collinson et al., 1997; Prigent-Combaret et al., 2000; Brown et al., 2001; Chapman et al., 2002). Curli fibers bind the Congo red dye and certain host proteins including fibronectin, laminin, plasminogen, and major histocompatibility complex class I molecules (Olsen et al., 1989; Olsen et al., 1998; Johansson et al., 2001). Curli possess characteristics of amyloid fibers, which in uropathogenic E. coli tend to form large aggregates during infection of the urinary tract by uropathogenic E. coli (Chapman et al., 2002). Cytokine activation during human sepsis has also been attributed to curli (Bian et al., 2000). In avian pathogenic E. coli isolates, curli promote adherence to avian intestinal cells, internalization of HeLa cells (Gophna et al., 2001), and persistence in the caecum of chickens (La Ragione et al., 1999). Curli are also involved in the colonization of abiotic surfaces, the development of biofilms, and bacterial auto-aggregation (Vidal et al., 1998; Dorel et al., 1999; Bian et al., 2000; Prigent-Combaret et al., 2000). Together with cellulose, curli form a honeycomb-like structure that enables biofilm development through bacteria-bacteria interactions and adhesion to biotic and abiotic surfaces, conferring protection against environmental foes (Gerstel and Romling, 2001; Zogaj et al., 2001; Solano et al., 2002; Jain and Chen, 2007; Jonas et al., 2007; Gualdi et al., 2008). Due to these features, curli are considered a virulence attribute in these bacteria (Provence and Curtiss, 1992; Bian et al., 2000).
The nucleotide sequence, structure, and regulation of curli operons in S. enterica serovar Typhimurium and E. coli are highly conserved (Romling et al., 1998). The biosynthesis and assembly of curli fibers are directed by proteins encoded within two divergently transcribed operons, csgDEFG and csgBA (Hammar et al., 1995). The CsgD protein (called AgfD in S. enterica) directly regulates the transcriptional activation of the csgBA operon (Hammar et al., 1995; Romling et al., 2000), while the expression of the csgDEFG operon is positively or negatively regulated by a myriad of transcriptional regulators such as Crl, MlrA, Rcs, and the two component systems OmpR/EnvZ and CpxA/CpxR (Vidal et al., 1998; Dorel et al., 1999; Brown et al., 2001), as well as by global regulators RpoS, IHF, and H-NS (Barnhart and Chapman, 2006). In addition, the production of the bacterial extracellular matrix is also under the control of this complex regulatory network, as CsgD, indirectly regulates cellulose production through the activation of the gene encoding AdrA, a protein involved in the synthesis of cyclic-di-GMP that is necessary for cellulose production (Romling et al., 2000).
The role of curli in the pathogenesis of enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC), two major causes of diarrheal disease in the world, has not been investigated in detail. Collectively, EHEC and EPEC are called attaching and effacing E. coli (AEEC) because they adhere closely and efface the intestinal epithelium due to the activity of type 3 secretion system effectors encoded on the locus of enterocyte effacement (LEE) (Staley et al., 1969; Moon et al., 1983; McDaniel et al., 1995; Frankel and Phillips, 2008). EPEC produces a type IV bundle-forming pilus (BFP) responsible for promoting bacterial interactions within the microcolony (Giron et al., 1991); an outer membrane protein called intimin, involved in the intimate attachment of the bacteria to the cell membrane (Jerse et al., 1990); and flagella, which contribute to the adherence properties of motile EPEC strains (Giron et al., 2002).
EHEC serotype O157:H7 is an emerging food-borne pathogen responsible for hemorrhagic colitis and the hemolytic uremic syndrome (HUS). A key aspect of EHEC infection is the colonization of the gut mucosa and the production of two potent Shiga toxins (Paton and Paton, 1998). So far, intimin is the only factor that has been demonstrated to play a role in intestinal colonization in vivo (McKee et al., 1995). Several fimbriae have been identified in EHEC O157:H7 and non-O157:H7 strains (Brunder et al., 2001; Srimanote et al., 2002; Torres et al., 2002; Uhlich et al., 2002; Kim and Kim, 2004; Low et al., 2006), but their role in human and bovine host colonization is unclear. Recently, we reported that pathogenic and non-pathogenic E. coli strains produce an adhesion appendage called “E. coli common pilus” or ECP, which confers the bacteria the ability to adhere to cultured epithelial cells (Rendon et al., 2007). EHEC O157:H7 strains also produce adhesive type IV pili called “hemorrhagic coli pilus” (HCP), which are proposed to be intestinal colonization factors (Xicohtencatl-Cortes et al., 2007). It was suggested that certain EHEC strains produce curli, which mediate attachment and invasion of human laryngeal epithelial cells (HEp-2) (Uhlich et al., 2002; Kim and Kim, 2004). In this report, we investigated the role and contribution of curli and cellulose in adherence of EPEC O127:H6 and EHEC O157:H7 to cultured mammalian cells and in biofilm formation. In addition, the participation of virulence and global regulators in the production of curli was studied.
Results
Production of curli by EPEC and EHEC strains at 37°C
Most pathogenic and commensal E. coli strains do not typically produce curli, especially at mammalian host temperature (37°C) (Olsen et al., 1989; Uhlich et al., 2001; Chapman et al., 2002). The production of curli and its role in epithelial cell adherence by prototypic EPEC and EHEC strains has not been investigated in detail. We began this study by investigating curli production in prototypic EPEC (O127:H6) strain E2348/69 and EHEC (O157:H7) strain EDL933 under growth conditions that are permissive for curli production, e.g. growth on low-salt medium (tryptone agar [T-medium]) at 37°C (Collinson et al., 1991). We observed the presence of fimbrial structures on EPEC E2348/69 and EHEC EDL933 (Fig. 1 and Table 1), which were recognized by anti-curli E. coli K-12 antibodies by immunogold labelling, confirming the identity of the fibers as curli (data not shown). For further characterization, these flexible fibers were purified from EPEC E2348/69 and found to be composed of a protein that migrated with an apparent molecular mass of 17 kDa, whose N-terminal amino acid sequence (GVVPQYGGGGGN) matched the N-terminus of CsgA, the major subunit of curli (Collinson et al., 1992). Purified EPEC curli was used to raise antibodies in rabbit by three weekly subcutaneous injections, and these antibodies were employed in subsequent studies.
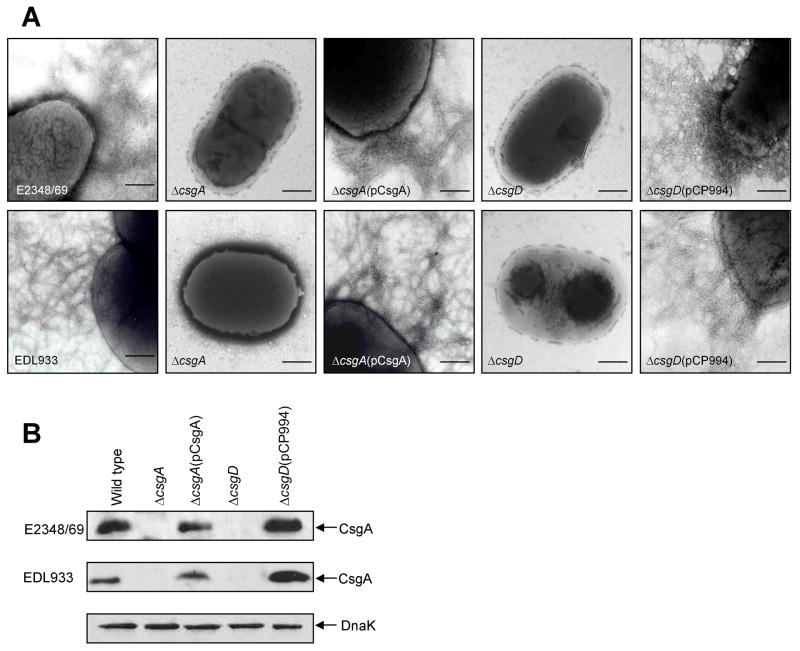
Fig. 1.
Analysis of curli fibers production in AEEC strains. (A) Electron micrographs of wild-type and derivative strains showing lack of curli fibers on the curli csgA and csgD mutants. The AEEC csgA and csgD mutants carrying pCsgA or pCP994 rshowed production of curli fibers. (B) Immunoblots of whole cell extracts of AEEC and derivative strains showing curlin, the samples were treated with formic acid before SDS-PAGE. Detection of DnaK with anti-DnaK antibody was used to ensure equal amounts of antigen tested. Scale bars, 250 μm.
Table 1.
