Abstract
Ciliated protozoa of the genus Euplotes have undergone genetic code reassignment, redefining the termination codon UGA to encode cysteine. In addition, Euplotes spp. genes very frequently employ shifty stop frameshifting. Both of these phenomena involve noncanonical events at a termination codon, suggesting they might have a common cause. We recently demonstrated that Euplotes octocarinatus peptide release factor eRF1 ignores UGA termination codons while continuing to recognize UAA and UAG. Here we show that both the Tetrahymena thermophila and E. octocarinatus eRF1 factors allow efficient frameshifting at all three termination codons, suggesting that UGA redefinition also impaired UAA/UAG recognition. Mutations of the Euplotes factor restoring a phylogenetically conserved motif in eRF1 (TASNIKS) reduced programmed frameshifting at all three termination codons. Mutation of another conserved residue, Cys124, strongly reduces frameshifting at UGA while actually increasing frameshifting at UAA/UAG. We will discuss these results in light of recent biochemical characterization of these mutations.
Keywords: Euplotes octocarinatus, Tetrahymena thermophila, codon reassignment, programmed frameshifting, translation termination, Saccharomyces cerevisiae
INTRODUCTION
A founding principle of molecular genetics was the universality of the genetic code. Crick (1968) proposed that the presumptive universal genetic code evolved from an early, error-prone code. That code must have used three-nucleotide codons, but each codon might not have discriminated among related amino acids, e.g., leucine, isoleucine, or valine. The early code evolved to the present code by increasing the specificity and accuracy of decoding and was frozen in its final form. We now recognize that the genetic code has continued to evolve. Nonstandard codes have evolved repeatedly in mitochondria, in some bacteria, and among the nuclear codes of some fungi, green algae, diplomonads, and ciliated protozoa (Knight et al. 2001).
How could the genetic code evolve to change meaning? Some nonstandard codes evolved in genomes of very low complexity as in mitochondria. The “codon capture” hypothesis (Osawa and Jukes 1989) predicts that in a small genome, genetic drift may cause a codon to stochastically become unused and then redefined with a novel identity. This is less likely in larger genomes, though extreme base composition bias could lead to codon loss, as with the loss of the codon CGG in Mycoplasma capricolum (Oba et al. 1991). Very large genomes are unlikely to completely abandon a codon, prompting an alternative explanation, the “ambiguous intermediate” hypothesis (Schultz and Yarus 1994). For example, certain Candida species decode CUG as both the canonical leucine and the noncanonical serine (Suzuki et al. 1997). This ambiguity may resolve back to a normal or to a strictly nonstandard code; many Candida species noncanonically decode CUG exclusively as serine (Suzuki et al. 1997).
Codon redefinition might be selectively neutral, or it could be selectively advantageous. For example, ambiguous decoding in Candida, leading to accumulation of misfolded proteins, constitutively activates the heat shock response providing stress tolerance (Santos et al. 1999, 2004). This change may have facilitated Candida in adopting a lifestyle as a human pathogen.
The reason for the evolution of other nonstandard genetic codes remains a mystery. In particular, it is unclear why nonstandard codes evolved so frequently among the ciliated protozoa (Tourancheau et al. 1995; Lozupone et al. 2001). All of these codes involve changing the meaning of canonical termination codons. Tetrahymena species, for example, recognize UGA as a stop codon but have reassigned UAA and UAG as glutamine (Horowitz and Gorovsky 1985). Multiple reassignments have occurred; for example, UGA is encoded as cysteine in Euplotes but as tryptophan in Colpoda (Meyer et al. 1991; Lozupone et al. 2001). The polyphyly of code reassignment is an especially striking result (Tourancheau et al. 1995). Osawa et al. (1992) proposed that termination codons might be more susceptible to genetic drift because they are far rarer in a genome than even the rarest sense codon.
Translation termination requires two classes of termination factor. Class I factors recognize the termination codons, and the Class II factors are GTP-binding stimulatory factors. Eukaryotes encode two factors, the Class I factor eukaryotic release factor 1 (eRF1) recognizing all three termination codons, and the Class II factor eRF3 (Stansfield et al. 1995; Zhouravleva et al. 1995). Bacteria encode three factors: the Class I factors RF1 and RF2, specific for UAA/UAG and UAA/UGA, respectively, and the Class II factor RF3 (Scolnick et al. 1968; Goldstein et al. 1970). The prokaryotic and eukaryotic Class I factors have no overall structural similarity despite their similar function (Vestergaard et al. 2001). eRF1 is composed of three functional domains (Frolova et al. 2000; Song et al. 2000). Domain 1 of eRF1 binds to the stop codon and initiates the termination process (Bertram et al. 2000; Chavatte et al. 2002; Ito et al. 2002; Seit-Nebi et al. 2002). Domain 2 interacts with the peptidyl transferase center of the ribosome and mediates release of the completed polypeptide chain from the peptidyl-tRNA molecule in the ribosomal P site (Frolova et al. 1999; Heurgue-Hamard et al. 2005). Domain 3 mediates the eRF1•eRF3 interaction (Ito et al. 1998; Ebihara and Nakamura 1999; Eurwilaichitr et al. 1999; Merkulova et al. 1999). GTP hydrolysis by eRF3 stimulates both polypeptide chain release and proper stop codon recognition by eRF1 (Frolova et al. 1998; Salas-Marco and Bedwell 2004; Alkalaeva et al. 2006).
Osawa et al. (1992) assumed that redefinition of termination codons required altering peptide release factor (RF) specificity and a nonsense suppressor tRNA. Supporting this mechanism, Ito et al. (2002) used chimeras between domain 1 of the Tetrahymena thermophila eRF1 and domains 2 and 3 of Schizosaccharomyces pombe eRF1 to show that the Tetrahymena factor recognizes only UGA. Tetrahymena also encodes two tRNAGln specific for UAG and UAA (Kuchino et al. 1985; Hanyu et al. 1986). Using an in vitro assay, Kervestin et al. (2001) showed that Euplotes aediculatus eRF1 recognizes UAA and UAG but not UGA as termination codons. We recently tested the mechanism of genetic code evolution in vivo using chimeric proteins in which domain 1 of the Euplotes octocarinatus or T. thermophila eRF1 is fused to domains 2 and 3 of the Saccharomyces cerevisiae protein (Salas-Marco et al. 2006). We confirmed the results of Kervestin et al. (2001) because the Euplotes/Saccharomyces (Eo/Sc) chimera allowed highly efficient read-through of UGA but not UAA and UAG. We did not confirm the results of Ito et al. (2002) since we found that the Tetrahymena/Saccharomyces (Tt/Sc) chimeric protein recognized all three stop codons. We suggested that reassignment of UAA and UAG to Gln in Tetrahymena actually did not require a change in eRF1 specificity but rather the endogenous UAA and UAG-recognizing tRNAGln isoacceptors supported sufficient read-through to impose the nonstandard code. A genome survey of Euplotes crassus revealed the presence of a putative UGA-recognizing tRNACys (V.N. Gladyshev, pers. comm.). The Euplotes suppressor might be less efficient, necessitating modifying the RF to reassign UGA as Cys.
