Abstract
In recent years, cell-based phenotypic assays have emerged as an effective and robust addition to the array of assay technologies available for drug discovery in the high throughput screening arena. Previously, biochemical target-based assays have been the technology of choice. With the emergence of stem cells as a basis for a new screening technology, it is important to keep in mind the lessons that have been learned from the adaptation of existing stable cell lines onto the high throughput screening drug discovery platform, with special consideration being given to assay miniaturization, liquid handling complications and instrument-introduced artifacts. We present an overview of the problems encountered with the implementation of multiple cell-based assays at the High Throughput Screening Center at Southern Research Institute as well as empirically defined effective solutions to these problems. These include examples of artifacts induced by temperature differences throughout the screening campaign, cell plating conditions including the effect of room temperature incubation on assay consistency, DMSO carry-over, and incubator induced artifacts.
Introduction
Mammalian cell-based assays, in which a physiological change within the cell is read as an endpoint, have become increasingly important to high throughput screening (HTS). While these assays can often provide higher value data than their target-based, or biochemical, counterparts, the inherent variability of cells presents problems with assay design, implementation and execution1,2. The readouts of these assays are dependent on a phenotypic response and this most often translates into longer incubation times than required for target-based assays. Any instrument-introduced artifacts in the shorter assays will only be exacerbated by these longer incubation times and in turn, previously unnoticed or minor artifacts or complications can become major problems. The ever increasing push towards higher density assay formats further complicates cell based assays because evaporation and gas exchange problems associated with microtiter plate cell culture are worsened by the lower assay volumes.
Throughout the implementation of multiple cell-based assays in the HTS Center at Southern Research Institute (SRI), we have encountered and resolved a wide range of assay problems. These have included reader induced artifacts, incubator related plate patterns, edge effects attributed to either evaporation or cell adhesion as well as liquid handling irregularities. We present an overview of a representative group of assays highlighting multiple endpoints and incubation times. Along with the problems encountered during implementation, a review of the diagnosis and solution to each problem is presented. This list of the potential problems and respective solutions has now been incorporated into the validation of every cell-based assay in the HTS Center at SRI. In addition, every new cell based assay provides a new opportunity to expand this list. All assays discussed below were performed in 384-well plate format.
EXPERIMENTAL ANALYSIS
Temperature and CO2 Sensitivity
A HeLa cell line stably transfected with a reporter construct containing the human hsp70.01 promoter fused upstream of a firefly luciferase reporter3 was the basis of this assay. Identification of novel compounds capable of upregulating the HSP70 promoter was the goal of the screening campaign. Promoter upregulation resulting in increased levels of intracellular luciferase were monitored using Bright-Glo by Promega (Madison, WI). The assay required a total incubation time of 48 h post plating. The primary problem observed during validation was increased background signal in the cell control in seemingly random plates throughout the batch run of 100 plates.
To determine the cause of the apparently random high background readings, a wide range of cell plating conditions were tested. These plating conditions included temperature sensitivity during plating, room temperature exposure post plating, and CO2 variation sensitivity during incubation. Temperature sensitivity during plating was tested first by dispensing the cells into the assay plates while the cell suspension was maintained on ice and at room temperature. Assay plates throughout the run still demonstrated elevated background values, though the cells maintained on ice during plating demonstrated this result later in the assay validation run. Hence, the temperature of the cell stock during plating was not the source of the problem nor did changing the temperature resolve the problem.