E. coli strains and plasmids used in this study.
| Strains | Notes | Source |
|---|---|---|
| EDL933 | EHEC (O157:H7) Wild type | (Riley et al., 1983) |
| EDL933ΔcsgA | csgA::km | This study |
| EDL933ΔbcsA | bcsA::cm | This study |
| EDL933ΔcsgD | csgD::km | This study |
| EDL933ΔcsgAΔbcsA | Double csgA::km/bcsA::cm mutant | This study |
| E2348/69 | EPEC (O127:H6) Wild type | (Levine et al., 1978) |
| E2348/69ΔcsgA | csgA::km | This study |
| E2348/69ΔbcsA | bcsA::cm | This study |
| E2348/69ΔcsgD | csgD::km | This study |
| E2348/69ΔfisΔcsgD | Double fis::cm/csgD::km mutant | This study |
| E2348/69ΔcsgAΔbcsA | Double csgA::km/bcsA::cm mutant | This study |
| E2348/69Δfis | fis::km | This study |
| E2348/69(pMLB1034) | Promoter-less control | This study |
| E2348/69ΔgrlA | grlA::km mutant | Bustamante et al., unpublished |
| E2348/69ΔgrlR | grlR::km mutant | Bustamante et al., unpublished |
| E2348/69ΔgrlRA | grlRA::km mutant | Bustamante et al., unpublished |
| E2348/69Δler | ler::km mutant | Bustamante et al., unpublished |
| E2348/69Δhns | hns::km mutant | Bustamante et al., unpublished |
| E2348/69Δihf | Δ himA::km mutant | Bustamante et al., unpublished |
| E2348/69ΔperA | Δ perA::km mutant | Bustamante et al., unpublished |
| E2348/69ΔperC | ΔperC::km mutant | Bustamante et al., unpublished |
| E2348/69ΔqseA | ΔqseA::km mutant | This study |
| Plasmids | ||
| pBAD-Topo | Cloning vector | Invitrogen |
| pCsgA | E2348/69csgBA in pBAD-Topo | This study |
| pBcsA | E2348/69bcsA in pUC19 | This study |
| pFis | E2348/69fis in pUC19 | This study |
| pCP994 | csgD in pKK233-2 | (Prigent-Combaret et al., 1999) |
| pMLB1034 | Promoter-less lacZ | (Uhlich et al., 2002) |
| pW5lac | csgD::lacZ in pMLB1034 | (Uhlich et al., 2002) |
Single csgA, bcsA, csgD and fis mutants were complemented with pCsgA, pBcsA, pCP994 or pFis, respectively.
To determine whether the ability to produce curli was a general feature of AEEC strains, the production of curli was monitored in a bacterial collection after growth on T-medium at 37°C by electron microscopy, Congo-red binding, and ELISA. This collection included a subset (n=13) of strains belonging to different EPEC serotypes (O111:NM, O86:H34, O86:NM, O55:H6, O55:H−, O119:H6, O127:H6, and O128:H2) (Gismero-Ordonez et al., 2002) and a subset of 12 EHEC strains of serotypes O157:H7, O55:H7, O111:H8, O117:H4, and O113:H21 (Table 2). We found that the majority of these strains, 11 of 13 EPEC and 9 of 12 EHEC, produced curli under the experimental conditions tested here (Table 2). These results are in contrast to previous reports showing that most EHEC strains do not produce curli (Uhlich et al., 2001).
Table 2.
Curli expression in different EPEC and EHEC serotypes.
| Serotype (Strain) | EM | Congo-red binding | ELISA (OD405±SD) |
|---|---|---|---|
| EPEC | |||
| O111:NM (E1056) | + | + | + (0.35±0.03) |
| O111:NM (E1317) | + | + | + (0.69±0.05) |
| O111:NM (E271) | − | − | − (0.04±0.01) |
| O86:NM (E428174) | + | + | + (0.58±0.15) |
| O55:H- (E55) | + | + | + (0.96±0.05) |
| O128:H2 (E18) | + | + | + (0.41±0.02) |
| O55:H6 (E21) | + | + | + (0.53±0.03) |
| O119:H6 (E10) | + | + | + (0.50±0.09) |
| O119:H6 (E49) | + | + | + (0.45±0.12) |
| O127:H6 (E2348/69) | + | + | + (0.62±0.06) |
| O86:H34 (E28) | + | + | + (0.46±0.07) |
| O86:H34 (E24) | + | + | + (0.54±0.10) |
| O86:H34 (E26) | − | − | − (0.15±0.02) |
| EHEC | |||
| O157:H7 (EDL933) | + | + | + (1.038±0.04) |
| O157:H7 [34(4)AKAN] | + | + | + (0.40±0.03) |
| O157:H7 (275F1) | + | + | + (0.51±0.09) |
| O157:H7 (23380-85) | − | − | − (0.05±0.01) |
| O157:H7 (37-1) | − | − | − (0.12±0.04) |
| O55:H7 (660-79) | + | + | + (0.88±0.07) |
| O111:H8 (H30C5) | + | + | + (1.32±0.04) |
| O111:H8 (202F1) | + | + | + (1.05±0.04) |
| O117:H4 (CL6L) | − | − | − (0.09±0.01) |
| O113:H21 (EH53) | + | + | + (1.17±0.09) |
| O113:H21 (EH71) | + | + | + (0.58±0.13) |
| O113:H21 (EH41) | + | + | + (1.16±0.03) |
Expression of curli was determined by EM, Congo red binding and ELISA. The absorbance values obtained with pre-immune serum were subtracted from absorbance values obtained with the curli antiserum. The cut-off value for the absorbance in ELISA experiment was more than 0.15, which was considered as positive. +, Positive expression of curli; −, negative expression of curli. Congo-red binding was determined by uptake of the dye by curliated bacteria when growing on Congo red plate. The results obtained by EM, Congo red binding and ELISA were consistent.
Analysis of curli-negative isogenic strains
To investigate the role of curli in adherence of AEEC to cultured epithelial cells, we constructed csgA (curli subunit gene) and csgD (activator gene of csgBA) mutants by allelic exchange (Datsenko and Wanner, 2000). The resulting csgA and csgD mutants did not produce the curli fibers seen in the wild-type strains (Fig. 1A). We also confirmed that the mutants lacked curli production as determined by immunoblotting of formic acid-treated whole bacterial cell extracts reacted with anti-EPEC curli antiserum (Fig. 1B). E2348/69ΔcsgA and EDL933ΔcsgA were complemented with E2348/69 csgBA carried on plasmid pCsgA, to yield strains E2348/69ΔcsgA(pCsgA) and EDL933ΔcsgA(pCsgA) (Table 1). In both trans-complemented strains the ability to produce curli was restored (Fig. 1A and B). E2348/69ΔcsgD and EDL933ΔcsgD were complemented with csgD carried on plasmid pCP994, which has been reported to over-express CsgD in E. coli K-12 (Prigent-Combaret et al., 1999). The resulting strains produced identical structures as the parental strains (Fig. 1).
Role of curli and cellulose in adherence to epithelial cells and cow intestinal explants
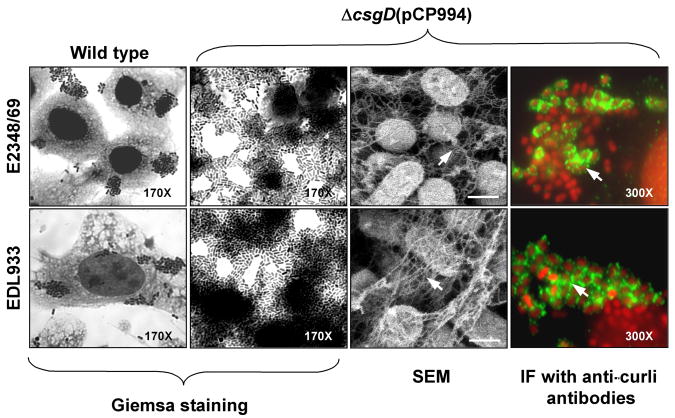
We tested the ability of AEEC csgA and csgD mutants to adhere to human cervix HeLa or colonic HT-29 cells for 3 and 6 h. The results obtained were similar with both cell lines at the different incubation times employed. The E2348/69-derived csgA and csgD mutants were still able to adhere forming microcolonies similar in size as those produced by the parental wild-type strains (data not shown). These observations were supported with comparative analysis in which the number of bacteria adhering to HT-29 cells was quantified by plating out serial dilutions (Table 3). EPEC and EHEC csgA and csgD mutants did not show a marked reduction in adhesion to epithelial cells, most likely because they are still able to produce other adhesins such as intimin, BFP and ECP in the case of EPEC, and intimin, ECP and HCP in the case of EHEC (Jerse et al., 1990; Giron et al., 1991; Donnenberg et al., 1992; Rendon et al., 2007; Xicohtencatl-Cortes et al., 2007). Complementation of the csgA mutant strains with plasmid pCsgA restored adherence, rendering strains slightly more adherent (Table 3). Interestingly, the adherence levels displayed by E2348/69ΔcsgD(pCP994) and EDL933ΔcsgD(pCP994) strains were significantly elevated (6.7-fold and 5.0-fold, respectively) compared to their parental strains (Table 3). Both of these strains adhered extensively to the cell surface and formed large aggregates of bacteria throughout the glass substratum and were highly curliated, as shown by high-resolution SEM and immunofluorescence using anti-curli antibody (Fig. 2).