Comparison of eRF1 sequences identified two highly conserved sequence motifs in domain 1 of canonical code proteins (TASNIKS and YxCxxxF) that show unusually high sequence divergence in variant code proteins, suggesting that they might be responsible for altering codon specificity (Knight and Landweber 2000; Song et al. 2000; Lozupone et al. 2001; Kim et al. 2005). Mutagenesis and protein cross-linking studies independently implicated these two motifs in codon recognition (Chavatte et al. 2002; Inagaki et al. 2002; Seit-Nebi et al. 2002; Kolosov et al. 2005). Fan-Minogue et al. (2008) tested the role of these two motifs in redefinition of UGA in E. octocarinatus using the E. octocarinatus/S. cerevisiae chimeric eRF1. The alternative form of the TASNIKS motif in Euplotes eRF1 (TAESIKS) diminishes UGA recognition. Mutating the cysteine in the YxCxxxF motif of Euplotes eRF1 restored UGA recognition, consistent with its apparent role in codon recognition. They proposed a conformational model for eRF3 activation of peptide release by eRF1 function. The model involves distinct conformations, one for UAA/UAG and one for UGA; the evidence suggests that the Euplotes factor recognizes UGA very poorly because it is specifically defective in the required conformational switch.
Euplotes genes employ programmed +1 translational frameshifting orders of magnitude more frequently than any other known species (Klobutcher and Farabaugh 2002; Klobutcher 2005). These frameshifts occur at “shifty stop” frameshift sites (Weiss et al. 1987) of the form AAA-UAA-A or sometimes AAA-UAG-A. Apparently the peptidyl-tRNALys decoding the AAA slips to the overlapping AAU codon and translation continues in the +1 frame with decoding of the following AAA or AGA codon. Klobutcher and Farabaugh (2002) proposed that altering eRF1 to ignore UGA impaired its recognition of the other termination codons. Because programmed frameshifting efficiency depends on a competition between normal decoding and noncanonical decoding, any decrease in the rate of normal decoding increases the rate of the competing noncanonical decoding event, in the case of Euplotes frameshifting the recognition of UAA or UAG.
We present evidence that indeed reassignment of UGA to Cys in E. octocarinatus did result in increased +1 slippery stop frameshifting at both UAA and UAG codons. Mutation of E. octocarinatus eRF1 to impair recognition of UGA had the pleiotropic effect of reducing recognition of UAA and UAG; this inefficiency appears to play a role in allowing frequent evolution of programmed shifty stop frameshift sites in this species. We also found that reassignment of UAA and UAG to encode glutamine in T. thermophila also allowed very efficient frameshifting on UAA, UAG, and UGA. This suggests that T. thermophila eRF1 recognition of these codons is also inefficient enough to stimulate significant frameshifting despite the lack of evidence of shifty stop frameshifting in T. thermophila.
RESULTS
A yeast genetic system to test the function of heterologous eRF1 proteins
We have developed an in vivo system for testing the codon specificity of recognition by eRF1 (Salas-Marco et al. 2006). The factors consist of three independently functioning domains that make few if any direct interactions (Song et al. 2000). We created genes expressing chimeras of the domain 1 of a E. octocarinatus or T. thermophila eRF1 fused to domains 2 and 3 of S. cerevisiae eRF1 with the linkage made at the middle of a 5 amino acid extended chain that links the two domains (Fig. 1). The S. cerevisiae gene encoding eRF1, SUP45, is essential. The Thermus thermophilus fusion protein (Tt/Sc eRF1) suppresses that lethality of sup45Δ but the E. octocarinatus protein (Eo/Sc eRF1) cannot, which is consistent with Tt/Sc eRF1 efficiently recognizing all three termination codons, whereas Eo/Sc eRF1 recognizes only UAA/UAG (Salas-Marco et al. 2006). Since the Eo/Sc eRF1 cannot support growth, we used a conditional expression strategy to test its function. Yeast expressing Sc eRF1 inducibly under control of the GAL1 promoter and Eo/Sc eRF1 constitutively from the SUP45 promoter were shifted from inducing (galactose) to repressing conditions (glucose). After the shift, synthesis of Sc eRF1 ceased and continued growth diluted the protein to 10% of its normal level (Salas-Marco et al. 2006). Continued protein synthesis depends on high levels of Eo/Sc eRF1. As controls, two additional strains were grown under identical carbon source shift conditions. One carried only the GAL1-driven Sc eRF1 so that after shift to glucose, the endogenous eRF1 would be depleted. A second carried only a constitutively expressed Sc eRF1 so that after the shift, the factor would continue to be present at normal levels. Using this method, we demonstrated that Eo/Sc eRF1 fails to recognize UGA (Salas-Marco et al. 2006). Thirty percent of yeast genes use UGA as terminator, explaining the failure of Eo/Sc eRF1 to replace Sc eRF1.
FIGURE 1.
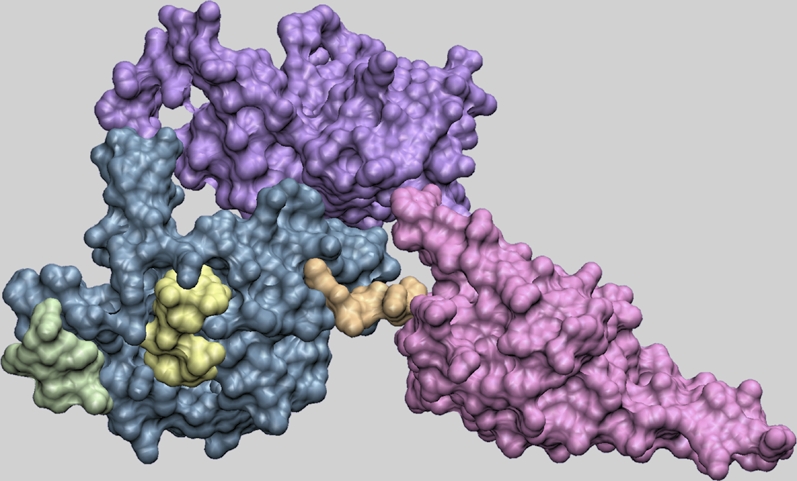
Structure of human eRF1. Domain 1 (blue), domain 2 (red), and domain 3 (violet) of human eRF1 are represented in a van der Waals surface format. The two conserved motifs in domain 1 are shown in contrasting colors: TASNIKS, green; YxCxxxF, yellow. The linker between domain 1 and domain 2 (orange) is the site of the fusion of the E. octocarinatus or T. thermophila domain 1 with domain 2 and 3 of the S. cerevisiae enzyme.
