Abstract
2-Amino-9H-pyrido[2,3-b]indole (AαC) is a carcinogenic heterocyclic aromatic amine (HAA) that is produced in high quantities in tobacco smoke, and also forms in charred meats. The bioactivation of AαC occurs by cytochrome P450-mediated (P450 1A2) N-oxidation of the exocyclic amine group, to form 2-hydroxyamino-9H-pyrido[2,3-b]indole (HONH-AαC). The HONH-AαC metabolite can then undergo further activation by phase II enzymes to form the penultimate ester species, which bind to DNA. Some epidemiological studies suggest a role for NAT2 genetic polymorphisms in human susceptibilities to various cancers from tobacco smoke and from consumption of well-done meats, where the exposures to AαC can be substantial. In this investigation, we have measured the genotoxicity of AαC in nucleotide excision repair-deficient Chinese hamster ovary (CHO) cells stably transfected with human P450 1A2 and either the NAT2*4 (rapid, wild-type acetylator) or the NAT2*5B (the most common slow acetylator) allele, to determine the role of NAT2 phenotype in the biological effects of AαC. Mutations at the hypoxanthine phosphoribosyl transferase (hprt) locus were induced in a dose-dependent manner by AαC, and were found to be highest in cells transfected with P450 1A2 and NAT2*4, followed by cells transfected with P450 1A2 and NAT2*5B. The level of formation of the deoxyguanosine (dG) adduct N-(deoxyguanosin-8-yl)-2-amino-9H-pyrido[2,3-b]indole (dG-C8-AαC) paralleled the mutagenic potency, in these cell lines. However, AαC did not form DNA adducts or induce mutations in untransfected CHO cells, or in cells only expressing P450 1A2. These findings clearly demonstrate that NAT2 genetic polymorphism plays a major role in the genotoxic potency of AαC.
Introduction
More than 20 heterocyclic aromatic amines1 (HAA) are formed in meats cooked well-done (1). Many of these compounds are carcinogenic and induce tumors at multiple sites, including lung, forestomach, liver, blood vessels, colorectum, prostate, and mammary gland, during long-term feeding studies of rodents (1). 2-Amino-9H-pyrido[2,3-b]indole (AαC) was one of the first HAAs to be identified in cooked meats. AαC is a pyrolysis product of protein; it was originally identified in a pyrolysate of soybean globulin (2). Thereafter AαC was detected in fried, broiled, and barbecued meats; pan scrapings of fried beef cooked very well-done (3-7); and diesel exhaust (8). A major source of exposure to AαC occurs through main-stream tobacco smoke, where levels up to 258 ng AαC /cigarette have been detected (9). These levels are ∼25 to 100-fold higher than the levels of 4-aminobiphenyl, 2-naphthylamine (10), or benzo(a)pyrene (11), and are comparable to the levels of tobacco-specific nitrosamines, such as 4-(methyl-nitrosamino)-1-(3-pyridyl)-1-butanone. The latter compounds are known human carcinogens (12). Thus, the exposure to AαC through tobacco smoke is significant.
AαC exerts its genotoxic effects following metabolic activation by P450-catalyzed N-oxidation of the exocylic amine group, to form 2-hydroxyamino-9H-pyrido[2,3-b]indole (HONH-AαC) (13). In humans, P450 1A2 is the most active P450 enzyme, followed by P450 1A1, and P450s 2C9/2C10 (14,15). HONH-AαC undergoes further metabolism by reaction with N-acetyltransferases (NATs) or sulfotransferases (SULTs) (16), to produce unstable esters which undergo heterolytic cleavage to form the nitrenium ion that reacts with DNA (17,18). The sole DNA adduct of AαC identified thus far is N-(deoxyguanosin-8-yl)-2-amino-9H-pyrido[2,3-b]indole (dG-C8-AαC) (19,20).
Genetic polymorphisms in the enzymes catalyzing the activation and/or detoxification of HAAs may account for inter-individual differences in susceptibility to these carcinogens (21). The constitutive P450 1A2 mRNA expression levels in human liver can vary by as much as 15-fold (22), and inter-individual expression of P450 1A2 protein can vary by 60-fold (23). Environmental and dietary factors, varying extents of CpG methylation (24), and genetic polymorphisms in the upstream 5′-regulatory region of the P450 1A2 gene (25,26), which affect the level of P450 1A2 mRNA expression, can all lead to variations in protein levels. Humans also exhibit common genetic polymorphism in arylamine N-acetyltransferase 2 (NAT2), giving rise to rapid and slow acetylator phenotypes. The NAT2*4 wild-type allele is associated with the rapid-acetylator phenotype, whereas the NAT2*5B allele is the most common haplotype associated with the slow-acetylator phenotype (27,28). The transfection of NAT2 into a bacterial or a mammalian cell line leads to profound increases in the genotoxic potency of many HAAs (29), demonstrating the critical role of this enzyme in HAA-mediated carcinogenesis. The reactive N-acetoxy HAA esters are believed to be the penultimate metabolites, which damage DNA (18).
The role of NAT2 genetic polymorphism in cancer risk has been studied extensively, and the elevated risk of urinary bladder cancer in cigarette smokers exposed to aromatic amines is well documented (30). This increased cancer risk has been attributed to the diminished capacity of slow N-acetylator individuals to detoxify aromatic amines, which are human bladder carcinogens (28). However, epidemiological data on the role of NAT2 genetic polymorphism in susceptibility to various cancers suggest that the role of NAT2 genetic polymorphism varies both with carcinogen and with organ site (27,31). In contrast to aromatic amines, many HAAs are not detoxified by NAT2; however, their N-hydroxylated metabolites are substrates for O-acetylation by NAT2, which catalyze formation of the reactive N-acetoxy intermediates (18). Since both phase I and II enzymes are required to bioactivate HAAs, risk may be markedly elevated in individuals who are both rapid N-oxidizers and rapid O-acetylators. Cigarette smoking is a known risk factor for colorectal cancer (32). Two epidemiological studies reported a marked increased risk of colorectal cancer among individuals who were tobacco smokers and frequently ate meats cooked well-done. Both tobacco smoke and well-done meats are sources of exposures to HAAs. The elevated cancer risk was only observed in individuals with high activities of both the P450 1A2 and NAT2 enzymes; these subjects had up to an 8.8-fold increased risk in colorectal cancer (33,34). It is noteworthy that AαC, a carcinogen formed at substantial amounts in tobacco smoke and well-done cooked meats, is a potent genotoxicant in colon of rodents (35,36).
Investigations on substrate specificities and catalytic activities of tissue fractions or recombinant NAT enzymes involved in metabolism of HAAs have largely been carried out at elevated HAA concentrations (16,37,38). Under these high substrate concentrations, enzymes may show catalytic activities, that do not occur in vivo, where the concentrations of HAAs remain well below the enzyme Km values (39). There is a critical need to characterize the genotype and associated phenotypic catalytic activities of NAT enzymes displaying common genetic polymorphisms in the bioactivation of HAAs in cell systems containing physiological concentrations of co-factors and environmental exposure levels of carcinogen. Such investigations should help us to identify specific HAA that undergo bioactivation by NAT2 and determine their roles in elevated risk of HAA-related cancers in rapid NAT2 acetylators. In this study, we have investigated the role of P450 1A2, and the contributions of the NAT2*4 rapid acetylator allele, and NAT2*5B, the most common slow acetylator allele (27), to the bioactivation of AαC in CHO cells, under dose concentrations almost as low as the in vivo exposure levels of AαC in tobacco smokers.