Table 3.
Comparative analysis of curli and cellulose production and adherence to HT-29 cells by EPEC E2348/69 and EHEC EDL933strains.
| Strain | Curli* | Cellulose** | Adherence to HT-29 cells CFUs × 107/ml ±SD (%) |
Adherence to bovine gut tissue CFUs × 107/ml ±SD (%) |
||
|---|---|---|---|---|---|---|
| E2348/69 | EDL933 | E2348/69 | EDL933 | |||
| Wild type | + | + | 4.9 ±0.3 (100) | 1.43±0.09 (100) | 1.09 ±0.09 (100) | 1.11 ±0.07 (100) |
| ΔcsgA | − | ++ | 2.2 ±0.2 (45) | 0.75±0.04 (52) | 0.49 ±0.04 (45) | 0.23 ±0.04 (21) |
| ΔcsgA(pCsgA) | + | + | 5.6 ±0.7 (114) | 2.55±0.04 (178) | ND | ND |
| ΔcsgA(pCP994) | − | ++ | 6.9 ±0.8 (141) | 1.22±0.08 (85) | ND | ND |
| ΔbcsA | + | − | 3.9 ±1.2 (81) | 0.34±0.07 (24) | 0.63 ±0.06 (58) | 0.62 ±0.14 (56) |
| ΔbcsA(pBcsA) | + | + | 4.8 ±0.5 (98) | 1.47±0.12 (103) | ND | ND |
| ΔbcsA(pCP994) | ++ | − | 3.6 ±0.3 (73) | 0.77±0.13 (54) | ND | ND |
| ΔcsgAΔbcsA | − | − | 7.0 ±0.3 (15) | 0.18±0.08 (12) | 0.10 ±0.04 (9) | 0.21 ±0.03 (20) |
| ΔcsgAΔbcsA(pBcsA) | − | + | 3.6 ±1.6 (74) | 1.42±0.55 (99) | ND | ND |
| ΔcsgD | − | − | 4.4 ±0.4 (89) | 0.92±0.02 (65) | 0.82 ±0.07 (75) | 0.88 ±0.10 (79) |
| ΔcsgD(pCP994) | ++ | ++ | 32.7 ±4.3 (667) | 7.22±0.63 (505) | ND | ND |
The experiments were performed three times in triplicate and the results represent the mean of the averages obtained from those three experiments ± standard deviations. The data correspond to 6 h of incubation. +, Positive expression; −, negative expression; ++ high levels of expression.
Phenotypic production of curli as demonstrated by EM, Congo red staining and immunoblotting;
Production of cellulose as determined by calcofluor binding.
Fig. 2.
Adherence of AEEC to cultured HT-29 epithelial cells. Cell monolayers were infected for 3 h with EPEC E2348/69, EHEC EDL933, and AEECΔcsgD(pCP994). Note the dramatic increase in adherence by the complemented strains, which produced abundant fibrillar meshwork by SEM and which contained curli (green) as determined by immunofluorescence (IF) using anti-curli antibody. Light microscopy micrographs are at 170X or 300X as indicated. Scale bars, 1 μ.
CsgD is a transcriptional activator of curli and indirectly of cellulose biosynthesis (Romling et al., 2000; Brombacher et al., 2006). To determine if the hyper-adherence phenotype observed in the ΔcsgD strains complemented with pCP994 was also related to the overproduction of cellulose, we constructed cellulose-negative (ΔbcsA) in AEEC strains (Table 1). Isogenic E2348/69ΔbcsA and EDL933ΔbcsA strains lacked production of cellulose as determined qualitatively by the calcofluor-binding assay (Table 3). EDL933ΔbcsA and E2348/69ΔbcsA showed 76% and 19% reduction in adherence respectively, in comparison to their parental strains (Table 3), suggesting that under the conditions tested cellulose might play a more important role in EHEC than in EPEC adherence to human cells. By restoring bcsA in these mutants with plasmid pBcsA (Table 1), adherence was brought back to wild-type levels (Table 3). In contrast, even when expression of CsgD from pCP994 in the ΔcsgA and ΔbcsA strains had a variable compensatory effect in adhesion for the ΔcsgA strains and for the EDL9333ΔbcsA strain, it did not render hyper-adherent strains as seen in E2348/69ΔcsgD(pCP994) and EDL933ΔcsgD(pCP994) (Table 3). According to these data, it is reasonable to propose that the hyper-adherence phenotype was the result of over-production of both curli and cellulose, which act synergistically to promote cell adherence. To confirm this hypothesis, double mutants csgAbcsA were constructed in AEEC strains (Table 1). The resulting double mutants were 85–88% reduced in adherence to HT-29 cells, in comparison to parental strains and showed the poorest adherence levels of all strains tested (Table 3).
Next, we wanted to know if curli and/or cellulose had any role in bovine gut tissue adherence, particularly for EHEC since this organism lives in cattle. In agreement with the results obtained above with tissue culture cells, adherence of E2348/69 and EDL933 csgD, csgA and bcsA mutants to cow colon explants was diminished, to different extent, in comparison with the wild-type strains. Furthermore, the ΔcsgAΔbcsA strains were much less adherent than any of the single mutants (Table 3). Taken together, our data strongly suggest that curli and cellulose are co-produced and function in concert promoting human and bovine epithelial cell adherence.
Over-expression of curli favors biofilm formation
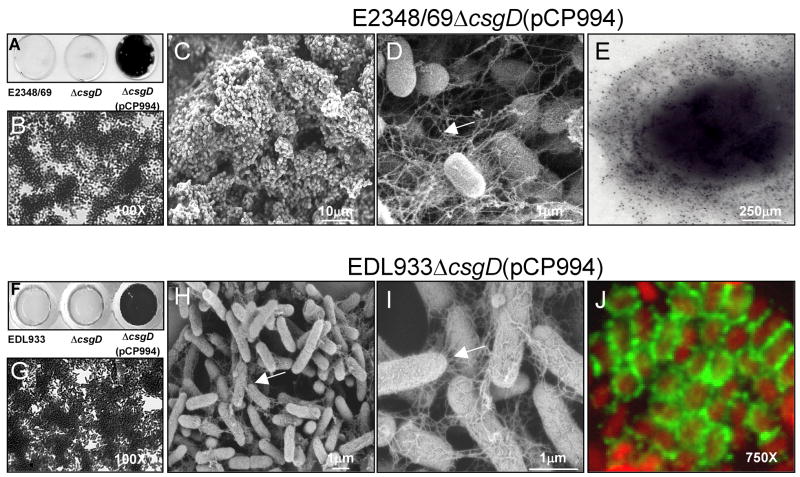
Curli has been associated with the formation of biofilms in S. enterica and E. coli strains including EHEC(Dorel et al., 1999; Prigent-Combaret et al., 2000). Here we tested wild type E2348/69 and EDL933 strains and their isogenic csgD mutants complemented or not with plasmid pCP994 for biofilm formation on glass coverslips after incubation for up to 24 h at 37°C. We found that both wild type and ΔcsgD strains adhered poorly to the glass surface and were unable to produce biofilms at 37°C in T-medium (Fig. 3A and 3F). In contrast, both of the AEEC ΔcsgD strains carrying (pCP994) produced a dense biofilm (Fig. 3A, 3B, 3F and 3G), which was most likely associated with over-production of curli and cellulose. The biofilm produced by these strains appeared by SEM as tri-dimensional aggregates of bacteria forming tall mushrooms with open water channels (Fig. 3C and 3H). Furthermore, when analyzed at higher magnifications, long fine filaments forming a meshwork resembling spider webs were observed (Fig. 3D and 3I) similar to those seen in the cell adherence assay (Fig. 2). The presence of curli filaments within the biofilm was confirmed by immunogold labelling (Fig. 3E) or immunofluorescence (Fig. 3J), using anti-curli antibodies; no reaction was seen with the pre-immune antiserum (data not shown).
Fig. 3.