The Euplotes/Saccharomyces eRF1 chimera promotes +1 shifty stop frameshifting at all three termination codons
Klobutcher and Farabaugh (2002) hypothesized that alteration of eRF1 function necessary for UGA codon reassignment required impairing UAA and UAG recognition, stimulating shifty stop frameshifting. We suspected that the Euplotes shifty stop frameshift sites would have little activity in yeast since we had shown that the last in-frame codon, AAA, is unable to stimulate frameshifting even under optimal conditions (Vimaladithan and Farabaugh 1994). We constructed reporter vectors including functional shifty stop frameshift sites in which the last codon decoded in frame is CUU, which efficiently stimulates frameshifting in yeast (Belcourt and Farabaugh 1990). RFs recognize a tetranucleotide consisting of the termination codon and its 3′ nearest neighbor (Brown et al. 1990a,b; Bonetti et al. 1995; Poole et al. 1995) and the identity of the 3′ neighbor influences shifty stop frameshifting (Tate and Mannering 1996). We tested constructs involving termination codons followed by U, C, A, or G to assess the influence of the fourth nucleotide.
Measuring frameshifting efficiency involves comparing the activity of β-galactosidase from a gene requiring frameshifting for its expression to that to of a gene not requiring frameshifting (Belcourt and Farabaugh 1990). Using matched reporter constructs, we eliminate all other transcriptional and translational effects on enzyme activity; the reporters also eliminate the effect of nonsense-mediated mRNA decay on frameshift efficiency measurements (Bidou et al. 2000; Stahl et al. 2000), which otherwise might reduce frameshift reporter expression relative to the in-frame control (Plant et al. 2004).
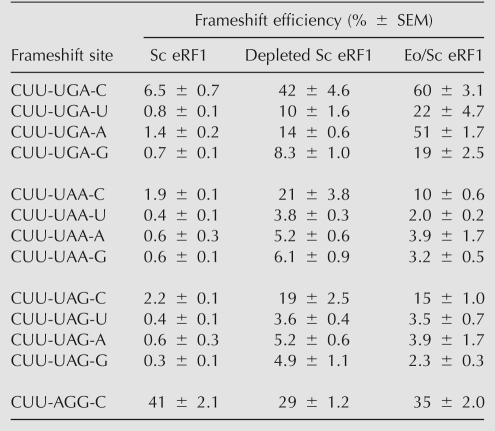
As expected, shifty stop frameshifting in the presence of Sc eRF1 varies with the identity of the stop codon and 3′ neighbor (Table 1, column 1). All constructs with 3′ C gave the highest frameshifting. Among constructs matched for the fourth nucleotide, UGA stimulated greater frameshifting than either UAG or UAA. These data are consistent with previous observations that Sc eRF1 recognizes UAA and UAG more efficiently than UGA (Bonetti et al. 1995; Pande et al. 1995). Sc eRF1 could be depleted to about 10% of normal before the growth rate significantly decreased (Salas-Marco et al. 2006). Depletion increased all shifty stop frameshifting about 10-fold regardless of termination codon or 3′ nucleotide (Table 1, column 2). This lack of codon-specific effect is consistent with a reduced rate of A site occupancy because the factor is limiting.
TABLE 1.
Effect of Eo/Sc eRF1 on shifty stop frameshifting
The effect of depleting Sc eRF1 in a strain constitutively expressing the chimeric Eo/Sc eRF1 was quantitatively different (Table 1, column 3). The level of frameshifting at UGA was significantly higher than in the strain depleted of Sc eRF1, while frameshifting at UAA and UAG was generally lower. This shows that the presence of the Eo/Sc eRF1 interfered with recognition of the UGA codon by the residual Sc eRF1 but that the Eo/Sc eRF1 accelerated the rate of UAA/UAG recognition. Compared with the depletion, UGA frameshifting increased an average of 2.4-fold, but UAA and UAG decreased 1.8- and 1.4-fold on average. As a control, we also tested the effect at a nonshifty stop frameshift site, CUU-AGG-C, which is the programmed +1 frameshift site of the Ty1 retrotransposon (Belcourt and Farabaugh 1990). This site does not involve a termination codon, and as expected, we observed little or no effect of changes in eRF1 availability (Table 1, last line). These data clearly indicate that the Eo/Sc eRF1 recognizes UGA much less efficiently than either UAA or UAG but also that it recognizes UAA and UAG much less efficiently than does Sc eRF1.
The Tetrahymena/Saccharomyces eRF1 chimera also promotes +1 shifty stop frameshifting at all three termination codons
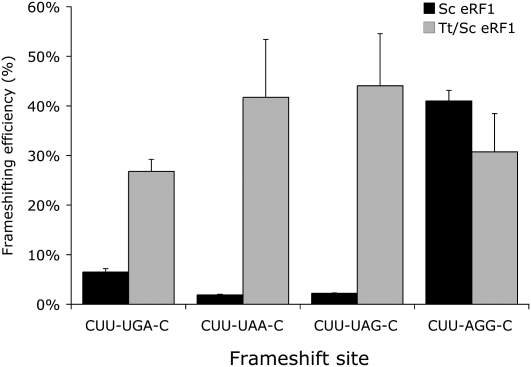
To extend our analysis, we tested whether Tt/Sc eRF1 stimulates frameshifting at shifty stop sites. This experiment was less complicated because a Tt/Sc eRF1 gene complements the lethality of sup45Δ yeast lacking Sc eRF1. Tt/Sc eRF1 suppresses read-through of all three translation codons when the cells are grown at 35°C, the normal growth temperature of T. thermophila. In contrast, at this temperature it allowed significant shifty stop frameshift at all three codons (Fig. 2). Moreover, in contrast to either Sc eRF1 or Eo/Sc eRF1, the Tt/Sc eRF1 stimulated frameshifting on UAA and UAG much more than on UGA both in terms of the absolute value and of the fold increase relative to Sc eRF1. Tt/Sc eRF1 had no significant effect on frameshifting at the nonshifty stop CUU-AGG-C site. These data suggest that Tt/Sc eRF1 recognizes all three termination codons significantly less efficiently than does Sc eRF1. This difference is most pronounced for UAA/UAG.