Experimental Procedures
Materials and Methods
AαC was purchased from Toronto Research Chemicals (Toronto, ON, Canada). [13C10]dG was purchased from Cambridge Isotopes (Andover, MA). Phosphodiesterase I (from Crotalus adamanteus venom) was purchased from GE Healthcare (Piscataway, NJ), and alkaline phosphatase (from E. coli), phosphodiesterase I (from Crotalus adamanteus venom), and nuclease P1 (from Penicillium citrinum) were all from Sigma (St. Louis, MO). All solvents used were high-purity B & J Brand® from Honeywell Burdick and Jackson (Muskegon, MI). ACS reagent grade HCO2H (88%) was purchased from J.T. Baker (Phillipsburg, NJ). Isolute C18(EC) (25 mg resin in 1 mL polypropylene cartridges) SPE cartridges containing additionally washed polyethylene frits were purchased from Biotage (Charlottesville, VA). 2-Nitro-9H-pyrido[2,3-b]indole (NO2-AαC) was a kind gift from Dr. Dwight Miller (National Center for Toxicological Research, Jefferson, AR). All other chemical reagents were ACS grade, and purchased from Sigma Aldrich.
Cell Culture
The UV5-CHO cell line, a nucleotide excision repair-deficient derivative of the AA8 line (40), was obtained from the ATCC (Catalog number: CRL-1865). Since UV5-CHO lacks nucleotide excision repair due to a mutation in the XPD (ERCC) gene (41), it is hypersensitive to bulky adduct mutagens and belongs to the excision repair cross complementation group 2. All cells were grown in alpha-modified minimal essential medium (Cambrex) without L-glutamine, ribosides, or deoxyribosides, supplemented with 10% fetal bovine serum (Hyclone), 100 units/mL penicillin, 100 units/mL streptomycin (Cambrex), and 2 mM L-glutamine (Cambrex) at 37°C in 5% CO2. Media were supplemented with selection agents as previously described (42) appropriate for maintenance of stable transfectants.
Construction of UV5-CHO Cells Expressing P450 1A2 and NAT2*4 or NAT2*5B
The construction of UV5-CHO cells expressing human P450 1A2 and NAT2*4 or NAT2*5B was recently reported and characterized (42). Briefly, the pFRT/lacZeo plasmid (Invitrogen) was transfected into nucleotide excision repair-deficient UV5 cell lines, to generate a UV5 cell line containing a single integrated FRT site (UV5FRT). Purified human NADPH cytochrome P450 reductase (POR) and P450 1A2 polymerase chain reaction (PCR) products were digested and ligated into similarly treated pIRES vector and transformed into DH5α competent cells. The pIRES plasmid containing cDNAs of human P450 1A2 and POR was transfected into the newly established UV5FRT cell line. Those UV5 cells expressing a single FRT site, P450 1A2, and POR are referred to as UV5/P450 1A2 cells. The colonies of these cells were expanded, and intact geneticin-resistant cells were assayed for P450 1A2 activity, through measurement of measuring 7-ethoxyresorufin O-deethylase activity as described previously (42). P450 1A2 catalytic activity in untransfected UV5 cells is undetectable (<0.2 pmoles/min/106 cells) whereas P450 1A2-transfected cells (with and without further transfection with NAT2) have P450 1A2 catalytic activities about 3 pmol/min/106 cells (42).
The open reading frames of NAT2*4 and NAT2*5B were amplified by PCR, digested with NheI and XhoI (New England Biolabs), and inserted into the pcDNA5/FRT vector (Invitrogen) as described earlier. The pcDNA5/FRT plasmid containing human NAT2*4 or NAT2*5B was co-transfected with pOG44, a Flp recombinase expression plasmid, into UV5FRT/P4501A2 cells. Integration of the pcDNA5/FRT construct into the FRT site was confirmed by PCR. The NAT2*4- and NAT2*5B-transfected cells were characterized for N-acetylation of sulfamethazine, a NAT2-selective substrate. NAT2 catalytic activities are undetectable (<20 pmol/min/mg total protein) in untransfected UV5 and UV5/P450 1A2 cells, about 1.5 nmol/min/mg total protein in P450 1A2/NAT2*4 cells and about 0.1 nmol/min/mg total protein in P450 1A2/NAT2*5B cells (42).
Cytotoxicity and Mutagenesis
Assays for cytotoxicity and mutagenesis were carried out as described previously (42,43). Briefly, cells were grown for 12 doublings, with selective agents in complete hypoxanthine-aminopterin-thymidine medium (30 μM hypoxanthine, 0.1 μM aminopterin, and 30 μM thymidine). Cells were plated at a density of 5 × 105 cells per T-25 flask and were incubated for 24 h, after which media were changed. The cell lines were treated for 48 h with various concentrations of AαC (0, 0.75, 1.5, 3.0 or 6 μM) or vehicle control (0.5% dimethyl sulfoxide). Survival was determined by colony-forming assay and was expressed as the percentage of the vehicle control. The remaining cells were replated and subcultured. After 7 days of growth, cultures were plated for cloning efficiency in complete media and for mutations in complete media containing 40 μM 6-thioguanine. Dishes were seeded at 1 × 105 cells per 100 mm dish (10 replicates) and were incubated for 7 days; cloning efficiency dishes were seeded with 100 cells per well per six-well plate in triplicate and were incubated for 6 days.
Isolation of DNA
Cells grown in 15-cm plates were treated with AαC, as described above for the cytotoxicity and mutagenesis assays. Cells were harvested after 48 h of treatment, and DNA was extracted and quantified as previously described (42,43). The cell pellet was resuspended in 500 μl of 20 mM sodium phosphate buffer. One-tenth volumes each of proteinase K solution (20 mg/mL) and 10% SDS were added to the cell lysate, and the mixture was incubated at 37°C for 60 min. One volume of phenol, equilibrated with 10 mM Tris HCl (pH 8.0), was added to the mixture, which was then vortexed and centrifuged at 3,600 × g for 15 min. The aqueous layer was removed and added to 1 volume of phenol/chloroform/isoamyl alcohol (25:24:1) saturated with 10 mM Tris-HCl (pH 8.0), which was vortexed and centrifuged. The aqueous layer was removed and added to 1 volume of cold (-20°C) isopropanol, and the mixture was vortexed and centrifuged. The DNA pellet was washed with 70% ethanol and redissolved in 5 mM Tris HCl (pH 7.4) containing 1 mM CaCl2, 1 mM ZnCl2, and 10 mM MgCl2. DNA was quantified by UV spectroscopy using A260 nm. DNA quality was monitored by UV spectroscopy using A260/280 nm, and this ratio was consistently above 1.9.