Role of curli in biofilm formation. (A) and (F) Crystal violet uptake by biofilms produced by wild-type, ΔcsgD, and ΔcsgD(pCP994) strains on glass coverslips. (B) and (G) Light microscopy micrographs AEEC ΔcsgD(pCP994) biofilms. (C), (D), (H) and (I) SEM micrographs of ΔcsgD(pCP994) biofilms at low and high magnification indicated by scale bars. (E) Immunogold labelling of curli on E2348/69ΔcsgD(pCP994) obtained from the biofilm assay. (J) Detection of curli (green peritrichous fibers) on EDL933ΔcsgD(pCP994) (red bacteria) by immunofluorescence. Biofilms were obtained on a glass substratum after growth of the bacteria at 37°C in T-medium for 24 h. Light microscopy micrographs are at 170X or 750X as indicated.
Role of global and virulence regulators in curli expression
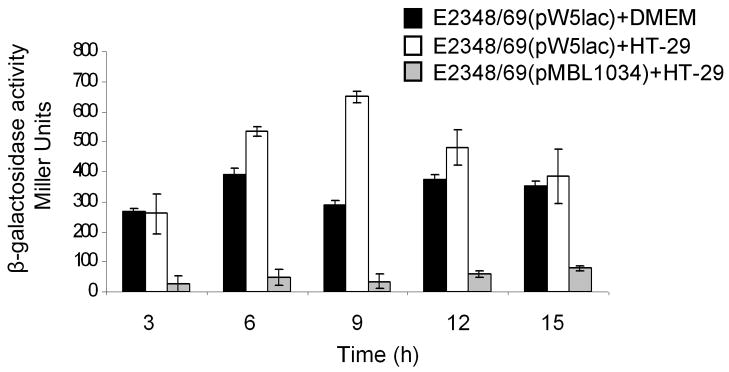
Considering the relevance that curli and cellulose may have during the interaction of AEEC with host cells, we first sought to determine if expression of the gene coding for CsgD was activated under the same growth conditions that trigger production of essential virulence factors such as T3SS products and BFP in EPEC, namely, growth in DMEM in the presence or absence of HT-29 epithelial cells at 37°C. A csgD::lacZ transcriptional fusion contained in plasmid pW5lac (Uhlich et al., 2001) was introduced in E2348/69 and β-galactosidase activity was measured after growth from 0 through 15 h. As shown in figure 4, csgD::lacZ was active in both conditions rendering slightly higher levels of expression in the presence of HT-29 cells (Fig. 4). No significant activity was observed in the negative control consisting of E2349/69 with the lacZ gene alone in pMLB1034. A range of pH between 6.8 and 8.4 was found to be optimal for csgD activation (data not shown). That CsgD is expressed under conditions that also favor the expression of other EPEC virulence factors is in agreement with the data showing that curli and cellulose are produced in contact of the bacteria with host cells at 37°C (Fig. 2 and data not shown) and that AEECΔcsgAΔbcsA strains are severely affected in adherence.
Fig. 4.
Transcriptional activation of csgD. A csgD::lacZ transcriptional fusion was tested for activity in E2348/69 grown in DMEM (black bars) and in the presence of HT-29 cells (white bars) from 0 through 15 h. The negative control (gray bars) is E2348/69 with pMLB1034 vector lacking csgD. The experiments were performed three times in triplicate on separate days and the results represent the mean of the averages obtained from those three experiments with standard deviations bars.
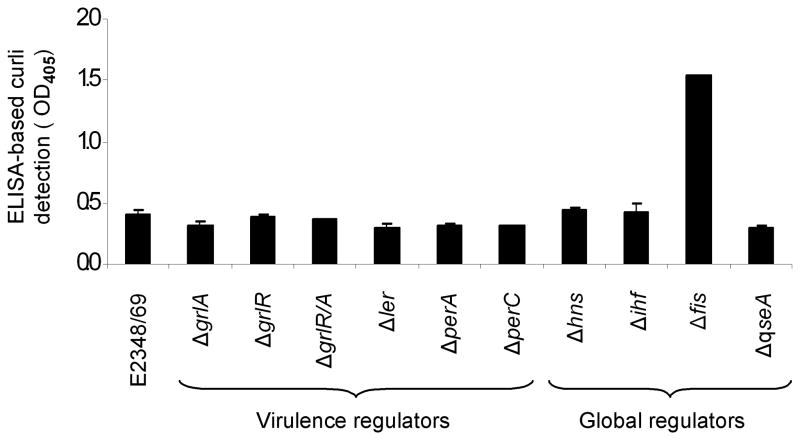
Studies in E. coli K-12 and S. enterica serovar Typhimurium have well established that global regulators such as H-NS, IHF, and RpoS among others, are part of a complex regulatory network that controls curli production (Olsen et al., 1993b; Barnhart and Chapman, 2006). In this context, we then sought to investigate what influence, if any, other global and AEEC-specific virulence regulators have on the expression of curli. The production of curli was measured in isogenic strains derived from EPEC E2348/69 mutated in global (H-NS, IHF, Fis, and QseA) and virulence (GrlA, GrlR, Ler, PerA, and PerC) regulatory genes by ELISA using anti-curli antibodies. In contrast to the important role that virulence regulators play in production of BFP or LEE-encoded genes (Mellies et al., 2007), we did not see any apparent effect of these regulators in the production of curli (Fig. 5). However, analysis of EPEC mutants in global regulatory genes identified Fis (factor for inversion stimulation) as a negative transcriptional regulator of curli, as demonstrated by the 3.9-fold increase in curli production in the EPECΔfis mutant with respect to the wild type (Fig. 5). Likewise, no effect was seen in the absence of IHF, which has been shown to play a positive role in csgD expression in S. Typhimurium (Gerstel et al., 2003).
Fig. 5.
Role of global and virulence regulators in production of curli. EPEC E2348/69 and sogenic mutants were monitored for production of curli fibers by ELISA using anti-curli antibodies. Pre-immune antiserum was used as negative control. The experiments were performed three times in triplicate on separate days with samples collected from T-medium after overnight growth and the results represent the mean of the averages obtained from those three experiments.
Fis is a negative regulator of curli
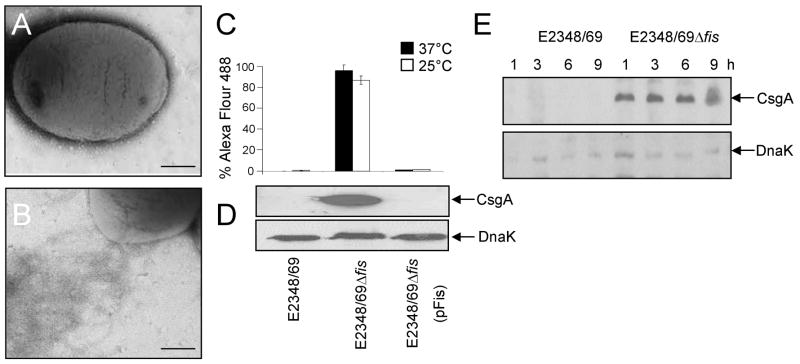
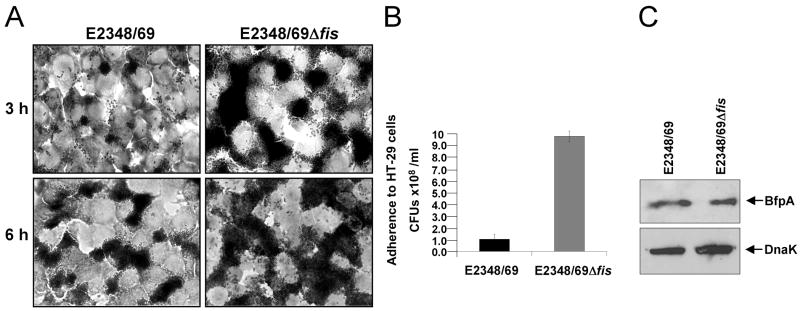
As described above, curli expression is enhanced in the absence of Fis in bacteria growing in T-medium (Fig. 5). In order to further characterize the role of Fis as a putative negative regulator of curli expression, wild type E2348/69 and its Δfis derivative were grown in LB broth, a non-permissive growth medium for curli production in E2348/69 (Fig. 6A–E). In contrast, expression of curli in E2348/69Δfis was derepressed in this medium, as demonstrated by the abundant assembly of curli fibers on the surface of this strain (Fig. 6B) and the presence of CsgA subunit in whole cell extracts (Fig. 6D), as well as for its temperature- (Fig. 6C) and growth phase-independent expression, which was observed from the first hour of growth (early phase) (Fig. 6E). Complementation of E2348/69Δfis with plasmid pFis reduced the production of curli to wild-type levels (Fig. 6C and D). Further, consistently with the role of Fis as a negative regulator, we observed a significant increase in cell adherence in the fis mutant (Fig. 7A and B), which was independent of BFP since BfpA production in this mutant was comparable to that of the wild-type strain (Fig. 7C).