Another parameter that was evaluated was the time assay plates spent at room temperature before being placed in the 37°C incubator. During the screen, assay plates were being removed from the cell dispenser and placed in the incubator in batches of ten to twenty. The time spent at room temperature after being dispensed to the assay plates varied between one and 20 minutes from the first to the last plate in a group of 20 plates. Because assay plates in all screens are processed in numeric order, it was possible to correlate the increased background signal with the order in which cells were added to the plates. The first plates spent the longest time at room temperature before being transferred to the 37°C incubator and the increased background signal appeared to correspond to the plate order. To confirm this, cells were dispensed to plates under controlled timing conditions and were left out at room temperature for increasing lengths of time. This experiment reproduced the effect that was observed during validation. Assay plates that were maintained at room temperature for greater than 10 min before placing them into the incubator showed elevated background levels, resulting in drastically reduced signal to background ratios. This problem was corrected by stirring the cells on ice while dispensing them to the assay plates and removing the plates from the dispenser in batches of five to minimize the amount of time the cells were exposed to room temperature. The implementation of these protocol changes resulted in a very robust assay. The robust nature of the assay was, in part, determined by the Z' factor, a commonly used assay quality factor.4
Several other interesting observations were made while troubleshooting this assay. To determine if the incubator itself was the source of the apparently random high background plates, identical plates were prepared and placed in three different incubators. Data from these plates showed distinct differences between the three incubators. All incubators were set at 37°C with 5% CO2 and high humidity and the digital displays on the instruments appeared to confirm these conditions. After observing different results from the incubators, each was evaluated with a NIST traceable thermometer and the CO2 level was checked using a Fyrite CO2 analyzer (Bacharach, New Kensington, PA). The results of this equipment check revealed that the temperature varied from 36.5 to 38°C and the CO2 varied from 4.5 to 6.5%. From this set of experiments, it was determined that if the cells were subject to a CO2 concentration greater than 6% there was no measurable luciferase activity present in the cells under any condition, not even in the control compound wells. As a result of this observation, the temperature and CO2 levels in the HTS incubators were monitored much more closely. In summary, if the cells spent too much time at room temperature, luciferase activity increased even in the absence of any other stimuli. The opposite response was observed if the CO2 level was too high. This resulted in no luciferase response at all. What was learned is that slight variation in environmental parameters can have a significant impact on assay performance.
TEMPERATURE SENSITIVITY REVISITED
Drug resistance is often achieved in cells by expression of an efflux pump. The pump transports compounds out of the cell reducing the intracellular concentration, thereby reducing the efficacy of compounds applied to the cells. The tumor cell line used in the assay discussed here (H69/AR) expresses the MRP-1 efflux pump protein and is very efficient at expelling compounds that have entered the cell and are substrates for this pump (PubChem AID: 602). Through this mechanism, H69/AR cells are highly resistant to the toxic effects of doxorubicin. All cells were treated with a concentration of doxorubicin at which treated control cells demonstrate 10% reduction in viability when compared with untreated cell control (EC10) and compounds were evaluated for their ability to increase the sensitivity of the cell to this treatment. Cytotoxicity was measured 96 h post compound treatment and 72 h post doxorubicin treatment using CellTiter-Glo (Promega, Madison, WI). These cells were difficult to adapt to high density assay plates. Early assay validation in 384-well format showed significant edge effect resulting in prohibitively high coefficients of variability (%CV) for these cells (Fig.1). Plating conditions were examined as the probable cause of the high variability. It was found that stirring the cells on ice during plating helped reduce the edge effect, but the % CV values were still unacceptably high (>15%). Based on the work of Lundholt et al2, allowing the assay plates to incubate at room temperature for various times prior to moving them to the 37°C incubator was evaluated. The rationale behind this strategy is that some cell lines need time to settle to the bottom of the well before being subjected to 37°C temperatures and therefore becoming metabolically active. The theory is that edge effect is caused by a temperature differential across the plate which is created when the plate is placed in the incubator. The outermost wells reach 37°C more rapidly than the inner wells and so the outer wells show a different response then the inner wells in an assay. When H69/AR assay plates were incubated for 1 h at room temperature prior to being placed the 37°C incubator, CV values fell from greater than 15% to less than 8%, making this assay much more amenable to HTS. Testing this plating parameter is now a regular part of assay validation for all cell based assays screened at SRI. It is interesting to note that while it was found that the previously discussed assay could not tolerate even moderate exposure to room temperature conditions, extended room temperature treatments were absolutely necessary in this assay. The lesson learned here is that every cell line is different and that the optimal conditions required for screening need to be determined empirically for each cell line.
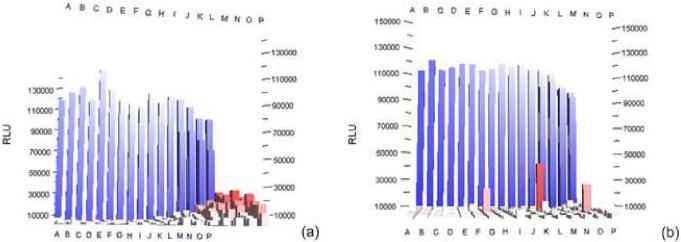
Figure 1.