Synthesis of N-(deoxyguanosin-8-yl)-2-amino-9H-pyrido[2,3-b]indole (dG-C8-AαC)
2-Hydroxy-amino-9H-pyrido[2,3-b]indole (HONH-AαC) was prepared by the reduction of NO2-AαC (2 mg) in tetrahydrofuran (1.5 mL) at −5 °C with 10% Pd/C (2 mg) with hydrazine (4 μL) as previously described (37,44). Following dilution with 10 ml of argon-purged distilled, deionized water, HONH-AαC was purified by solid phase extraction (37). The purified HON-AαC was reacted with dG (5 mg) or [13C10]dG (5 mg) in the presence of acetic anhydride (30 mol excess relative to HONH-AαC) in 100 mM K2HPO4 buffer (pH 7.5) containing 0.5 mM EDTA as previously described (20). The dG-C8-AαC adduct was purified by HPLC (45).
DNA Digestion Conditions
Assays were done in triplicate with 50 μg of DNA in 50 μL of 10 mM Bis-Tris buffer (pH 7.1). The enzymatic digestion for the hydrolysis of DNA to the 2′-deoxynucleosides was done with a cocktail of four enzymes as described (46). The [13C10]dG-C8-AαC internal standard was added prior to enzyme digestion at a level of 5 adducts per 108 bases (3.47 pg per 50 μg DNA). Cold C2H5OH (200 proof) was added to the hydrolysis mixture (3× the total vol) to terminate the reaction, and the C2H5OH /DNA solution was stored at -80°C until sample analyses were performed.
Solid-phase Extraction (SPE) DNA Adduct Enrichment
The proteins from the C2H5OH/DNA digest solution were precipitated by centrifugation at 15,000 × g for 5 min. The DNA adduct-containing supernatant was removed and dried by vacuum centrifugation. Samples were dissolved in 0.1% HCO2H containing 10% CH3OH (0.25 mL), and were purified by SPE using Isolute C18(EC) 25 mg SPE cartridges containing additionally washed polyethylene frits. The SPE cartridge was washed 2× with 0.1% HCO2H containing 10% CH3OH (0.25 mL), and the adducts were eluted with CH3OH containing 0.1% HCO2H. The eluent was collected into total recovery capLC vials (Waters, New Milford, MA), evaporated to dryness by vacuum centrifugation, and reconstituted in 1:1 DMSO:H2O (30 μL) (46).
LC-ESI/MS Analyses
Mass spectral data were acquired on either a Finnigan™ Quantum Ultra Triple Stage Quadrupole MS (TSQ/MS) or a Finnigan™ LTQ™ 2-D linear quadrupole ion trap mass spectrometer (LTQ/MS) (Thermo Fisher, San Jose, CA). Both instruments were equipped with an Ion Max™ electrospray ionization source operated in positive ionization mode. Xcalibur software (version 1.4; Thermo Electron) was used for system operation and data manipulation. Data from on-line analyses were acquired in centroid mode. Initial characterization of the synthetic adducts and MS/MS product ion spectra were carried out with the TSQ/MS. Representative optimized instrument tune parameters used on the TSQ/MS were as follows: capillary temperature 275°C; source spray voltage 4.0 kV; tube lens offset 95 V; capillary offset 35 V; source fragmentation offset 10 V. Nitrogen was used as the sheath gas, and was set at 35 units. Data acquisition in the product ion scan mode was performed using the following conditions: collision energy 40 V; peak width (in Q1 and Q3) 0.7 Da; scan width set at 0.7 Da; and scanning from m/z 150 to 500, at a rate of 250 Da/s. The collision gas was argon, set at 1.5 mTorr.
Ensuing analyses and adduct measurements of AαC adducts in CHO cells were conducted on the LTQ/MS. Representative optimized instrument tune parameters were as follows: capillary temperature 350 °C; source spray voltage 2.5 kV; sheath gas setting 3; auxilliary gas setting 6; capillary voltage 15 V; tube lens voltage 75 V; and in-source fragmentation 0 V. Helium was used as the collision damping gas in the ion trap, set at a pressure of 1 mTorr. One μscan was used for data acquisition, and the maximum injection time was 50 ms. The automatic gain control settings were: full MS target 30,000; MSn target 10,000. The instrument method used for the acquisition of on-line data consisted of a single segment that contained the consecutive reaction monitoring scan events (up to the MS3 stage of the aglycone ion [BH2]+). In full scan mode, the ion trap was scanning from m/z 240 to 500 Da; in MS/MS the isolation width was 3.0 Da; normalized collision energy 22, activation Q = 0.35, and 30 ms activation time; and in MS3 scan mode the activation Q = 0.35, the width 1.5 Da, normalized collision energy 32, and 30 ms activation time. The MS3 product ion spectra were acquired on aglycone ion adducts [BH2]+ scanning from m/z 140 to 400 Da.
Chromatography was performed on an Agilent 1100 Series capillary LC system (Agilent Technologies, Palo Alto, CA) equipped with an Aquasil C18 column (0.5 mm i.d. × 250 mm, 5 μm particle size; Thermo Electron). All of the samples were prepared in Waters (Milford, MA) borosilicate total recovery capillary LC vials. Samples were stored at 5°C in an Agilent autosampler throughout the analyses. The flow rate was set to 12 μL/min, using mixtures of (A) 9:1 H2O:CH3CN with 0.05% HCO2H and (B) CH3CN with 0.05% HCO2H and a gradient starting at 0% B for the first minute followed by a linear gradient to 100% B over 29 min, with a hold at 100% B for 3 min, back to starting conditions (0% B) over 2 min, and finally a 14-min period to allow the column to re-equilibrate to the starting conditions (0% B). Samples (8 μL) were injected through an 8-μL injection loop into a six-port switching valve injector (Rheodyne) that diverted the column eluent to waste for the first 4 min of the gradient.
Calibration Curves
External calibration curves were produced in triplicate by the addition of a fixed amount of [13C10]dG-C8-AαC as the internal standard, with varying amounts of unlabeled dG-C8-AαC at levels of modification equivalent to 0, 0.3, 1.0, 3.0, 9.0 and 30 adducts per 108 bases. The slope of the curve was 0.95 when expressed as area ratio (dG-C8-AαC/internal standard) versus the amount ratio (dG-C8-AαC/internal standard) with an r2 > 0.99.
Results
AαC-induced Cytotoxicity
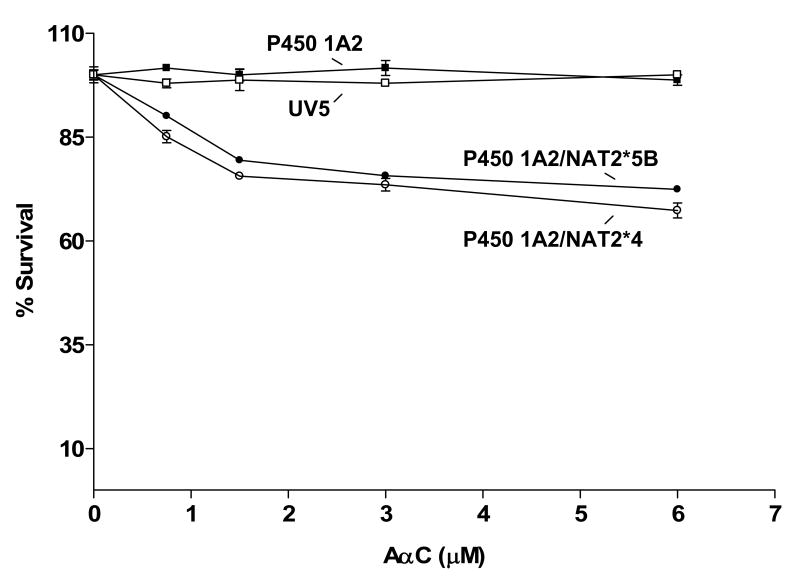
As shown in Figure 1, only the UV5/P450 1A2/NAT2*4 and UV5/P450 1A2/NAT2*5B cell lines showed concentration-dependent cytotoxicity following AαC treatment. Dose-dependent cytotoxicity from AαC was slightly greater in the UV5/P450 1A2/NAT2*4 than in the UV5/P450 1A2/NAT2*5B cell line, but the difference was not significant (p > 0.05).