Fig. 6.
Fis is a negative regulator of curli. (A) and (B) Electron microscopy micrographs showing lack of curli in E2348/69 (A) and presence of curli in E2348/69Δfis (B) after growth in LB broth. (C) Detection of curli by flow cytometry and (D) immunoblotting of E2348/69, E2348/69Δfis and E2348/69Δfis(pFis) using anti-curli antibody. (E) Kinetics of curli expression in E2348/69 and E2348/69Δfis, between 1–9 h of growth in LB as determined by immunoblotting. Detection of DnaK was used as a loading control.
Fig. 7.
EPEC E2348/69 adheres more efficiently to HT-29 cells in the absence of Fis. (A) Light microscopy micrographs comparing the adherence between E2348/69 and E2348/69Δfis to cultured HT-29 epithelial cells (magnification 100X). Cell monolayers were infected for 3 h and 6 h. (B) Quantification of bacterial adherence to HT-29 cells. (C) Immunoblotting of E2348/69 and E2348/69Δfis with anti-BFP and anti-DnaK antibodies.
Based on these results, we sought to investigate if this negative effect was the result of repression of the gene encoding the positive regulator CsgD. Thus, the csgD::lacZ transcriptional fusion contained in plasmid pW5lac (Uhlich et al., 2001) was introduced in E2348/69 and in its different regulatory mutants (Table 1) and β-galactosidase activity was measured after growth in T-medium at 37°C (data not shown). Transcription analysis of the csgD promoter indicated that only in the hns mutant was there an increase in csgD expression; while the mutations in fis had no effect (data not shown). These results strongly suggest that the negative effect of Fis on curli expression is at not at the level of csgD, but of csgA transcription.
Furthermore, in order to determine if there is an antagonist effect between Fis and CsgD in curli expression, a double mutant (ΔfisΔcsgD) was created. This mutant yielded no detectable values of curli production (data not shown), indicating that even in the absence of Fis, the CsgD regulator is essential for curli expression. This is to our knowledge the first report in which Fis is shown to play a significant role in curli regulation.
Discussion
It is generally thought that although all E. coli K-12 strains carry csgA, only a subset of them can transcribe the gene and that growth at 37°C represses production of curli in most E. coli strains (Olsen et al., 1989; Olsen et al., 1993b; Uhlich et al., 2001). In the present study, we found reproducibly that the majority of EPEC and EHEC strains of heterologous serotypes tested were able to assemble curli fibers while growing on salt-less T-medium at 37°C (Table 2), suggesting that the fimbriae could be produced in their human or bovine hosts. Genetic differences amongst E. coli strains and the use of different temperature and bacteriological growth media, e.g. CFA, Yesca or T-medium by several workers to monitor the presence of curli on E. coli isolates may account for the differences in curli production observed between strains (Olsen et al., 1989; Collinson et al., 1991; Uhlich et al., 2001; Bokranz et al., 2005).
Despite the potential role of curli in the adherence mechanisms of AEEC strains, the lack of curli production in E2348/69 and EDL933 csgA and csgD mutants did not significantly affect the ability of the strains to adhere to host cells under the conditions tested here (Table 3). That AEEC curli mutants are still able to adhere to epithelial cells may be explained considering the repertoire of redundant adhesins produced by these organisms (Giron et al., 1991; McKee et al., 1995; Phillips and Frankel, 2000; Rendon et al., 2007; Xicohtencatl-Cortes et al., 2007). Interestingly, AEEC csgD mutants complemented with csgD in trans were much more adherent to epithelial cells and efficient in biofilm formation than the wild-type strains, due most likely to hyper-production of curli and cellulose. These strains generated a dense fibrillar meshwork containing curli, in which the adhering bacteria were embedded, was demonstrated by several immunoassays (Fig. 2 and Fig. 3). It is also expected that cellulose be present within this fibrillar meshwork as cellulose is the second component of bacterial extracellular matrix (White et al., 2003) and its biosynthesis is also positively regulated by CsgD (Romling et al., 2000). In fact, the complementation of single csgA and bcsA mutants with csgD in trans did not render hyper-adherent strains, indicating that neither component alone is sufficient to promote adherence. This notion is also supported by the significant decrease in adherence to HT-29 cells and cow bovine tissue in vitro observed for AEEC double mutants (ΔcsgAΔbcsA) (Table 3). These results suggest a synergistic effect between curli and cellulose, which under wild-type conditions would favor adherence to epithelial cells in a host. In all, these observations indicate that curli and cellulose may function simultaneously as accessory adhesive factors of AEEC by promoting tight interactions between bacteria favoring host colonization or biofilm development. It is apparent though, that this may not be a generalized feature among the E. coli, since in E. coli K-12 MG1655 cellulose seems to affect curli-mediated adhesion (Gualdi et al., 2008).
Here we have shown that AEEC strains produce curli during adhesion to cultured epithelial cells. Since CsgD also enhances cellulose production, most likely through its effect on the expression of the adrA gene (Zogaj et al., 2001) reviewed in Barnhart and Chapman 2006), it was then expected that regulation of curli expression in AEEC strains was mainly exerted by controlling csgD expression. In agreement with our phenotypic analysis of curli expression in AEEC, by measuring csgD::lacZ transcriptional activity in EPEC in the absence (e.g. DMEM alone) or presence of epithelial cells and at different pHs, we found that csgD is significantly activated in the presence of cultured epithelial cells and in a pH range of 6.2 to 8.4 (Fig. 4 and data not shown). We found that beyond 9 h of infection of cultured cells, the levels of csgD appeared to be down-regulated. It was previously reported that CsgD does not regulate its own expression and it was suggested that when cultures are in high osmolarity medium, the Rcs and Cpx pathways negatively regulate csgD (Hung et al., 2001; Ferrieres and Clarke, 2003; Jubelin et al., 2005; Barnhart and Chapman, 2006). After 9 h of infection, the composition in the cell culture medium changes due to bacterial overgrowth, and this might allow the expression of Cpx or Rcs pathways and consequently down-regulation of csgD expression may occur. In agreement with our data, growth in the presence of epithelial cells trigger the expression of other adhesins in AEEC strains (Donnenberg et al., 1992; Puente et al., 1996; Kenny et al., 1997; Mellies et al., 2007), suggesting that curli and cellulose may be expressed under conditions that might mimic, in part, those prevailing in the human gut mucosa. Based on this notion, we explored the possibility that curli expression could be coordinated with other virulence attributes by controlling csgD expression through specific virulence gene regulatory proteins, such as Ler, GrlA, GrlR, Per A, and PerC, as well as via global regulators that have been involved in virulence gene control, such as H-NS, IHF, and Fis (Mellies et al., 2007). However, none of the regulators tested had an effect on csgD or curli expression, except for H-NS that has a negative role (data not shown), as has been previously shown for the expression of the csgBA promoter (Arnqvist et al., 1994) or of EPEC virulence factors (Mellies et al., 2007). We then explored a possible regulatory link between curli regulator CsgD and EPEC virulence factors. However, neither the mutation nor the over-expression of csgD affected the production of BFP, intimin, flagellin, Tir, or secreted proteins (EspA, EspB, and EspD) or the formation of AE lesions (data not shown).
Thus, we then evaluated if any of the the regulators investigated could have a potential role in the regulation of curli at a different level and so curli production and assembly in the EPEC isogenic mutant strains was analyzed by ELISA (Fig. 5). As for csgD, AEEC specific virulence gene regulators did not show an effect on curli expression; however, curli production was significantly enhanced in the fis mutant (Fig. 5 and 6), which was also hyper-adherent and developed a biofilm much more efficiently (Fig. 7). As the mutation of the fis gene does not affect the expression of csgD (data not shown), but strongly enhances the expression of CsgA, we sought to investigate the possibility that CsgD acts as a Fis antagonist to control curli expression. However, the csgD mutation in the Δfis background completely abrogated curli production and the hyper-adherence phenotype (data not shown), indicating that with or without Fis, CsgD is essential for curli expression in EPEC.
Fis is considered the most abundant nucleoid-associated protein in exponentially growing E. coli, reaching up to 50,000 molecules per cell that dramatically drop during stationary phase (Ball et al., 1992; Ali Azam et al., 1999). Fis binds DNA as a homodimer to a degenerate 15-base pair sequence (Shao et al., 2008) and has been shown to regulate, in a growth phase-dependent fashion, different processes including site-specific DNA inversions, as well as the transcription of more than 200 genes including rRNA and tRNA genes (Bradley et al., 2007; Shao et al., 2008).