The formation of temperature gradients within cell-based assay plates can cause drastic edge effects as described by Lundholt et al2. Assay plates that were placed directly into the incubator after plating demonstrate this edge effect (%CV = 16.4%) (a). A one hour incubation at room temperature before being placed in the 37°C incubator dramatically reduces this effect (%CV = 8.6%) (b) The numbers and lettering are column and row indicators. The Z-axis is Relative Luminescence Units (RLU)
INCUBATOR INDUCED ARTIFACTS
An anti-viral screen was conducted with MDCK cells being incubated for a total of 96 h post plating and 72 h post viral infection5. During this assay, cells were plated and incubated 24 h before the addition of test compounds and the immediate addition of virus. The assay plates were then incubated for an additional 72 h before adding the endpoint reagent. Edge effect from evaporation, usually evident in differential cell viability when comparing the outermost wells of a microtiter plate to the inner wells, is always suspect in an assay with an incubation time of this length, but the edge effect observed in this assay was not typical of that caused by evaporation (Fig. 2). This pattern occurred throughout the screening run to varying degrees resulting in 5-7% of the assay plates being rejected and repeated. Identifying the source of this plate pattern was further complicated by the nature of this screen; cells were plated and compounds were added in the primary high-throughput screening lab, but because of the BSL-2 containment requirements of the virus, virus addition and post virus incubation was performed in a separate BSL-2 HTS lab. Because of this transfer from one lab to another, the order of treatment of each assay plate throughout the assay process was not kept consistent from lab to lab resulting in an apparent random distribution of the affected plates.
Figure 2.
Assay plates placed in the right-most front position in the incubator demonstrated unusual plate patterns (a). The orientation of the plates was such that column 1 faced the front of the incubator. Assay plates that were not placed in these positions in the incubator did not demonstrate this incubator induced artifact (b). The letters are column indicators.
The most likely candidates for the cause of this pattern were the incubation steps. Therefore, plate positions in each incubator were tracked and recorded. Additionally, each step of the assay was performed in numerical order with relation to the barcode on the plate. Once these steps were put in place, the apparent random distribution of the effected plates disappeared and it was observed that the unacceptable edge effects appeared in an 18 - 20 plate-pattern. When this numeric distribution was compared to the plate position in the incubators, it became clear that the right-most front of the shelves in one of the incubators was the problem area. When the incubator shelves were filled with assay plates, those plates that were positioned nearest the right incubator wall in the front position of each shelf demonstrated the observed plate pattern. The incubator was decontaminated and serviced. This included a thorough check of all of the door gaskets, the CO2 solenoid, the circulation fans, the temperature control, and the humidity control. Nothing was found to be out of order but the pattern still appeared on plates placed in those positions. The ultimate solution was simply to avoid that position on the shelves when placing assay plates in the incubator. The lesson learned in this assay was the importance of tracking order and position of every plate throughout the assay process in order to facilitate trouble shooting when needed. These parameters are now tracked routinely for all assays and plates are still not placed in the right-most front position of any shelf in this incubator.
LIQUID HANDLING CONSIDERATIONS
HEK293 cells were stably transfected with a construct consisting of an SMN2 promoter, cDNA and exon 7 splicing reporter cassette on a selectable replicating vector using luciferase as the reporter6. This assay was validated without many of the problems discussed previously in this publication. The screening campaign was begun with all assay parameters optimized to give low %CV's and excellent Z' scores. Analysis of the first 30,000 compound screening batch revealed a dramatic four-plate repeating pattern in the values for the positive control (Fig. 3). The positive control signal for the first plate of each four plate group was ~30% higher than the subsequent three plates in the group.
Figure 3.
Compounds were added to assay plates in groups of four. The control well values show a distinct four plate pattern in which the first plate of each group of four demonstrated a significantly higher positive control signal. This was attributed to DMSO carry over into the positive control wells resulting from compound addition tips being washed in DMSO between assay plate additions (a). After removing the tips from the compound addition tips that enter the control wells, the control wells on each assay plate gave a much more consistent response (b).