Figure 1.
AαC-induced cytotoxicity in UV5/P450 1A2/NAT2*4 (○), UV5/P450 1A2/NAT2*5B (●), UV5/P450 1A2 (■), and untransfected UV5 (□) cell lines. Each data point represents the mean ± S.E.M. from three experiments. Dose-dependent cytotoxicity from AαC was slightly greater in the UV5/ P450 1A2/NAT2*4 than in the UV5/P450 1A2/NAT2*5B cell line, but the difference was not significant (p > 0.05).
AαC-induced Mutagenesis
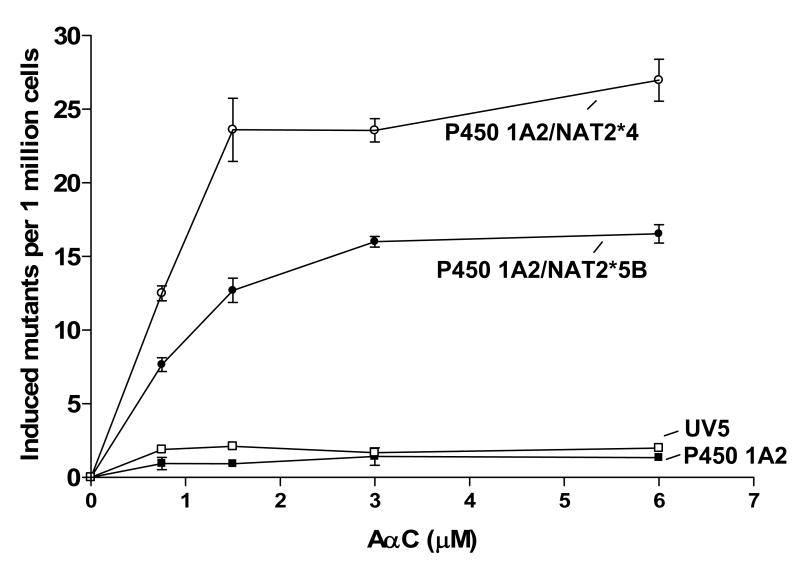
The UV5/P450 1A2/NAT2*4 and UV5/P450 1A2/NAT2*5B cell lines showed concentration-dependent mutagenicity after AαC treatment (Figure 2). However, the UV5/P450 1A2/NAT2*4 cell line showed significantly higher (p<0.001) levels of dose-dependent AαC-induced hprt mutants than did the UV5/P450 1A2/NAT2*5B cell line at all concentrations tested. AαC treatment did not result in cytotoxicity or mutations induced above background levels in either cell lines transfected with only P450 1A2 or in untransfected cells.
Figure 2.
AαC-induced mutagenesis UV5/P450 1A2/NAT2*4 (○), UV5/P450 1A2/NAT2*5B (●), UV5/P450 1A2 (■), and untransfected UV5 (□) cell lines. Each data point represents the mean ± S.E.M. from three experiments. The UV5/P450 1A2/NAT2*4 cell line showed significantly higher (p<0.001) dose-dependent AαC-induced hprt mutants than did the UV5/P450 1A2/NAT2*5B cell line, at all concentrations tested.
Mass Spectral Characterization of dG-C8-AαC
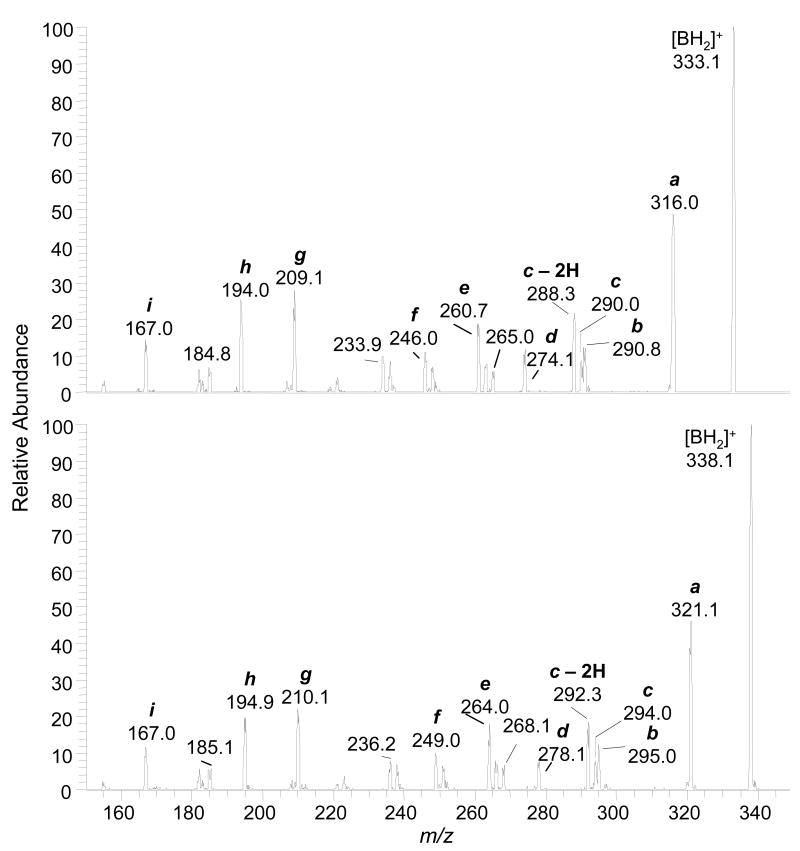
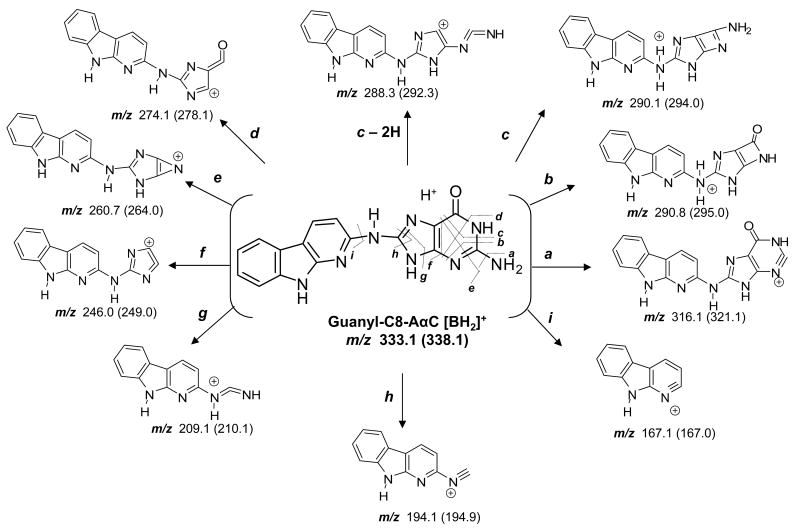
The product ion spectra of synthetic dG-C8-AαC and [13C10]dG-C8-AαC, obtained with the TSQ/MS under elevated collision-induced-dissociation (CID) conditions, are shown in Figure 3. The base peaks for the unlabeled and 13C-labeled adducts were observed at m/z 333 and 339, respectively. These ions are attributed to [M + H - 116]+ and [M + H - 121]+, and arise from a loss of dR or [13C5]dR, respectively, with the back-transfer of a hydrogen from the sugar moiety to the base. The elevated CID conditions resulted in secondary fragmentations of the agyclone adducts [BH2]+. Many of the pathways of CID fragmentation of dG-C8-AαC are similar to those reported for dG-C8-HAA adducts (46-48), and other dG-C8-arylamine adducts (Scheme 1) acquired by LC-ESI/MS employing either a TQ/MS/MS or quadrupole ion trap mass spectrometry at the MS3 scan stage. The proposed structures of the fragment ions and neutral losses are tentative; confirmation will require exact mass measurements and the use of adducts containing stable isotopes at defined positions of the molecule.