Curli genes are maximally expressed during stationary phase and their expression is dependent upon RpoS (Arnqvist et al., 1994), which means that the transcription of csgA is under the control of an environmental program that responds positively to low temperature, low osmolarity, and stationary phase growth conditions (Olsen et al., 1993a; Pratt and Silhavy, 1998). The fact that curli are mainly produced during the stationary phase of growth when Fis levels are low is in line with our finding that Fis acts as a negative regulator of curli expression. Based on our data, it is tempting to speculate that during early exponential growth, Fis and H-NS repress the expression of the csgBA operon, which is consistent with the notion that many promoters regulated by Fis are also regulated by H-NS (Dorman, 2007). During the transition to stationary phase, a drop in Fis concentration may allow CsgD, together with other regulatory factors, to overcome H-NS repression on the csgBA promoter and in consequence, curli assembly. In the absence of Fis, curli is abundantly expressed during early growth phase (Fig. 6); thus, in the context of the intestinal environment, it is likely that once the bacteria have colonized the small intestine the expression of curli and cellulose would occur leading to stabilization of the bacterial microcolony. Overall, adding Fis to the complex regulatory network of curli expression was of particular interest because, to our knowledge, this is the first report linking Fis to the regulation of curli and in general of fimbrial genes.
The ability of EPEC and EHEC strains to express adhesive curli and cellulose under intestinal and/or extra-intestinal environments suggests an adaptive advantage for pathogenic bacteria to grow and survive in diverse biological niches. Certainly, the development of curli-mediated adherence and biofilm formation in the intestinal tract could contribute to colonization and disease. It remains to be elucidated if production of curli in vivo, in humans or in bovines, is a pre-requisite for infection and colonization.
Experimental procedures
Bacterial strains and growth conditions
AEEC strains and isogenic derivatives employed are listed in Table 1. The production of curli on these strains was studied after growth of the bacteria at 37°C on 1% tryptone medium (T-medium, pH 7.2) (Collinson et al., 1991), unless otherwise stated.. When required, antibiotics kanamycin (50 μg/ml), ampicillin (200 μg/ml) and chloramphenicol (30 μg/ml) were added to the media.
Ultrastructural studies
The presence of pili on bacterial cells was visualized by transmission electron microscopy (TEM) and immunogold labelling. For TEM, 10 μl of bacterial samples were negatively stained on Formvar carbon–coated grid with 1% sodium phosphotungstic acid (pH 7.2), and examined under a Philips transmission electron microscope. For immunogold-EM, primary rabbit anti-E. coli K-12 curli antibody (kindly donated by Corinne Dorel) and goat anti-rabbit IgG labeled with 10-nm gold particles (BBI International) diluted 1:10 in PBS containing 1% (wt/vol) bovine serum albumin (BSA) were used (Giron et al., 2002). For scanning electron microscopy (SEM), infected cells were fixed with 2% formalin in PBS and postfixed with 1% osmium tetraoxide. Samples were dehydrated by sequential ethanol concentrations, dried to critical point, and coated with a mixture of gold and palladium (de Oliveira-Garcia et al., 2002). Specimens were examined in a high-resolution Leo scanning electron microscope.
Curli purification
EPEC E2348/69 was grown on T-medium agar at 37°C to induce curli production. The fibers were detached by vigorous shaking and purified by differential centrifugation followed by ultracentrifugation in a cesium chloride-1% sarkosyl gradient (Tacket et al., 1987). After extensive dialysis the samples were treated with 90% formic acid (Collinson et al., 1991) and analyzed by sodium dodecyl-sulphate polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli, 1970). The presence of curli was confirmed by TEM. A protein band of 17-kDa was excised and subjected to N-terminal sequence analysis (Protein and Nucleic Acid Facility at Stanford University). The purified EPEC curli were used to raise antibodies in rabbit by injection of 4 weekly doses of the antigen (Lampire Biological Laboratories).
SDS-PAGE and immunoblotting analysis
Bacterial suspensions were adjusted to an absorbance of 0.7 at OD600. Equal numbers of bacteria were used to prepare whole cell extracts using formic acid and electrophoresed in 16% SDS-PAGE gels (Collinson et al., 1991). Anti-DnaK was used as a control for protein loading. The gels were stained with Coomasie blue or electroblotted onto PVDF membranes and the immobilized proteins were reacted with primary antibodies against curli, followed by incubation with goat anti-rabbit IgG conjugated to peroxidase (Sigma Aldrich). The substrate was a chemo-luminescent reagent (Amersham).
Flow Cytometry
Flow cytometry was used to detect the expression of curli. The strains were growth in LB media overnight at 37°C and 45-μl aliquots were incubated with 25 μl of anti-curli antibodies using a dilution of 1:500 for 1 h on ice. After three gentle washes with PBS, the bacteria were resuspended in 25 μl of a dilution 1:500 of goat anti-rabbit IgG (H+L) Alexa Fluor conjugate (Invitrogen). After 1 h incubation at 4°C, the bacteria were gently washed three times with PBS and resuspended in 800-μl final volume of PBS. For the analysis, the bacteria were labelled with 3 μl of a propidium iodide solution (Sigma Aldrich). Propidium iodide was visualized through a 42-nm band pass centered at 585. The FITC fluorescence emission was collected through a 30-nm band-pass filter centered at 530 in which 50,000 events were measured. The samples were analysed at the ARL Biotechnology/ACCC Cytometry Core Facility at the University of Arizona, by using a FACScan (Becton Dickinson, Franklin Lakes, NJ). The experiments were performed in triplicate at least 3 times on separate days and the data are expressed as the mean of the averages of the results obtained from the 3 experiments performed. Standard deviations are represented by error bars and represent the average of all the results obtained from the 3 experiments performed.
Construction of isogenic mutants
Non-polar deletion mutants in csgA, bcsA, fis, and csgD genes were generated by the lambda Red recombinase method previously described (Datsenko and Wanner, 2000). The primers employed for DNA amplification are listed in Table 4. Primers csgA/H1P1 and csgA/H2P2 and primers bcsA/H1P1 and bcsA/H2P2 were employed to mutate csgA and bcsA, respectively. Primers csgD/H1P1 and csgD/H2P2 and primers fis/H1P1 and fis/H2P2 were employed to mutate csgD and fis, respectively. Primers flanking the csgA, bcsA, fis, and csgD genes as well as primers inside the kanamycin and chloramphenicol resistance gene were used to confirm the required gene replacement by PCR. To complement the fis mutation, fis gene was amplified from E2348/69 using the primers G260 and G261 and cloned into the BamHI and HindIII sites in pUC19 vector. Primers G74 and G75 were used to amplify a region of 1,645 bp of the csgBA operon of E2348/69 including the promoter region, using Taq polymerase (Invitrogen). The amplicon obtained was cloned into pBAD TOPO TA (Invitrogen) yielding plasmid pCsgA, and since transcription of csgBA is under control of the native promoter, no induction with arabinose was necessary. To complement the bcsA mutation, primers G445 and G446 were used to amplify a region of 2,767 bp of the bcsA operon of EPEC and cloned into the EcoRI and HindIII sites in pUC19 vector. Mutants in csgA, bcsA, csgD and fis were complemented with pCsgA, pBcsA, pCP994 (Prigent-Combaret et al., 1999) or pFis, respectively (Table 1).
Table 4.
Primers employed.
| Primer name | Sequence (5′ to 3′) |
|---|---|
| csgA/H1P1 | ATCCGATGGGGGTTTTACATG* AAACTTTTAAAAGTAGCAGCAtgtaggctggagctgcttcg |
| csgA/H2P2 | TTTAATACAGAGGATGTATTA* GTACTGATGAGCGGTCGCGTTcatatgaatatcctccttaggagctgcttcg |
| csgD/H1P1 | AAAAAGTGGGGTTTCATCATG* TTTAATGAAGTCCATAGTATTtgtaggctggagctgcttcg |
| csgD/H2P2 | TTA* TCGCCTGAGGTTATCGTTTGCCCAGGAAACCGCTTGTGTcatatgaatatcctccttag |
| fis/H1P1 | TAAAGAGCTGACAGAACTATG* TTCGAACAACGCGTAAATgtgtaggctggagctgcttc |
| fis/H2P2 | ATTTAGCTAAAACCTGAATTA* GTTCATGCCGTATTTTTTcatatgaatatcctccttag |
| bcsA/H1P1 | CGGTGGTTGCTTATCCCGCCGGTCAACGAGCGGCTTATCgtgtaggctggagctgcttc |
| bcsA/H2P2 | CTTTTCATCGCGTTATCATCA* TTGTTGAGCCAAAGCCTGcatatgaatatcctccttag |
| G74 | CCCGAATTCGTGTAGTAATAAATCAGCC |
| G75 | CCCAAGCTTCCCTGTTTCTTTAATACAG |
| G260 | GACAGGATCCACTATGTTCGAA |
| G261 | GCTAAGCTTCTGAATTAGTTCATG |
| G445 | GAAGAATTCCTGACGCTGGCTAA |
| G446 | TGAAAGCTTGGAACGCACTCATC |
Indicates the start or stop codon of the gene.