FIGURE 2.
Tt/Sc eRF1 stimulates programmed +1 frameshifting at all termination codons. The efficiency of programmed frameshifting on three shifty stop frameshift sites and one nonshifty stop frameshift site, as indicated, was measured in the presence of S. cerevisiae eRF1 (Sc eRF1) or the T. thermophila/S. cerevisiae chimeric enzyme (Tt/Sc eRF1). Error bars, SEM.
Changes to the phylogenetically conserved TASNIKS motif in E. octocarinatus eRF1 cause increased frameshifting at UAA and UAG codons
Fan-Minogue et al. (2008) identified point mutations in Eo/Sc eRF1 that allow it to support growth of a sup45Δ S. cerevisiae strain and that partially restore UGA recognition. The mutations target the conserved sequence motifs TAESIKS (TASNIKS in canonical code proteins) and YxCxxxF. The C124S mutation, which alters the conserved cysteine in the YxCxxxF motif, reduces UGA read-through 76-fold, suggesting that it increases the efficiency of UGA recognition (Fan-Minogue et al. 2008). A double mutant of the TAESIKS motif, E57S/S58N, which creates the canonical TASNIKS motif, reduces UGA read-through 15-fold. Each mutant appeared to retain efficient termination at UAA and UAG. In vitro peptide release assays showed that Eo/Sc eRF1 recognized UGA significantly more slowly than UAA/UAG, that the two motif mutations accelerated UGA recognition, but that neither mutant altered the rate of UAA/UAG recognition, consistent with in vivo read-through results (Fan-Minogue et al. 2008).
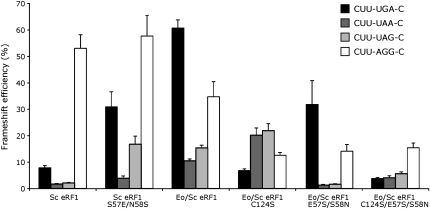
The programmed frameshift assays reported here suggest that Eo/Sc eRF1 recognizes UAA/UAG significantly less efficiently than Sc eRF1, whereas previous read-through results suggested that the factors were approximately equally efficient (Fan-Minogue et al. 2008). To understand the source of this difference, we determined if the mutations targeting the TASNIKS and YxCxxxF motifs of Eo/Sc eRF1 affect shifty stop frameshifting. As shown in Figure 3, the E57S/S58N double mutation reduced UGA frameshifting about twofold, but it decreased frameshifting at UAA/UAG to the level seen with Sc eRF1. In contrast, the C124S mutation reduced frameshifting on UGA to the Sc eRF1 level, but it had the opposite effect on frameshifting at UAA/UAG, causing a significant increase to a level far above that seen with Sc eRF1. In the triple mutant, C124S/E57S/S58N, UGA frameshifting was slightly below and UAA/UAG frameshifting slightly above Sc eRF1 levels. These data suggest that the TASNIKS to TAESIKS mutation of E. octocarinatus eRF1 decreased recognition of all three termination codons, not just UGA as implied by the termination read-through assays. Altering the phylogenetically conserved C124 appears to have increased UGA recognition at the expense of UAA/UAG recognition, which also fails to confirm the results of the read-through assays.
FIGURE 3.
Effect of mutations of phylogenetically conserved motifs of Eo/Sc eRF1 on shifty stop frameshifting. The efficiency of programmed +1 frameshifting on the same programmed frameshift sites represented in Figure 2 (identified in the legend) was measured in the presence of six variant forms of eRF1 as shown. Error bars, SEM.
These mutational data suggest that the TASNIKS motif plays a major role in UAA/UAG recognition but that, in the context of the Eo/Sc eRF1, it has little effect on UGA recognition. We tested whether loss of the motif in the context of the Sc eRF1 would have a reciprocal effect, reducing UAG/UAA recognition more than UGA. As shown in Figure 3, a mutation introducing the E. octocarinatus sequence TAESIKS (Sc eRF1 S57E/N58S) did not have this effect. Frameshifting increased on all three shifty stop sites though the magnitude of the effect varied: 2.3-fold at UAA, fourfold on UGA, and eightfold on UAG. The change from TASNIKS to TAESIKS, isolated from other changes introduced into the E. octocarinatus enzyme, appears to reduce the efficiency of stop codon recognition in general. Given that the fold reduction in UAG recognition was greater than for UGA, this change cannot by itself explain the change in codon specificity that occurred during genetic code redefinition.
We observed that the motif mutations of the Eo/Sc hybrid eRF1 caused a reproducible decrease in frameshifting at the control nonshifty stop frameshift site, CUU-AGG-C (Fig. 3). Each mutation caused more than a twofold decrease in frameshifting. Given that this frameshift site does not involve a nonsense codon, the effect is unexpected. However, we have shown previously that frameshifting at this site is very sensitive to cell physiology, decreasing drastically as cells enter post-diauxic phase (Stahl et al. 2004). At this stage, cellular growth rate decreases strongly; we argued that the decrease in this case reflects the fact that slowly growing cells experience less demand for the rare tRNAArgCCU because of reduced rates of protein synthesis. The Eo/Sc eRF1 strains grow much more slowly than strains expressing Sc eRF1 or Tt/Sc eRF1, and the level of total protein synthesis is also reduced (data not shown). We suspect that the reduction in frameshifting at CUU-AGG-C, therefore, does not directly relate to Eo/Sc eRF1 activity. We note that whereas frameshifting at the three shifty stop sites varies strongly among the three mutants, CUU-AGG-C frameshifting is very consistent. This result is inconsistent with any alternative model in which mutant Eo/Sc eRF1 incorrectly recognizes the AGG sense codon.
DISCUSSION
Termination codon reassignment in two ciliates results in increased frameshifting
Our data clearly suggest that in Euplotes, a genus in which UGA has been reassigned to a sense codon, eRF1 inefficiently recognizes all three canonical termination codons. The decreased efficiency results independently from mutations in two conserved regions of eRF1 domain 1. Inefficient termination also allows frequent frameshifting at shifty stop codons. The mechanism of genetic code reassignment in T. thermophila involved creation of UAA and UAG suppressor tRNAGln isoacceptors (Schüll and Beier 1994). Nonsense suppressors can decode inefficiently because of competition for the termination codon by RF (for example, see Martin et al. 1988). The codon capture model thus predicts that reassignment requires altering eRF1 codon specificity. Salas-Marco et al. (2006) showed that Tetrahymena reassignment did not result in a change in codon specificity by eRF1 since it recognized all three terminators equally well. The programmed frameshifting data presented confirm that the factor has less than a twofold preference for UGA codons, which is probably insufficient in itself to explain reassignment. The read-through data also imply that Tt/Sc chimeric eRF1 is about as efficient as the endogenous Sc eRF1; however the programmed frameshifting results presented here suggest that it is much less efficient. These data suggest that the sensitivity of the read-through assay is too low to reveal this decrease in relative activity of the Tt/Sc eRF1. The factor appears to have undergone a codon-nonspecific reduction in eRF1 activity, which, along with the elaboration of abundant tRNA suppressors, could explain how UAA/UAG reassignment was accomplished.