Figure 3.
LC-ESI/MS product ion spectra of synthetic dG-C8-AαC (upper panel) and [13C10]dG-C8-AαC (lower panel) at a CID of 40 eV, obtained with the TSQ/MS.
Scheme 1.
Proposed pathways of fragmentation of dG-C8-AαC and [13C10]dG--C8-AαC by ESI with the TSQ MS, under elevated CID conditions.
Identification and Quantitation of DNA Adducts
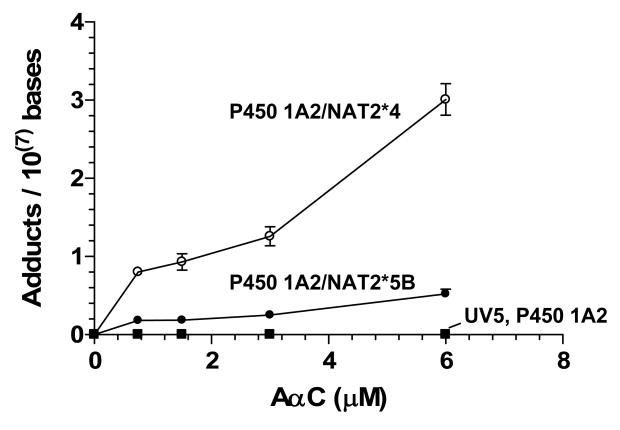
The dG-C8-AαC adduct was readily detected in CHO cell lines expressing human P450 1A2 with either NAT2*4 or NAT2*5B. In contrast to the cell lines expressing P450 1A2 and either NAT2*4 or NAT2*5B, the untransfected cell lines, and the cell lines expressing only P450 1A2 did not have detectable levels of DNA adduct (limit of detection <3 adducts per 109 bases), consistent with the absence of induced mutants in these cell lines. The high level of sensitivity conferred by the LTQ/MS enabled DNA adduct measurements (1.8 ± 0.2 adducts per 108 bases) and acquisition of the MS3 product ion spectrum of the agyclone adduct [BH2]+ at the lowest dose tested (0.75 μM), in DNA isolated from the NAT2*5B slow acetylator cell line (Figure 4).
Figure 4.
LC-ESI/MS/MS3 chromatograms obtained with the LIT/MS on (A) UV5 cells and (B) CHO cell lines expressing P450 1A2 and NAT2*5B treated with AαC (0.75 μM), and (C) [13C10]dG-C8-AαC added to DNA at a level of 5 adducts per 108 bases. Ions were extracted at m/z 291.1 and 316.1 for dG-C8-AαC and at m/z 295.1 and 321.1 for [13C10]dG-C8-AαC. The level of the dG-C8-AαC adduct was estimated at 1.8 adducts per 108 bases. Product ion spectra (MS3) of the guanyl-C8-AαC adduct from CHO cells (Panel B) and internal standard (Panel C) were acquired to confirm adduct identity.
The level of DNA adduct formation in the different cell lines as a function of AαC dose is presented in Figure 5. The levels of mutants were very low in the UV5 and P450 1A2-transfected cells and DNA adducts were undetectable (< 3 adducts per 109 bases). Both adducts and induced mutants were detected in the P450 1A2/NAT2*5B cells, which suggests that NAT2 contributes to both adducts and mutants; and still higher levels of mutants and adducts were detected in P450 1A2/NAT2*4 cells. The level of DNA adducts was about 6-fold higher in cell lines expressing P450 1A2 and NAT2*4 than in cell lines expressing P450 1A2 and NAT2*5B, at all dose concentrations (p<0.0001). These findings suggest that NAT2 4 protein contributes more than the NAT2 5B protein to adduct formation and induction of mutants, and that dG-C8-AαC is a mutagenic lesion.
Figure 5.
AαC-DNA adduct formation UV5/P450 1A2/NAT2*4 (○), UV5/P450 1A2/NAT2*5B (●), UV5/P450 1A2 (■), and untransfected UV5 (□) cell lines. Each data point represents the mean ± S.E.M. from three independent analyses. Linear regression analysis revealed that the slope of the curve (adduct formation/AαC concentration) was significantly higher for the UV5/P450 1A2/NAT2*4 cell line than for the UV5/P450 1A2/NAT2*5B cell line (p<0.0001).
Discussion
CHO cells stably transfected with P450 1A2 and either NAT2*4 or NAT2*5B have been established, to allow us to examine the genotoxic potential of HAAs (42). The phase I and II enzyme activities of non-transfected cells were 10-fold lower than the activities measured in transfected cells. AαC was shown to be a strong mutagen in these CHO cells transfected with P450 1A2 and NAT2. The cytotoxic and genotoxic effects induced by AαC required the expression of both enzymes. These findings strongly suggest that the N-acetoxy-AαC intermediate is the penultimate metabolite that reacts with DNA and is responsible for the mutagenicity of AαC. Lower levels of DNA adduct formation and frequency of hprt mutants occurred in cells transfected with P450 1A2 and NAT2*5B. Thus, the changes in the nucleotide sequence of the NAT2*4 → NAT2*5B (341T>C; 481C>T; 803A>G) and the resultant amino acid changes in the protein (I114T; K268R) (28) resulted in a diminished catalytic activity toward HONH-AαC.