Interaction with eukaryotic cells
The adherence and immunofluorescence assays were performed using HeLa and HT-29 cells (Giron et al., 2002). The cell monolayers were infected for 3 or 6 h, washed with phosphate buffer saline (PBS, pH 7.4), fixed with 2% formalin, and then stained with Giemsa. For immunofluorescence, rabbit anti-EPEC curli antibodies were added for 1 h and secondary anti-rabbit IgG Alexa-conjugated antibodies (Molecular Probes) were added for an additional hour. The samples were mounted on glass slides to be observed under UV light and phase contrast using an AX10 Imager. A1 Zeiss microscope. To quantify adherence, the infected cells were washed and lysed with PBS-0.5% Triton X-100, serial diluted, and plated onto LB agar with appropriate antibiotics to count the number of adhering bacteria, represented as colony-forming-units (CFUs). The experiments were performed in triplicate at least 3 times on separate days and the data are expressed as the mean of the averages of the results obtained from the 3 experiments performed. Standard deviations are represented by error bars and represent the average of all the results obtained from the 3 experiments performed. Replica samples were stained with Giemsa and examined by light microscopy.
Adherence to cow intestinal explants
Bovine intestinal tissue obtained from a cow (kind gift of the Meat Science Laboratory, University of Arizona) was rinsed thoroughly with PBS (pH 7.4), cut into 8 × 8 mm squares (0.8 g of weight), split open with a scalpel, and then flushed with HEPES-Hanks buffer (pH 7.4) to remove debris. The tissues in 1 ml of DMEM were incubated at 37°C for 6 h with 106 bacteria; previously grown overnight in T-medium broth to induce activation of curli. After washing with PBS, the attached bacteria were detached by vortexing for 10 minutes with glass beads and then serially diluted and plated onto MacConkey agar to obtain CFUs. The results shown are the mean of the averages of 3 experiments performed in triplicate on separate days. Standard deviations are represented by error bars and represent the average of all the results obtained from the 3 experiments performed.
ELISA
A standard ELISA was performed to determine and quantitate the presence of curli using 96-well plates coated with 100 μl of bacterial suspension of E2348/69 wild-type strain and isogenic mutants in different global and virulence regulator genes adjusted to OD600 0.7 in carbonate buffer (pH 9.6). After overnight incubation at 4°C, the plates were washed and blocked for 2 h with PBS-BSA. After washing with PBS-0.5% Tween-20, the plates were first incubated for 1 h with 1:5,000 dilutions of rabbit anti-EPEC curli antiserum. Goat anti-rabbit IgG conjugated to alkaline phosphatase was added for 1 h followed by phosphatase substrate. The development of color was read at an OD405 in an ELISA reader (Multiskan EX). Pre-immune antiserum was used as negative control. The experiments were performed in triplicate at least 3 times on separate days and the data are expressed as the mean of the averages of the results obtained from the 3 experiments performed. Standard deviations (SD) are represented by error bars and represent the average of all the results obtained from the 3 experiments performed.
Congo Red dye and calcofluor staining
Curliated bacteria stain red when grown on plates supplemented with the diazo dye Congo red (100 μg/ml) (Collinson et al., 1993). EPEC and EHEC strains and isogenic derivatives employed were grown in T-medium agar containing Congo red. Indication for cellulose production was obtained when fluorescent colonies were observed under a 366-nm UV light source after growth on T-medium containing 50 μm Calcofluor (fluorescent brightener 28) (Romling et al., 2003).
Biofilm assay
Adhesion to abiotic surfaces was analyzed at 37°C using a 40 μl aliquot of an overnight culture of bacteria grown in LB broth that was added to the wells containing 500 μl of T-medium and incubated for 24 h using 24-well plates (Nunc) with or without glass coverslips (Vidal et al., 1998). After washing three times with PBS, the bound bacteria were fixed with 2% formalin, stained with Crystal Violet and visualized under a light microscope or processed for SEM as described above.
β-Galactosidase assays
E2348/69 wild-type strain and mutants in different global and virulence regulator genes containing the csgD::lacZ fusion were grown with shaking for 21 h at 37°C in LB, diluted 1:100 and grown at 37°C to mid-log phase (OD600 of 0.5–1) in fresh T-medium, DMEM or DMEM with HT-29 cells. The bacterial pellets were diluted in Z buffer and assayed for β-galactosidase activity using ONPG (Sigma-Aldrich) as a substrate, as described previously, and expressed in Miller units (Miller, 1972, 1992). The β-galactosidase experiments were repeated at least three times in triplicate on separate days and the data are expressed as the mean of the averages of the results obtained from the 3 experiments performed. Standard deviations are represented by error bars and represent the average of all the results obtained from the 3 experiments performed.
Statistical Analysis
Data corresponding to adherence assays were compared using ANOVA and then the T-student test. The significance level was 5% in all tests. The SPSS statistical package was used.
Acknowledgments
We thank Corinne Dorel and Arne Olsen for providing antiserum against E. coli K-12 curli used at the beginning of this study, Gaylen Uhlich for providing plasmids pMLB1034 and pCP994. This work was supported by NIH grants AI063211 to JAG, NIH AI21657 to JBK, and DGAPA (IN224107) and CONACyT (60796) to JLP.
References
- Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist A, Olsen A, Normark S. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- Ball CA, Osuna R, Ferguson KC, Johnson RC. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Z, Normark S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. Embo J. 1997;16:5827–5836. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Z, Brauner A, Li Y, Normark S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis. 2000;181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- Bokranz W, Wang X, Tschape H, Romling U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Microbiol. 2005;54:1171–1182. doi: 10.1099/jmm.0.46064-0. [DOI] [PubMed] [Google Scholar]
- Bradley MD, Beach MB, de Koning AP, Pratt TS, Osuna R. Effects of Fis on Escherichia coli gene expression during different growth stages. Microbiology. 2007;153:2922–2940. doi: 10.1099/mic.0.2007/008565-0. [DOI] [PubMed] [Google Scholar]
- Brombacher E, Baratto A, Dorel C, Landini P. Gene expression regulation by the Curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J Bacteriol. 2006;188:2027–2037. doi: 10.1128/JB.188.6.2027-2037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss R., 3rd MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol Microbiol. 2001;41:349–363. doi: 10.1046/j.1365-2958.2001.02529.x. [DOI] [PubMed] [Google Scholar]
- Brunder W, Khan AS, Hacker J, Karch H. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H(−) Infect Immun. 2001;69:4447–4457. doi: 10.1128/IAI.69.7.4447-4457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson SK, Emody L, Trust TJ, Kay WW. Thin aggregative fimbriae from diarrheagenic Escherichia coli. J Bacteriol. 1992;174:4490–4495. doi: 10.1128/jb.174.13.4490-4495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson SK, Clouthier SC, Doran JL, Banser PA, Kay WW. Characterization of the agfBA fimbrial operon encoding thin aggregative fimbriae of Salmonella enteritidis. Adv Exp Med Biol. 1997;412:247–248. doi: 10.1007/978-1-4899-1828-4_37. [DOI] [PubMed] [Google Scholar]
- Collinson SK, Doig PC, Doran JL, Clouthier S, Trust TJ, Kay WW. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J Bacteriol. 1993;175:12–18. doi: 10.1128/jb.175.1.12-18.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira-Garcia D, Dall’Agnol M, Rosales M, Azzuz AC, Martinez MB, Giron JA. Characterization of flagella produced by clinical strains of Stenotrophomonas maltophilia. Emerg Infect Dis. 2002;8:918–923. doi: 10.3201/eid0809.010535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg MS, Giron JA, Nataro JP, Kaper JB. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- Dorel C, Vidal O, Prigent-Combaret C, Vallet I, Lejeune P. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol Lett. 1999;178:169–175. doi: 10.1111/j.1574-6968.1999.tb13774.x. [DOI] [PubMed] [Google Scholar]
- Dorman CJ. H-NS, the genome sentinel. Nat Rev Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- Ferrieres L, Clarke DJ. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol Microbiol. 2003;50:1665–1682. doi: 10.1046/j.1365-2958.2003.03815.x. [DOI] [PubMed] [Google Scholar]
- Frankel G, Phillips AD. Attaching effacing Escherichia coli and paradigms of Tir-triggered actin polymerization: getting off the pedestal. Cell Microbiol. 2008;10:549–556. doi: 10.1111/j.1462-5822.2007.01103.x. [DOI] [PubMed] [Google Scholar]