The possible problems with carry-over and contamination related to washing pipette tips between compound transfers has been well documented7,8 and must be weighed against the additional cost incurred by using a new box of pipette tips for each unique compound transfer. When the assay process was reviewed step by step starting at the very beginning, it appeared that the cause of this pattern was the compound addition step. Compounds were added to the assay plates in groups of four with a Beckman Coulter BioMek FX using sterile Beckman 384-well pipette tips. The first plate received compounds using a new sterile box of pipette tips and the remaining plates in the group were drugged with the same pipette tips after washing in 70% EtOH followed by a wash in 100% DMSO to remove any residual EtOH. The cause of the erroneous data was indeed determined to be from the compound addition step and was attributed to the carryover of dimethyl sulfoxide (DMSO) into the positive control wells from the DMSO rinse following the washing of the pipette tips. This was confirmed by performing an experiment in which the positive control was added to assay plates both with and without the washing of pipette tips in ethanol and DMSO. The plates into which the positive control was added after the wash demonstrated a 30% lower signal than those into which the control was added with dry tips. The solution for the remainder of the screen was to remove the tips from the tip racks used for compound addition that entered the control wells. This eliminated the additional carry-over of DMSO that was pushing the assay beyond its threshold for DMSO tolerance. The removal of control tips from compound addition tip racks is now routinely conducted for all cell based screens.
PLATE COATING CONSIDERATIONS AND MORE CO2 ISSUES
An assay that presented more obstacles than any other assay implemented at the SRI HTS center was a cell growth/viability assay utilizing human umbilical vein endothelial cells (HUVECs) (PubChem AID: 648). HUVECs are primary cells that require specialty media (Endothelial Cell Basal Medium supplemented with 2% fetal bovine serum, 12 μg/mL bovine brain extract, 1 μg/mL hydrocortisone, and 1 μg/mL GA-1000) which are particularly sensitive to passage number and environmental conditions. In addition to the difficulties associated with the cell type, the assay ran for an extended period of time, a total of 96 h. All of the lessons that were learned through the implementation of the previous cell based assays conducted in the HTS Center were applied to this assay. Numerous combinations of plating conditions were tried; including plating with and without stirring, maintaining cells on ice or at room temperature during plating and incubating plates for various times at room temperature prior to moving them to the 37°C incubator. Close monitoring of timing, plate order and plate position in the incubator was maintained throughout all steps of the assay. After this series of experiments, the best CVs observed were still greater than 15% in 384-well format. Because of cost and instrumentation considerations, adapting the assay to 384-well format was imperative. Therefore, troubleshooting and validation continued.
It has been observed that the pre-treatment of assay plates with collagen, fibronectin and other proteins can increase cell adherence and mitigate cell detachment in cell-based screens9. Microtiter plates that have been pre-treated with collagen are available from multiple commercial sources. However, the increased cost of coated plates can be significant. In order to minimize screening cost, a method for pre treating plates with collagen was developed in-house. Corning 384-well tissue culture treated plates were pre-treated with a 0.5 mg/mL collagen solution dissolved in 1 mM acetic acid. The plates were incubated for 1 h at room temperature in a biological safety cabinet and then the collagen solution was removed using the BioMek FX. These treated plates were then allowed to dry for 1 h and stored at 4°C for up to two weeks before use. Assay validation continued using the collagen coated plates. Cells were dispensed to the pre-treated plates and handled according to the procedures producing the best results to date. In summary, the plates were incubated at room temperature for 1 h and then placed in the incubator. The in-house prepared collagen treated plates produced better results than standard tissue culture treated plates and better results than collagen plates purchased commercially. However, the CVs were still around 12%, which was not sufficient to run the screen. To ensure that equipment problems were not contributing to the high CVs, every piece of equipment used in the assay was checked for proper operation and set points. All equipment in the HTS lab was found to be working properly and temperature and CO2 settings on the incubators were verified with calibrated thermometers and a Fyrite. Nothing was outside of an acceptable range. This process was then extended to the lab responsible for maintaining the cells and providing cells to the HTS for assay validation. It was found that the incubator used to maintain the cells was actually at 8% CO2 when checked with a Fyrite, even though the incubator display indicated 5% CO2. When the incubator was calibrated and the actual CO2 level reduced to 5%, the CV dropped to 8%. The assay was now ready for the HTS screening campaign.
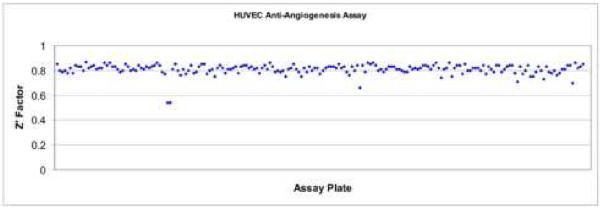
For the screen, compounds were delivered with the control well tips removed from the compound addition tip racks based on our previous experience. This combination of correct instrument settings, collagen treatment of the plates, and optimized conditions for cell plating allowed for a screening of 100,000 compounds to be conducted utilizing primary cells. Z' scores of ~ 0.8 (Fig. 4) were maintained during the entire screen and there were no additional failures observed.