The mutagenic potency of AαC, at a concentration of 1.5 μM, in CHO cells was about half the potency that was previously observed for MeIQx in this CHO cell-line transfected with P450 1A1 and NAT2*4 (43), and about one-tenth the activity previously observed for PhIP in this CHO cell-line transfected with P450 1A2 and NAT2*4 (42). On the basis of estimates of dG-C8-HAA adduct formation and assuming that these adducts are the principal genotoxic lesions, we estimate that one dG-C8-HAA adduct per 107 bases in the CHO genome induced about 1.5 hprt mutants for MeIQx; 2.5 hprt mutants for AαC; and 2.8 hprt mutants for PhIP per one million cells, at the dose concentrations of 1.5 μM (MeIQx and AαC) (43) and 1.2 μM (PhIP) (42). The potential of these dG-HAA adducts to induce mutations at the hprt locus appears similar; the different levels of mutation induced by these three HAAs in CHO cells are driven by the differing levels of adduct formation, which are dependent upon the levels of N-oxidation of HAAs and ensuing activation by phase II enzymes. The kinetic parameters (kcat and Km) of P450 1A2-mediated N-oxidation of PhIP and MeIQx are comparable (49), but the kinetic parameters of N-oxidation of AαC have not been reported. Moreover, the kinetic parameters of NAT2 (or other phase II enzymes) implicated in catalysis of DNA binding of these HONH-HAAs have not been determined and are likely to influence the levels of DNA adducts and genotoxicity of these respective HAAs
The genotoxicity and level of DNA adducts of AαC were highest in cell lines expressing both P450 1A2 and NAT2 4, but the protein derived from the slow NAT2*5B allele also catalyzed DNA binding and induction of mutants at a significant rate. In the case of MeIQx, the NAT2 4 enzyme was critical for the catalysis of DNA adduct formation; the levels of adducts and induced mutants were decreased by 100-fold or more in cells expressing the slow acetylator NAT2 5B protein. The biological effects of PhIP were only marginally enhanced by expression of either NAT2 4 or NAT2 5B protein in cells co-transfected with P450 1A2 (29,42,50). Thus, there are clear differences in the relative effect of NAT2 genetic polymorphism among AαC, MeIQx, and PhIP, suggesting that the identification of the specific HAA exposure is very important in molecular epidemiological investigations that seek to assess the importance of HAAs and NAT2 genetic polymorphism in cancer risk.
The genotoxic potencies of AαC, MeIQx, and PhIP in Salmonella typhimurium TA98 (frame-shift specific) tester strain are strikingly different from the activities observed in CHO cells. In TA98 cells, which also lack nucleotide excision repair (51), the revertants induced by MeIQx were about 80-fold more numerous than the revertants induced by PhIP, and almost 500-fold more numerous than the revertants induced by AαC (1). The discrepancies in the biological potencies of HAAs in these in vitro assays are likely due to differing metabolic activation systems, differing gene locus endpoints for mutagenicity, and different base sequence contexts and neighboring base effects on the HAA-DNA lesions, all of which affect mutation frequencies.
The focus of this study was to investigate the impact of NAT2 acetylator genotype on the bioactivation of AαC; however, N-acetyltransferase 1, which also displays genetic polymorphisms and is expressed in extrahepatic tissues at high levels (31), catalyzes the bioactivation of HONH-AαC (16,38). Moreover, P450s 1A1 and 2C9, which are highly expressed in the colon and the respiratory tract, N-hydroxylate AαC at appreciable levels (14). Thus, AαC has the potential to form DNA adducts in human liver and a number of extrahepatic tissues. Given the high levels of AαC reported in tobacco smoke (3,9) and the recent discovery of AαC in urine of cigarette smokers at high frequency (52), the role of AαC, and the modulating effects of N-acetyltransferase genetic polymorphisms, in tobacco-associated cancer risk warrant further investigation.
Acknowledgments
The project was supported by grant R21ES-014438 (RJT and IY) from the National Institute of Environmental Health Sciences, grant R01CA-122320 from the National Cancer Institute (RJT), and grants R01-CA034627 and P30-ES014443 (DWH).
Footnotes
Abbreviations: AαC, 2-Amino-9H-pyrido[2,3-b]indole; HONH-AαC, 2-hydroxyamino-9H-pyrido[2,3-b]indole; MeIQx, 2-amino-3,8-dimethylimdazo[4,5-f]quinoxaline; PhIP, 2-amino-1-methyl--6-phenylimidazo[4,5-b]pyridine; dG, 2′-deoxyguanosine; dG-C8-AαC, N-(deoxyguanosin-8-yl)-2-amino-9H-pyrido[2,3-b]indole; dG-C8-MeIQx, N-(deoxyguanosin-8-yl)-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline; dG-C8-PhIP, N-(deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine; dR, deoxyribose; CHO, Chinese hamster ovary cells; CID, collision-induced dissociation; HAA, heterocyclic aromatic amine; hprt, hypoxanthine phosphoribosyl transferase; LC-ESI/MS, liquid chromatography-electrospray ionization/mass spectrometry; LTQ/MS, Finnigan LTQ 2-D linear quadrupole ion trap mass spectrometer; NAT, N-acetyltransferase; ppb, parts-per-billion; SPE, solid phase extraction; SRM, selected reaction monitoring; SULTs, sulfotransferases; TQ/MS/MS, triple quadrupole tandem mass spectrometry; TSQ, Finnigan TSQ Quantum Ultra triple quadrupole mass spectrometer.
References
- 1.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida D, Matsumoto T, Yoshimura R, Matsuzaki T. Mutagenicity of amino-alpha-carbolines in pyrolysis products of soybean globulin. Biochem Biophys Res Commun. 1978;83:915–920. doi: 10.1016/0006-291x(78)91482-1. [DOI] [PubMed] [Google Scholar]
- 3.Matsumoto T, Yoshida D, Tomita H. Determination of mutagens, amino-alpha-carbolines in grilled foods and cigarette smoke condensate. Cancer Lett. 1981;12:105–110. doi: 10.1016/0304-3835(81)90045-8. [DOI] [PubMed] [Google Scholar]
- 4.Holder CL, Preece SW, Conway SC, Pu YM, Doerge DR. Quantification of heterocyclic amine carcinogens in cooked meats using isotope dilution liquid chromatography/atmospheric pressure chemical ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 1997;11:1667–1672. doi: 10.1002/(SICI)1097-0231(19971015)11:15<1667::AID-RCM58>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 5.Gross GA, Gruter A. Quantitation of mutagenic/carcinogenic heterocyclic aromatic amines in food products. J Chromatogr. 1992;592:271–278. doi: 10.1016/0021-9673(92)85095-b. [DOI] [PubMed] [Google Scholar]