- Gerstel U, Romling U. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella typhimurium. Environ Microbiol. 2001;3:638–648. doi: 10.1046/j.1462-2920.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- Gerstel U, Park C, Romling U. Complex regulation of csgD promoter activity by global regulatory proteins. Mol Microbiol. 2003;49:639–654. doi: 10.1046/j.1365-2958.2003.03594.x. [DOI] [PubMed] [Google Scholar]
- Giron JA, Ho AS, Schoolnik GK. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- Giron JA, Torres AG, Freer E, Kaper JB. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol Microbiol. 2002;44:361–379. doi: 10.1046/j.1365-2958.2002.02899.x. [DOI] [PubMed] [Google Scholar]
- Gismero-Ordonez J, Dall’Agnol M, Trabulsi LR, Girón JA. Expression of the bundle-forming pilus by enteropathogenic Escherichia coli strains of heterologous serotypes. J Clin Microbiol. 2002;40:2291–2296. doi: 10.1128/JCM.40.6.2291-2296.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U, Barlev M, Seijffers R, Oelschlager TA, Hacker J, Ron EZ. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect Immun. 2001;69:2659–2665. doi: 10.1128/IAI.69.4.2659-2665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdi L, Tagliabue L, Bertagnoli S, Ierano T, De Castro C, Landini P. Cellulose modulates biofilm formation by counteracting curli-mediated colonization of solid surfaces in Escherichia coli. Microbiology. 2008;154:2017–2024. doi: 10.1099/mic.0.2008/018093-0. [DOI] [PubMed] [Google Scholar]
- Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- Hung DL, Raivio TL, Jones CH, Silhavy TJ, Hultgren SJ. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. Embo J. 2001;20:1508–1518. doi: 10.1093/emboj/20.7.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Chen J. Attachment and biofilm formation by various serotypes of Salmonella as influenced by cellulose production and thin aggregative fimbriae biosynthesis. J Food Prot. 2007;70:2473–2479. doi: 10.4315/0362-028x-70.11.2473. [DOI] [PubMed] [Google Scholar]
- Jerse AE, Yu J, Tall BD, Kaper JB. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson C, Nilsson T, Olsen A, Wick MJ. The influence of curli, a MHC-I-binding bacterial surface structure, on macrophage-T cell interactions. FEMS Immunol Med Microbiol. 2001;30:21–29. doi: 10.1111/j.1574-695X.2001.tb01545.x. [DOI] [PubMed] [Google Scholar]
- Jonas K, Tomenius H, Kader A, Normark S, Romling U, Belova LM, Melefors O. Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol. 2007;7:70. doi: 10.1186/1471-2180-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubelin G, Vianney A, Beloin C, Ghigo JM, Lazzaroni JC, Lejeune P, Dorel C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol. 2005;187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny B, Abe A, Stein M, Finlay BB. Enteropathogenic Escherichia coliprotein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect Immun. 1997;65:2606–2612. doi: 10.1128/iai.65.7.2606-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kim YH. Escherichia coli O157:H7 adherence to HEp-2 cells is implicated with curli expression and outer membrane integrity. J Vet Sci. 2004;5:119–124. [PubMed] [Google Scholar]
- La Ragione RM, Collighan RJ, Woodward MJ. Non-curliation of Escherichia coli O78:K80 isolates associated with IS1 insertion in csgB and reduced persistence in poultry infection. FEMS Microbiol Lett. 1999;175:247–253. doi: 10.1111/j.1574-6968.1999.tb13627.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, Sotman S. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;1:1119–1122. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- Low AS, Dziva F, Torres AG, Martinez JL, Rosser T, Naylor S, et al. Cloning, expression, and characterization of fimbrial operon F9 from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2006;74:2233–2244. doi: 10.1128/IAI.74.4.2233-2244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee ML, Melton-Celsa AR, Moxley RA, Francis DH, O’Brien AD. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect Immun. 1995;63:3739–3744. doi: 10.1128/iai.63.9.3739-3744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellies JL, Barron AM, Carmona AM. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect Immun. 2007;75:4199–4210. doi: 10.1128/IAI.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y.: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Moon HW, Whipp SC, Argenzio RA, Levine MM, Giannella RA. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun. 1983;41:1340–1351. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- Olsen A, Arnqvist A, Hammar M, Normark S. Environmental regulation of curli production in Escherichia coli. Infect Agents Dis. 1993a;2:272–274. [PubMed] [Google Scholar]
- Olsen A, Wick MJ, Morgelin M, Bjorck L. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect Immun. 1998;66:944–949. doi: 10.1128/iai.66.3.944-949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol. 1993b;7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- Paton JC, Paton AW. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998;11:450–479. doi: 10.1128/cmr.11.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AD, Frankel G. Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J Infect Dis. 2000;181:1496–1500. doi: 10.1086/315404. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Silhavy TJ. Crl stimulates RpoS activity during stationary phase. Mol Microbiol. 1998;29:1225–1236. doi: 10.1046/j.1365-2958.1998.01007.x. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Vidal O, Dorel C, Lejeune P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999;181:5993–6002. doi: 10.1128/jb.181.19.5993-6002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol. 2000;2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- Provence DL, Curtiss R., 3rd Role of crl in avian pathogenic Escherichia coli: a knockout mutation of crl does not affect hemagglutination activity, fibronectin binding, or Curli production. Infect Immun. 1992;60:4460–4467. doi: 10.1128/iai.60.11.4460-4467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente JL, Bieber D, Ramer SW, Murray W, Schoolnik GK. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol. 1996;20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- Rendon MA, Saldana Z, Erdem AL, Monteiro-Neto V, Vazquez A, Kaper JB, et al. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc Natl Acad Sci U S A. 2007;104:10637–10642. doi: 10.1073/pnas.0704104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Rohde M, Olsen A, Normark S, Reinkoster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- Romling U, Bokranz W, Rabsch W, Zogaj X, Nimtz M, Tschape H. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int J Med Microbiol. 2003;293:273–285. doi: 10.1078/1438-4221-00268. [DOI] [PubMed] [Google Scholar]
- Shao Y, Feldman-Cohen LS, Osuna R. Functional characterization of the Escherichia coli Fis-DNA binding sequence. J Mol Biol. 2008;376:771–785. doi: 10.1016/j.jmb.2007.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano C, Garcia B, Valle J, Berasain C, Ghigo JM, Gamazo C, Lasa I. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol Microbiol. 2002;43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- Srimanote P, Paton AW, Paton JC. Characterization of a novel type IV pilus locus encoded on the large plasmid of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect Immun. 2002;70:3094–3100. doi: 10.1128/IAI.70.6.3094-3100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley TE, Jones EW, Corley LD. Attachment and penetration of Escherichia coli into intestinal epithelium of the ileum in newborn pigs. Am J Pathol. 1969;56:371–392. [PMC free article] [PubMed] [Google Scholar]
- Tacket CO, Maneval DR, Levine MM. Purification, morphology, and genetics of a new fimbrial putative colonization factor of enterotoxigenic Escherichia coli O159:H4. Infect Immun. 1987;55:1063–1069. doi: 10.1128/iai.55.5.1063-1069.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Giron JA, Perna NT, Burland V, Blattner FR, Avelino-Flores F, Kaper JB. Identification and characterization of lpfABCC’DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2002;70:5416–5427. doi: 10.1128/IAI.70.10.5416-5427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlich GA, Keen JE, Elder RO. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl Environ Microbiol. 2001;67:2367–2370. doi: 10.1128/AEM.67.5.2367-2370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlich GA, Keen JE, Elder RO. Variations in the csgD promoter of Escherichia coli O157:H7 associated with increased virulence in mice and increased invasion of HEp-2 cells. Infect Immun. 2002;70:395–399. doi: 10.1128/IAI.70.1.395-399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AP, Gibson DL, Collinson SK, Banser PA, Kay WW. Extracellular polysaccharides associated with thin aggregative fimbriae of Salmonella enterica serovar enteritidis. J Bacteriol. 2003;185:5398–5407. doi: 10.1128/JB.185.18.5398-5407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicohtencatl-Cortes J, Monteiro-Neto V, Ledesma MA, Jordan DM, Francetic O, Kaper JB, et al. Intestinal adherence associated with type IV pili of enterohemorrhagic Escherichia coli O157:H7. J Clin Invest. 2007;117:3519–3529. doi: 10.1172/JCI30727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]