In the case of reassignment in Euplotes spp., the results of Salas-Marco et al. (2006) suggested that reassignment resulted from a change in codon specificity. Eo/Sc eRF1 recognized UGA codons much less efficiently than it did UAA or UAG. The data presented here confirm that conclusion; frameshifting was much more efficient on UGA than on UAA or UAG. However, Eo/Sc eRF1 clearly allows much more frameshifting on UAA or UAG than does the endogenous yeast factor. This is consistent with the prediction that altering the factor to eliminate recognition of one termination codon might require impairing recognition of the other two codons (Klobutcher and Farabaugh 2002). A recent genome survey of E. crassus, a species that also decodes UGA as cysteine, identified a gene encoding a cysteine-inserting UGA-specific tRNA suppressor (V.N. Gladyshev, pers. comm.). Apparently, Euplotes UGA codon reassignment required both a tRNA suppressor and an eRF1 with altered codon specificity. It remains to be seen if the two styles of termination codon reassignment, UAA/UAG and UGA, necessarily use these distinct mechanisms in other species.
Comparison of many eRF1 sequences identified two highly conserved sequence motifs in domain 1 of canonical code proteins (TASNIKS and YxCxxxF) that show unusually high sequence divergence in variant code proteins, suggesting that they might be responsible for altering codon specificity (Knight and Landweber 2000; Song et al. 2000; Lozupone et al. 2001; Kim et al. 2005). Mutagenesis and protein cross-linking studies independently implicated these two motifs in codon recognition (Chavatte et al. 2002; Inagaki et al. 2002; Seit-Nebi et al. 2002; Kolosov et al. 2005). Fan-Minogue et al. (2008) tested the role of these two motifs in redefinition of UGA in E. octocarinatus. The Euplotes eRF1 substitutes TASNIKS with TAESIKS; restoring the canonical TASNIKS sequence by creating the double mutation E57S/S58N greatly increased UGA recognition without significantly decreasing recognition of UAA/UAG. They also showed that a mutation to the conserved cysteine in the YxCxxxF motif (C124S) had a similar effect, although the efficiency of UGA recognition by the C124S mutant was greater than by the E57S/S58N mutant. Using an in vitro translation system, they demonstrated that the Eo/Sc chimeric eRF1 recognizes UGA much more slowly than it does UAA or UAG and that mutating the motifs eliminate this distinction. The E57S/S58N mutant also eliminated the stimulatory effect of eRF3 on recognition of UAA or UAG but not on UGA. They proposed a conformational model for eRF1 function in which eRF3•GDP induces a conformational change in eRF1, leading to peptide release; GTP hydrolysis by eRF3 stimulates peptide release in vitro (Alkalaeva et al. 2006). The model suggests that there are two such conformations, one for UAA/UAG and one for UGA; the Euplotes factor adopts the UAA/UAG conformation rapidly but the UGA conformation much more slowly. The TAESIKS motif appears to be largely responsible for the slow acquisition of the UGA conformation.
Frameshifting is a common feature of gene expression in Euplotes spp.; in excess of 10% of Euplotes genes incorporate a frameshift site (for review, see Klobutcher and Farabaugh 2002). Most of these sites are AAA-UAA-A, but there are a few variant AAA-UAG-A sites. The presence of the nonstandard TAESIKS motif in Eo eRF1 partly explains the increased frameshifting at UAA/UAG. The change from the canonical TASNIKS to this variant was necessary to minimize recognition of UGA (Fan-Minogue et al. 2008), but according to our data, it also resulted in a large increase in programmed +1 frameshifting at UAA/UAG. The decreased efficiency of UAA/UAG recognition was no doubt a precondition for evolving the many AAA-UAR-A frameshift sites found in Euplotes spp. It may have been inevitable that reassigning UGA as a sense codon would do so, or it may have been fortuitous and other modifications of eRF1 might have resulted in higher activity with UAA/UAG. It will be necessary to characterize more UGA reassignments to be certain which is true.
Poor recognition of the terminators is necessary but not sufficient to evolve efficient frameshifting. Two facts imply that one or more other factors must have been involved. First, Tetrahymena eRF1 appears to recognize UAA/UAG less efficiently than does the Euplotes factor, but there is no evidence of its using shifty stop frameshifting. Second, the frameshift sites in Euplotes are surprisingly unvarying; at every site identified the shift codon, immediately upstream of the pause-inducing stop codon, is AAA encoding lysine. The well-characterized +1 frameshift sites of S. cerevisiae uniformly employ a shift codon for which the cognate tRNA is either in very low abundance or is entirely absent (Sundararajan et al. 1999). The lack of the cognate tRNA for this codon allows a near-cognate tRNA to decode it, creating a usually illegal wobble interaction, for example, a U•U pair. The T. thermophila genome sequence reveals a full complement of 47 standard tRNAs able to decode all 63 sense codons, including the tRNAGln specific for UAA/UAG (Eisen et al. 2006). Ten of these tRNAs are encoded by one or two structural genes, suggesting they may be low abundance; eight of these decode G-ending codons. This resembles the situation in S. cerevisiae, which has 12 tRNAs encoded by one or two genes, eight decoding G-ending codons. Only three S. cerevisiae genes employ programmed frameshifting, ABP140 (Asakura et al. 1998), EST3 (Morris and Lundblad 1997), and OAZ1 (Palanimurugan et al. 2004). By analogy, T. thermophila might encode a small number of unrecognized frameshift products employing one of the poorly decoded codons and a poorly recognized UGA stop codon. Since the OAZ1 frameshift went unrecognized for almost a decade after the completion of the yeast genome, despite its using a well-characterized frameshift mechanism, the idea that genes have been missed in Tetrahymena is not unexpected.