- 6.Turesky RJ, Taylor J, Schnackenberg L, Freeman JP, Holland RD. Quantitation of carcinogenic heterocyclic aromatic amines and detection of novel heterocyclic aromatic amines in cooked meats and grill scrapings by HPLC/ESI-MS. J Agric Food Chem. 2005;53:3248–3258. doi: 10.1021/jf048290g. [DOI] [PubMed] [Google Scholar]
- 7.Brockstedt U, Pfau W. Formation of 2-amino-α-carbolines in pan-fried poultry and 32P-postlabelling analysis of DNA adducts. Z Lebensm Unters Forsch A. 1998;207:472–476. [Google Scholar]
- 8.Manabe S, Izumikawa S, Asakuno K, Wada O, Kanai Y. Detection of carcinogenic amino-alpha-carbolines and amino-gamma-carbolines in diesel-exhaust particles. Environ Pollut. 1991;70:255–265. doi: 10.1016/0269-7491(91)90013-m. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida D, Matsumoto T. Amino-alpha-carbolines as mutagenic agents in cigarette smoke condensate. Cancer Lett. 1980;10:141–149. doi: 10.1016/0304-3835(80)90037-3. [DOI] [PubMed] [Google Scholar]
- 10.Patrianakos C, Hoffmann D. Chemical studies on tobacco smoke LXIV. On the analysis of aromatic amines in cigarette smoke. J Assoc Off Anal Chem. 1979;3:150–154. [Google Scholar]
- 11.Zha Q, Qian NX, Moldoveanu SC. Analysis of polycyclic aromatic hydrocarbons in the particulate phase of cigarette smoke using a gas chromatographic-high-resolution mass spectrometric technique. J Chromatogr Sci. 2002;40:403–408. doi: 10.1093/chromsci/40.7.403. [DOI] [PubMed] [Google Scholar]
- 12.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 13.Niwa T, Yamazoe Y, Kato R. Metabolic activation of 2-amino-9H-pyrido[2,3-b]indole by rat-liver microsomes. Mutat Res. 1982;95:159–170. doi: 10.1016/0027-5107(82)90254-8. [DOI] [PubMed] [Google Scholar]
- 14.Raza H, King RS, Squires RB, Guengerich FP, Miller DW, Freeman JP, Lang NP, Kadlubar FF. Metabolism of 2-amino-alpha-carboline. A food-borne heterocyclic amine mutagen and carcinogen by human and rodent liver microsomes and by human cytochrome P4501A2. Drug Metab Dispos. 1996;24:395–400. [PubMed] [Google Scholar]
- 15.Frederiksen H, Frandsen H. Impact of five cytochrome p450 enzymes on the metabolism of two heterocyclic aromatic amines, 2-amino-9H-pyrido[2,3-b]indole (AalphaC) and 2-amino-3-methyl-9H-pyrido[2,3-b]indole (MeAalphaC) Pharmacol Toxicol. 2003;92:246–248. doi: 10.1034/j.1600-0773.2003.920508.x. [DOI] [PubMed] [Google Scholar]
- 16.King RS, Teitel CH, Kadlubar FF. In vitro bioactivation of N-hydroxy-2-amino-alpha-carboline. Carcinogenesis. 2000;21:1347–1354. [PubMed] [Google Scholar]
- 17.Turesky RJ. DNA adducts of heterocyclic aromatic amines, arylazides and 4-nitroquinoline 1-oxide. In: Hemminki K, Dipple A, Shuker DEG, Kadlubar FF, Segerbäck D, Bartsch H, editors. DNA Adducts: Identification and Biological Significance. International Agency for Research on Cancer; Lyon: 1994. pp. 217–228. [PubMed] [Google Scholar]
- 18.Schut HA, Snyderwine EG. DNA adducts of heterocyclic amine food mutagens: implications for mutagenesis and carcinogenesis. Carcinogenesis. 1999;20:353–368. doi: 10.1093/carcin/20.3.353. [DOI] [PubMed] [Google Scholar]
- 19.Pfau W, Schulze C, Shirai T, Hasegawa R, Brockstedt U. Identification of the major hepatic DNA adduct formed by the food mutagen 2-amino-9H-pyrido[2,3-b]indole (A alpha C) Chem Res Toxicol. 1997;10:1192–1197. doi: 10.1021/tx9701182. [DOI] [PubMed] [Google Scholar]
- 20.Frederiksen H, Frandsen H, Pfau W. Syntheses of DNA-adducts of two heterocyclic amines, 2-amino-3-methyl-9H-pyrido[2,3-b]indole (MeA{alpha}C) and 2-amino-9H-pyrido[2,3-b]indole (A{alpha}C) and identification of DNA-adducts in organs from rats dosed with MeA{alpha}C. Carcinogenesis. 2004;25:1525–1533. doi: 10.1093/carcin/bgh156. [DOI] [PubMed] [Google Scholar]
- 21.Turesky RJ. The role of genetic polymorphisms in metabolism of carcinogenic heterocyclic aromatic amines. Curr Drug Metab. 2004;5:169–180. doi: 10.2174/1389200043489036. [DOI] [PubMed] [Google Scholar]
- 22.Eaton DL, Gallagher EP, Bammler TK, Kunze KL. Role of cytochrome P450 1A2 in chemical carcinogenesis: implications for human variability in expression and enzyme activity. Pharmacogenetics. 1995;5:259–274. doi: 10.1097/00008571-199510000-00001. [DOI] [PubMed] [Google Scholar]
- 23.Turesky RJ, Constable A, Richoz J, Varga N, Markovic J, Martin MV, Guengerich FP. Activation of heterocyclic aromatic amines by rat and human liver microsomes and by purified rat and human cytochrome P450 1A2. Chem Res Toxicol. 1998;11:925–936. doi: 10.1021/tx980022n. [DOI] [PubMed] [Google Scholar]
- 24.Hammons GJ, Yan-Sanders Y, Jin B, Blann E, Kadlubar FF, Lyn-Cook BD. Specific site methylation in the 5′-flanking region of CYP1A2 interindividual differences in human livers. Life Sci. 2001;69:839–845. doi: 10.1016/s0024-3205(01)01175-4. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima M, Yokoi T, Mizutani M, Kinoshita M, Funayama M, Kamataki T. Genetic polymorphism in the 5′-flanking region of human CYP1A2 gene: effect on the CYP1A2 inducibility in humans. J Biochem (Tokyo) 1999;125:803–808. doi: 10.1093/oxfordjournals.jbchem.a022352. [DOI] [PubMed] [Google Scholar]
- 26.Sachse C, Bhambra U, Smith G, Lightfoot TJ, Barrett JH, Scollay J, Garner RC, Boobis AR, Wolf CR, Gooderham NJ. Polymorphisms in the cytochrome P450 CYP1A2 gene (CYP1A2) in colorectal cancer patients and controls: allele frequencies, linkage disequilibrium and influence on caffeine metabolism. Br J Clin Pharmacol. 2003;55:68–76. doi: 10.1046/j.1365-2125.2003.01733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002;506-507:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- 28.Hein DW. N-acetyltransferase 2 genetic polymorphism: effects of carcinogen and haplotype on urinary bladder cancer risk. Oncogene. 2006;25:1649–1658. doi: 10.1038/sj.onc.1209374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glatt H. Metabolic factors affecting the mutagenicity of heterocyclic amines. In: Skog K, Alexander J, editors. Acrylamide and other hazardous compounds in heat-treated foods. Woodhead Publishing Ltd.; Cambridge, England: 2006. pp. 358–404. [Google Scholar]