Figure 4.
After the application of the lessons learned including pre-coating the assay plates with collagen, incubating the assay plates at room temperature for an hour and strict monitoring of environmental conditions during the screen, this resulted in a very robust, highly reproducible assay with Z' factors averaging 0.8.
Summary/Conclusion
Based on these and many other experiences in the adaptation of cell-based assays to automation platforms at SRI, the HTS Center tries to approach cell-based assays without prejudice. Every cell line or construct is assumed to be unique and every cell line is optimized based on its behavior in an assay plate. “Dogma” is often the enemy of conducting a successful HTS campaign because assumptions are made based on general knowledge which may or may not apply to the specific cell line being used. The same approach is used in troubleshooting and no assumptions are made concerning any variable or condition until it is evaluated experimentally. To avoid wasting time and money on incorrect assumptions, troubleshooting during assay development and validation always begins at the first step and proceeds through all steps of the assay to determine the cause of any problems. This procedure has saved time and money on numerous occasions when the initial assessment of the assay would have eliminated certain steps as unlikely contributors to the problem.
We have found that mammalian cells are a useful, but often problematic tool for the HTS scientist. Cell plating conditions, incubation times, compound addition parameters and assay endpoint considerations can all affect the quality of data produced when conducting a screening campaign utilizing mammalian cells. The testing of all of the variables described is now a routine part of the assay validation process at SRI. The utilization of stem cells as a screening platform represents an exciting new frontier in screening technology. Since these cells are significantly different from the established cell lines commonly used in research, much work is still required to define the conditions for optimal cell behavior. To be useful in HTS campaigns any cell line must produce a consistent response to experimental conditions. As more work is done with stem cells, researchers will discover many variables that affect these cells. Some may be in common with established cell lines and include those listed here and other will be unique to stem cells. Advances in stem cell culture will be made by identifying and controlling critical variables in screening assays and these advances will expand the use and usefulness of stem cells.
ACKNOWLEDGEMENTS
The projects described were supported in part by grant number U54HG003912 (Gary A. Piazza, P.I.) from the National Human Genome Research Institute and grant number NO1-NS-22348 (Joseph A. Maddry, P.I.) from the National Institute of Neurological Disorders and Stroke. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or any of its institutes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Beske OE, Goldbard S. High-throughput cell analysis using multiplexed array technologies. Drug Discovery Today. 2002;7:S131–S135. doi: 10.1016/s1359-6446(02)02388-7. [DOI] [PubMed] [Google Scholar]
- 2.Lundholt BK, Scudder KM, Pagliaro L. A simple technique for reducing edge effect in cell-based assays. J. Biomol. Screen. 2003;10:566–570. doi: 10.1177/1087057103256465. [DOI] [PubMed] [Google Scholar]
- 3.Westerheide SD, et al. Celastrols as Inducers of the Heat Shock Response and Cytoprotection. J. Biol. Chem. 2004;12:56053–56060. doi: 10.1074/jbc.M409267200. 279(53) [DOI] [PubMed] [Google Scholar]
- 4.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 5.Noah JW, Severson W, Noah DL, Rasmussen L, White EL, Jonsson CB. A cell-based luminescence assay is effective for high-throughput screening of potential influenza antivirals. Antiviral Res. 2007;73(1):50–9. doi: 10.1016/j.antiviral.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Singh NN, Androphy EJ, Singh RN. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2004;315(2):381–8. doi: 10.1016/j.bbrc.2004.01.067. [DOI] [PubMed] [Google Scholar]
- 7.Astle TW, Akowitz A. Accuracy and Tip Carryover Contamination in 96-Well Pipetting. J. Biomol. Screen. 1996;6(1):211–216. [Google Scholar]
- 8.Taylor PB, et al. A Standard Operating Procedure for Assessing Liquid Handler Performance in High-Throughput Screening. J. Biomol. Screen. 2002;12(7):555–569. doi: 10.1177/1087057102238630. [DOI] [PubMed] [Google Scholar]
- 9.Galietta LVJ, Jayaraman S, Verkman AS. Cell-based assay for high-throughput quantitative screening of CFTR chloride transport agonists. Am. J. Physiol. Cell Physiol. 2001;281:C1734–C1742. doi: 10.1152/ajpcell.2001.281.5.C1734. [DOI] [PubMed] [Google Scholar]