Frequent use of frameshifting in Euplotes spp. implies high efficiency because of the deleterious effects of inefficient frameshifts, which would reduce gene output when present. To the extent that efficient frameshifts do not greatly reduce gene output, they should be neutral events, which would explain their frequent occurrence (Klobutcher and Farabaugh 2002). Given that Eo/Sc eRF1 recognizes UAA/UAG more efficiently than Tt/Sc eRF1, its reduced UAA/UAG efficiency in and of itself cannot explain the presumed high efficiency. As with other programmed events (for review, see Farabaugh 1996), Euplotes +1 programmed frameshifting should depend on the coincidence of several factors. We can imagine several possible explanations. The Euplotes programmed frameshifts involve a tRNALys decoding the codon immediately upstream of the poorly recognized UAA or UAG terminator. Either the tRNA or the lysine amino acid itself could stimulate frameshifting. There are precedents for unusual tRNAs stimulating frameshifting. In Escherichia coli an unconventional anticodon loop structure of tRNALys stimulates −1 frameshifting (Agris et al. 1997). The E. crassus genome survey, however, identified 23 copies of a gene for the AAA decoding tRNALysUUU and 17 copies of the AAG decoding tRNALysCUU with no evidence of any unusual structure (V.N. Gladyshev, pers. comm.). Hypomodification of tRNAs can stimulate frameshifting (Hagervall et al. 1993; Brierley et al. 1997; Carlson et al. 1999; Urbonavicius et al. 2001, 2003; Leipuviene and Bjork 2005; Waas et al. 2007), so in Euplotes, frameshifting could require an undermodified tRNA. The C-terminal lysine on the nascent peptide chain could also stimulate frameshifting since the amino acid at this position may decrease eRF1 efficiency by reducing the rate of peptide release (Brown et al. 1990b; Arkov et al. 1993; Mottagui-Tabar and Isaksson 1997). A further possibility is that the sequence AAA in the mRNA influences recognition by eRF1 directly as a context effect although how that would function is not clear.
MATERIALS AND METHODS
Strains and hybrid eRF1 constructs
The S. cerevisiae strain carrying a deletion of the gene encoding eRF1 (SUP45), YDB447, MATα ura3-52 leu2-3,112 ade1-14 lys2 trp1 his3 sup45Δ∷HIS3 [psi–], was used in all experiments. The Euplotes and Tetrahymena hybrid eRF1 gene constructs have been described (Salas-Marco et al. 2006).
S. cerevisiae eRF1 depletion experiments were done essentially as described (Salas-Marco et al. 2006). The cells containing the eRF1 plasmids and the frameshift reporter plasmids were grown in medium containing galactose as carbon source for several generations. The cells at mid-log stage were washed and resuspended in glucose medium. The cells were harvested for β-galactosidase assays at mid-exponential stage (OD600 ≈ 1).
Frameshift constructs and frameshift assays
In order to test the effect of the Euplotes eRF1 on frameshifting, we transformed the above strains with frameshift reporter plasmids that are derivatives of pMB38 (Belcourt and Farabaugh 1990), a shuttle vector with a URA3 marker. The plasmid contains the first 33 codons of the HIS4 gene fused to a lacZ gene via an intervening programmed frameshift site derived from the Ty1 retrotransposon CUU-AGG-C. To assess the effect of eRF1 on frameshifting, we replaced the AGG pause codon of this site with each of the three nonsense codons using the QuikChange II XL site directed mutagenesis kit (Stratagene). Because the nucleotide 3′ of the termination codon influences recognition by eRF1, we constructed sites using a 3′ A, G, or T using QuikChange. In order to measure the frameshifting efficiency, we compared expression from the frameshift reporter genes with expression from a frame fusion control plasmid in which the HIS4 gene and lacZ gene are in the same translation frame; the frame fusion plasmid lacks the second U in the CUU codon. The ratio of the expression of the β-galactosidase from frameshift control to the frame fusion control gives the frameshifting efficiency. Triplicate β-galactosidase assays of at least three independent transformants were performed as described (Farabaugh et al. 1989)
ACKNOWLEDGMENTS
This work was supported by grants from the National Institute of General Medical Science to P.J.F. (GM029480) and to D.M.B. (GM068854).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1508109.
REFERENCES
- Agris P.F., Guenther R., Ingram P.C., Basti M.M., Stuart J.W., Sochacka E., Malkiewicz A. Unconventional structure of tRNALysSUU anticodon explains tRNA's role in bacterial and mammalian ribosomal frameshifting and primer selection by HIV-1. RNA. 1997;3:420–428. [PMC free article] [PubMed] [Google Scholar]
- Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Arkov A.L., Korolev S.V., Kisselev L.L. Termination of translation in bacteria may be modulated via specific interaction between peptide chain release factor 2 and the last peptidyl-tRNASer/Phe. Nucleic Acids Res. 1993;21:2891–2897. doi: 10.1093/nar/21.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura T., Sasaki T., Nagano F., Satoh A., Obaishi H., Nishioka H., Imamura H., Hotta K., Tanaka K., Nakanishi H., et al. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene. 1998;16:121–130. doi: 10.1038/sj.onc.1201487. [DOI] [PubMed] [Google Scholar]
- Belcourt M.F., Farabaugh P.J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram G., Bell H.A., Ritchie D.W., Fullerton G., Stansfield I. Terminating eukaryote translation: Domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidou L., Stahl G., Hatin I., Namy O., Rousset J.P., Farabaugh P.J. Nonsense-mediated decay mutants do not affect programmed −1 frameshifting. RNA. 2000;6:952–961. doi: 10.1017/s1355838200000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti B., Fu L., Moon J., Bedwell D.M. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995;251:334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- Brierley I., Meredith M.R., Bloys A.J., Hagervall T.G. Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: Influence of tRNA anticodon modification on frameshifting. J. Mol. Biol. 1997;270:360–373. doi: 10.1006/jmbi.1997.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.M., Stockwell P.A., Trotman C.N.A., Tate W.P. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990a;18:6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.M., Stockwell P.A., Trotman C.N.A., Tate W.P. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990b;18:2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson B.A., Kwon S.Y., Chamorro M., Oroszlan S., Hatfield D.L., Lee B.J. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology. 1999;255:2–8. doi: 10.1006/viro.1998.9569. [DOI] [PubMed] [Google Scholar]