- 30.Garcia-Closas M, Malats N, Silverman D, Dosemeci M, Kogevinas M, Hein DW, Tardon A, Serra C, Carrato A, Garcia-Closas R, Lloreta J, Castano-Vinyals G, Yeager M, Welch R, Chanock S, Chatterjee N, Wacholder S, Samanic C, Tora M, Fernandez F, Real FX, Rothman N. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366:649–659. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hein DW, Doll MA, Fretland AJ, Leff MA, Webb SJ, Xiao GH, Devanaboyina US, Nangju NA, Feng Y. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev. 2000;9:29–42. [PubMed] [Google Scholar]
- 32.Giovannucci E. An updated review of the epidemiological evidence that cigarette smoking increases risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:725–731. [PubMed] [Google Scholar]
- 33.Lang NP, Butler MA, Massengill JP, Lawson M, Stotts RC, Hauer-Jensen M, Kadlubar FF. Rapid metabolic phenotypes for acetyltransferase and cytochrome P4501A2 and putative exposure to food-borne heterocyclic amines increase the risk for colorectal cancer or polyps. Cancer Epidemiol Biomarkers Prev. 1994;3:675–682. [PubMed] [Google Scholar]
- 34.Le Marchand L, Hankin JH, Pierce LM, Sinha R, Nerurkar PV, Franke AA, Wilkens LR, Kolonel LN, Donlon T, Seifried A, Custer LJ, Lum-Jones A, Chang W. Well-done red meat, metabolic phenotypes and colorectal cancer in Hawaii. Mutat Res. 2002;506-507:205–14. doi: 10.1016/s0027-5107(02)00167-7. 205-214. [DOI] [PubMed] [Google Scholar]
- 35.Zhang XB, Felton JS, Tucker JD, Urlando C, Heddle JA. Intestinal mutagenicity of two carcinogenic food mutagens in transgenic mice: 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and amino(alpha)carboline. Carcinogenesis. 1996;17:2259–2265. doi: 10.1093/carcin/17.10.2259. [DOI] [PubMed] [Google Scholar]
- 36.Okonogi H, Ushijima T, Shimizu H, Sugimura T, Nagao M. Induction of aberrant crypt foci in C57BL/6N mice by 2-amino-9H-pyrido[2,3-b]indole (A alphaC) and 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx) Cancer Lett. 1997;111:105–109. doi: 10.1016/s0304-3835(96)04505-3. [DOI] [PubMed] [Google Scholar]
- 37.Turesky RJ, Lang NP, Butler MA, Teitel CH, Kadlubar FF. Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis. 1991;12:1839–1845. doi: 10.1093/carcin/12.10.1839. [DOI] [PubMed] [Google Scholar]
- 38.Minchin RF, Reeves PT, Teitel CH, McManus ME, Mojarrabi B, Ilett KF, Kadlubar FF. N- and O-acetylation of aromatic and hetercyclic amine carcinogens by human monomorphic and polymorphic acetyltransferases expressed in COS-1 cells. Biochem and Biophys Res Comm. 1992;185:839–844. doi: 10.1016/0006-291x(92)91703-s. [DOI] [PubMed] [Google Scholar]
- 39.Turesky RJ. Interspecies metabolism of heterocyclic aromatic amines and the uncertainties in extrapolation of animal toxicity data for human risk assessment. Mol Nutr Food Res. 2005;49:101–117. doi: 10.1002/mnfr.200400076. [DOI] [PubMed] [Google Scholar]
- 40.Thompson LH, Rubin JS, Cleaver JE, Whitmore GF, Brookman K. A screening method for isolating DNA repair-deficient mutants of CHO cells. Somatic Cell Genet. 1980;6:391–405. doi: 10.1007/BF01542791. [DOI] [PubMed] [Google Scholar]
- 41.Weber CA, Salazar EP, Stewart SA, Thompson LH. Molecular cloning and biological characterization of a human gene, ERCC2, that corrects the nucleotide excision repair defect in CHO UV5 cells. Mol Cell Biol. 1988;8:1137–1146. doi: 10.1128/mcb.8.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metry KJ, Zhao S, Neale JR, Doll MA, States JC, McGregor WG, Pierce WM, Jr, Hein DW. 2-amino-1-methyl-6-phenylimidazo [4,5-b] pyridine-induced DNA adducts and genotoxicity in chinese hamster ovary (CHO) cells expressing human CYP1A2 and rapid or slow acetylator N-acetyltransferase 2. Mol Carcinog. 2007;46:553–563. doi: 10.1002/mc.20302. [DOI] [PubMed] [Google Scholar]
- 43.Bendaly J, Zhao S, Neale JR, Metry KJ, Doll MA, States JC, Pierce WM, Jr, Hein DW. 2-Amino-3,8-dimethylimidazo-[4,5-f]quinoxaline-induced DNA adduct formation and mutagenesis in DNA repair-deficient Chinese hamster ovary cells expressing human cytochrome P4501A1 and rapid or slow acetylator N-acetyltransferase 2. Cancer Epidemiol Biomarkers Prev. 2007;16:1503–1509. doi: 10.1158/1055-9965.EPI-07-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westra JG. A rapid and simple synthesis of reactive metabolites of carcinogenic aromatic amines in high yield. Carcinogenesis. 1981;2:355–357. doi: 10.1093/carcin/2.4.355. [DOI] [PubMed] [Google Scholar]
- 45.Turesky RJ, Rossi SC, Welti DH, Lay JO, Jr, Kadlubar FF. Characterization of DNA adducts formed in vitro by reaction of N-hydroxy-2-amino-3-methylimidazo[4,5-f]quinoline and N-hydroxy-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline at the C-8 and N2 atoms of guanine. Chem Res Toxicol. 1992;5:479–490. doi: 10.1021/tx00028a005. [DOI] [PubMed] [Google Scholar]
- 46.Goodenough AK, Schut HA, Turesky RJ. Novel LC-ESI/MS/MSn method for the characterization and quantification of 2′-deoxyguanosine adducts of the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by 2-D linear quadrupole ion trap mass spectrometry. Chem Res Toxicol. 2007;20:263–276. doi: 10.1021/tx0601713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turesky RJ, Vouros P. Formation and analysis of heterocyclic aromatic amine-DNA adducts in vitro and in vivo. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;802:155–166. doi: 10.1016/j.jchromb.2003.10.053. [DOI] [PubMed] [Google Scholar]
- 48.Chiarelli MP, Lay JO., Jr Mass spectrometry for the analysis of carcinogen-DNA adducts. Mass Spectrom Rev. 1992;11:447–493. [Google Scholar]
- 49.Turesky RJ, Constable A, Fay LB, Guengerich FP. Interspecies differences in metabolism of heterocyclic aromatic amines by rat and human P450 1A2. Cancer Lett. 1999;143:109–112. doi: 10.1016/s0304-3835(99)00137-8. [DOI] [PubMed] [Google Scholar]
- 50.Wu RW, Tucker JD, Sorensen KJ, Thompson LH, Felton JS. Differential effect of acetyltransferase expression on the genotoxicity of heterocyclic amines in CHO cells. Mutat Res. 1997;390:93–103. doi: 10.1016/s0165-1218(97)00005-0. [DOI] [PubMed] [Google Scholar]
- 51.Maron DM, Ames BN. Revised methods for the Salmonella mutagenicity test. Mutat Res. 1983;113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- 52.Turesky RJ, Yuan JM, Wang R, Peterson S, Yu MC. Tobacco Smoking and Urinary Levels of 2-Amino-9H-Pyrido[2,3-b]Indole in Men of Shanghai, China. Cancer Epidemiol Biomarkers Prev. 2007;16:1554–1560. doi: 10.1158/1055-9965.EPI-07-0132. [DOI] [PubMed] [Google Scholar]