- Chavatte L., Seit-Nebi A., Dubovaya V., Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F.H. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Ebihara K., Nakamura Y. C-terminal interaction of translational release factors eRF1 and eRF3 of fission yeast: G-domain uncoupled binding and the role of conserved amino acids. RNA. 1999;5:739–750. doi: 10.1017/s135583829998216x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A., Coyne R.S., Wu M., Wu D., Thiagarajan M., Wortman J.R., Badger J.H., Ren Q., Amedeo P., Jones K.M., et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eurwilaichitr L., Graves F.M., Stansfield I., Tuite M.F. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol. 1999;32:485–496. doi: 10.1046/j.1365-2958.1999.01346.x. [DOI] [PubMed] [Google Scholar]
- Fan-Minogue H., Du M., Pisarev A.V., Kallmeyer A.K., Salas-Marco J., Keeling K.M., Thompson S.R., Pestova T.V., Bedwell D.M. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol. Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J. Programmed translational frameshifting. Microbiol. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P., Liao X.-B., Belcourt M., Zhao H., Kapakos J., Clare J. Enhancer and silencerlike sites within the transcribed portion of a Ty2 transposable element of Saccharomyces cerevisiae. Mol. Cell. Biol. 1989;9:4824–4834. doi: 10.1128/mcb.9.11.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L.Y., Simonsen J.L., Merkulova T.I., Litvinov D.Y., Martensen P.M., Rechinsky V.O., Camonis J.H., Kisselev L.L., Justesen J. Functional expression of eukaryotic polypeptide chain release factors 1 and 3 by means of baculovirus/insect cells and complex formation between the factors. Eur. J. Biochem. 1998;256:36–44. doi: 10.1046/j.1432-1327.1998.2560036.x. [DOI] [PubMed] [Google Scholar]
- Frolova L.Y., Tsivkovskii R.Y., Sivolobova G.F., Oparina N.Y., Serpinsky O.I., Blinov V.M., Tatkov S.I., Kisselev L.L. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L.Y., Merkulova T.I., Kisselev L.L. Translation termination in eukaryotes: Polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA. 2000;6:381–390. doi: 10.1017/s135583820099143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J., Milman G., Scolnick E., Caskey T. Peptide chain termination. VI. Purification and site of action of S. Proc. Natl. Acad. Sci. 1970;65:430–437. doi: 10.1073/pnas.65.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagervall T., Tuohy T., Atkins J., Björk G. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J. Mol. Biol. 1993;232:756–765. doi: 10.1006/jmbi.1993.1429. [DOI] [PubMed] [Google Scholar]
- Hanyu N., Kuchino Y., Nishimura S., Beier H. Dramatic events in ciliate evolution: Alteration of UAA and UAG termination codons to glutamine codons due to anticodon mutations in two Tetrahymena tRNAs. EMBO J. 1986;5:1307–1311. doi: 10.1002/j.1460-2075.1986.tb04360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V., Champ S., Mora L., Merkoulova-Rainon T., Kisselev L.L., Buckingham R.H. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 2005;280:2439–2445. doi: 10.1074/jbc.M407252200. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Gorovsky M.A. An unusual genetic code in nuclear genes of Tetrahymena. Proc. Natl. Acad. Sci. 1985;82:2452–2455. doi: 10.1073/pnas.82.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y., Blouin C., Doolittle W.F., Roger A.J. Convergence and constraint in eukaryotic release factor 1 (eRF1) domain 1: The evolution of stop codon specificity. Nucleic Acids Res. 2002;30:532–544. doi: 10.1093/nar/30.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Ebihara K., Nakamura Y. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 1998;4:958–972. doi: 10.1017/s1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Frolova L., Seit-Nebi A., Karamyshev A., Kisselev L., Nakamura Y. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc. Natl. Acad. Sci. 2002;99:8494–8499. doi: 10.1073/pnas.142690099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S., Frolova L., Kisselev L., Jean-Jean O. Stop codon recognition in ciliates: Euplotes release factor does not respond to reassigned UGA codon. EMBO Rep. 2001;2:680–684. doi: 10.1093/embo-reports/kve156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O.T., Yura K., Go N., Harumoto T. Newly sequenced eRF1s from ciliates: The diversity of stop codon usage and the molecular surfaces that are important for stop codon interactions. Gene. 2005;346:277–286. doi: 10.1016/j.gene.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Klobutcher L.A. Sequencing of random Euplotes crassus macronuclear genes supports a high frequency of +1 translational frameshifting. Eukaryot. Cell. 2005;4:2098–2105. doi: 10.1128/EC.4.12.2098-2105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher L.A., Farabaugh P.J. Shifty ciliates. Frequent programmed translational frameshifting in euplotids. Cell. 2002;111:763–766. doi: 10.1016/s0092-8674(02)01138-8. [DOI] [PubMed] [Google Scholar]
- Knight R.D., Landweber L.F. The early evolution of the genetic code. Cell. 2000;101:569–572. doi: 10.1016/s0092-8674(00)80866-1. [DOI] [PubMed] [Google Scholar]
- Knight R.D., Freeland S.J., Landweber L.F. Rewiring the keyboard: Evolvability of the genetic code. Nat. Rev. Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- Kolosov P., Frolova L., Seit-Nebi A., Dubovaya V., Kononenko A., Oparina N., Justesen J., Efimov A., Kisselev L. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res. 2005;33:6418–6425. doi: 10.1093/nar/gki927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y., Hanyu N., Tashiro F., Nishimura S. Tetrahymena thermophila glutamine tRNA and its gene that corresponds to UAA termination codon. Proc. Natl. Acad. Sci. 1985;82:4758–4762. doi: 10.1073/pnas.82.14.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Leipuviene R., Bjork G.R. A reduced level of charged
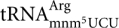 triggers the wild-type peptidyl-tRNA to frameshift. RNA. 2005;11:796–807. doi: 10.1261/rna.7256705. [DOI] [PMC free article] [PubMed] [Google Scholar]
triggers the wild-type peptidyl-tRNA to frameshift. RNA. 2005;11:796–807. doi: 10.1261/rna.7256705. [DOI] [PMC free article] [PubMed] [Google Scholar] - Lozupone C.A., Knight R.D., Landweber L.F. The molecular basis of nuclear genetic code change in ciliates. Curr. Biol. 2001;11:65–74. doi: 10.1016/s0960-9822(01)00028-8. [DOI] [PubMed] [Google Scholar]
- Martin R., Hearn M., Jenny P., Gallant J. Release factor competition is equivalent at strong and weakly suppressed nonsense codons. Mol. Gen. Genet. 1988;213:144–149. doi: 10.1007/BF00333411. [DOI] [PubMed] [Google Scholar]
- Merkulova T.I., Frolova L.Y., Lazar M., Camonis J., Kisselev L.L. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 1999;443:41–47. doi: 10.1016/s0014-5793(98)01669-x. [DOI] [PubMed] [Google Scholar]
- Meyer F., Schmidt H.J., Plumper E., Hasilik A., Mersmann G., Meyer H.E., Engstrom A., Heckmann K. UGA is translated as cysteine in pheromone 3 of Euplotes octocarinatus. Proc. Natl. Acad. Sci. 1991;88:3758–3761. doi: 10.1073/pnas.88.9.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D.K., Lundblad V. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S., Isaksson L.A. Only the last amino acids in the nascent peptide influence translation termination in Escherichia coli genes. FEBS Lett. 1997;414:165–170. doi: 10.1016/s0014-5793(97)00978-2. [DOI] [PubMed] [Google Scholar]
- Oba T., Andachi Y., Muto A., Osawa S. CGG: An unassigned or nonsense codon in Mycoplasma capricolum. Proc. Natl. Acad. Sci. 1991;88:921–925. doi: 10.1073/pnas.88.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S., Jukes T.H. Codon reassignment (codon capture) in evolution. J. Mol. Evol. 1989;28:271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T., Watanabe K., Muto A. Recent evidence for evolution of the genetic code. Microbiol. Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanimurugan R., Scheel H., Hofmann K., Dohmen R.J. Polyamines regulate their synthesis by inducing expression and blocking degradation of ODC antizyme. EMBO J. 2004;23:4857–4867. doi: 10.1038/sj.emboj.7600473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande S., Vimaladithan A., Zhao H., Farabaugh P.J. Pulling the ribosome out of frame +1 at a programmed frameshift site by cognate binding of aminoacyl-tRNA. Mol. Cell. Biol. 1995;15:298–304. doi: 10.1128/mcb.15.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant E.P., Wang P., Jacobs J.L., Dinman J.D. A programmed −1 ribosomal frameshift signal can function as a cis-acting mRNA destabilizing element. Nucleic Acids Res. 2004;32:784–790. doi: 10.1093/nar/gkh256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole E.S., Brown C.M., Tate W.P. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995;14:151–158. doi: 10.1002/j.1460-2075.1995.tb06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J., Bedwell D.M. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J., Fan-Minogue H., Kallmeyer A.K., Klobutcher L.A., Farabaugh P.J., Bedwell D.M. Distinct paths to stop codon reassignment by the variant-code organisms Tetrahymena and Euplotes. Mol. Cell. Biol. 2006;26:438–447. doi: 10.1128/MCB.26.2.438-447.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M.A., Cheesman C., Costa V., Moradas-Ferreira P., Tuite M.F. Selective advantages created by codon ambiguity allowed for the evolution of an alternative genetic code in Candida spp. Mol. Microbiol. 1999;31:937–947. doi: 10.1046/j.1365-2958.1999.01233.x. [DOI] [PubMed] [Google Scholar]
- Santos M.A., Moura G., Massey S.E., Tuite M.F. Driving change: The evolution of alternative genetic codes. Trends Genet. 2004;20:95–102. doi: 10.1016/j.tig.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Schüll C., Beier H. Three Tetrahymena tRNAGln isoacceptors as tools for studying unorthodox codon recognition and codon context effects during protein synthesis in vitro. Nucleic Acids Res. 1994;22:1974–1980. doi: 10.1093/nar/22.11.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D.W., Yarus M. Transfer RNA mutation and the malleability of the genetic code. J. Mol. Biol. 1994;235:1377–1380. doi: 10.1006/jmbi.1994.1094. [DOI] [PubMed] [Google Scholar]
- Scolnick E., Tompkins R., Caskey T., Nirenberg M. Release factors differing in specificity for terminator codons. Proc. Natl. Acad. Sci. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit-Nebi A., Frolova L., Kisselev L. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002;3:881–886. doi: 10.1093/embo-reports/kvf178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F., Hemmings B.A., Barford D. The crystal structure of human eukaryotic release factor eRF1—Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Stahl G., Bidou L., Hatin I., Namy O., Rousset J.P., Farabaugh P. The case against the involvement of the NMD proteins in programmed frameshifting. RNA. 2000;6:1687–1688. doi: 10.1017/s1355838200001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl G., Salem S.N., Chen L., Zhao B., Farabaugh P.J. Translational accuracy during exponential, postdiauxic, and stationary growth phases in Saccharomyces cerevisiae. Eukaryot. Cell. 2004;3:331–338. doi: 10.1128/EC.3.2.331-338.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield I., Jones K.M., Kushnirov V.V., Dagkesamanskaya A.R., Poznyakovski A.I., Paushkin S.V., Nierras C.R., Cox B.S., Ter-Avanesyan M.D., Tuite M.F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan A., Michaud W.A., Qian Q., Stahl G., Farabaugh P.J. Near-cognate peptidyl-tRNAs promote +1 programmed translational frameshifting in yeast. Mol. Cell. 1999;4:1005–1015. doi: 10.1016/s1097-2765(00)80229-4. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Ueda T., Watanabe K. The “polysemous” codon—A codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 1997;16:1122–1134. doi: 10.1093/emboj/16.5.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate W.P., Mannering S.A. Three, four or more: The translational stop signal at length. Mol. Microbiol. 1996;21:213–219. doi: 10.1046/j.1365-2958.1996.6391352.x. [DOI] [PubMed] [Google Scholar]
- Tourancheau A.B., Tsao N., Klobutcher L.A., Pearlman R.E., Adoutte A. Genetic code deviations in the ciliates: Evidence for multiple and independent events. EMBO J. 1995;14:3262–3267. doi: 10.1002/j.1460-2075.1995.tb07329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J., Qian Q., Durand J.M., Hagervall T.G., Bjork G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J., Stahl G., Durand J.M., Ben Salem S.N., Qian Q., Farabaugh P.J., Bjork G.R. Transfer RNA modifications that alter +1 frameshifting in general fail to affect −1 frameshifting. RNA. 2003;9:760–768. doi: 10.1261/rna.5210803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard B., Van L.B., Andersen G.R., Nyborg J., Buckingham R.H., Kjeldgaard M. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell. 2001;8:1375–1382. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- Vimaladithan A., Farabaugh P.J. Special peptidyl-tRNA molecules promote translational frameshifting without slippage. Mol. Cell. Biol. 1994;14:8107–8116. doi: 10.1128/mcb.14.12.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waas W.F., Druzina Z., Hanan M., Schimmel P. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J. Biol. Chem. 2007;282:26026–26034. doi: 10.1074/jbc.M703391200. [DOI] [PubMed] [Google Scholar]
- Weiss R.B., Dunn D.M., Atkins J.F., Gesteland R.F. Slippery runs, shifty stops, backward steps, and forward hops: –2, –1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]