Abstract
Mitochondrial RNAs in trypanosomes are edited by the insertion and deletion of uridine (U) nucleotides to form translatable mRNAs. Editing is catalyzed by three distinct editosomes that contain two related U-specific exonucleases (exoUases), KREX1 and KREX2, with the former present exclusively in KREN1 editosomes and the latter present in all editosomes. We show here that repression of KREX1 expression leads to a concomitant reduction of KREN1 in ∼20S editosomes, whereas KREX2 repression results in reductions of KREPA2 and KREL1 in ∼20S editosomes. Knockdown of KREX1 results in reduced cell viability, reduction of some edited RNA in vivo, and a significant reduction in deletion but not insertion endonuclease activity in vitro. In contrast, KREX2 knockdown does not affect cell growth or editing in vivo but results in modest reductions of both insertion and deletion endonuclease activities and a significant reduction of U removal in vitro. Simultaneous knockdown of both proteins leads to a more severe inhibition of cell growth and editing in vivo and an additive effect on endonuclease cleavage in vitro. Taken together, these results indicate that both KREX1 and KREX2 are important for retention of other proteins in editosomes, and suggest that the reduction in cell viability upon KREX1 knockdown is likely a consequence of KREN1 loss. Furthermore, although KREX2 appears dispensable for cell growth, the increased inhibition of editing and parasite viability upon knockdown of both KREX1 and KREX2 together suggests that both proteins have roles in editing.
Keywords: RNA editing, Trypanosoma brucei, KREX1, KREX2, endonuclease, exonuclease, phosphatase domain
INTRODUCTION
Trypanosomes are protozoan parasites that are the causative agents of African sleeping sickness and Chagas' disease in humans and nagana in cattle. RNA editing in these parasites is a unique and essential process that remodels most mitochondrial (mt) pre-mRNAs into functional mRNAs by the guided insertion and deletion of uridine (U) nucleotides. The amount of editing in each RNA varies from just a few sites to hundreds, in some cases specifying more than half of the mature mRNA sequences (Stuart et al. 1997). Editing at each site is achieved by a series of coordinated catalytic events in which the RNA is cleaved, Us are added or removed, and the processed RNA fragments are religated as directed by the sequence of trans-acting guide RNAs (gRNAs) (Madison-Antenucci et al. 2002; Simpson et al. 2003; Stuart et al. 2005).
RNA editing is catalyzed by multiprotein ∼20S editosomes that contain ∼20 proteins in aggregate (Rusche et al. 1997; Madison-Antenucci and Hajduk 2001; Panigrahi et al. 2001a,b, 2003; Aphasizhev et al. 2003a). The proteins that perform the catalytic steps have been identified: the KREN1 deletion site endonuclease, and the KREN2 and KREN3 insertion site endonucleases that cleave the RNA (Carnes et al. 2005, 2008; Trotter et al. 2005; Kang et al. 2006), the KRET2 TUTase that adds Us (Aphasizhev et al. 2003b; Ernst et al. 2003), the KREX1 and KREX2 U-specific exonucleases (exoUases) that are the focus of this study (Kang et al. 2005; Rogers et al. 2007), and finally the KREL1 and KREL2 ligases that rejoin the processed RNA fragments (McManus et al. 2001; Rusché et al. 2001; Schnaufer et al. 2001). A second TUTase, KRET1, is present in another complex, adds the gRNA (U) tails, and is also important for editing (Aphasizhev et al. 2002, 2003b). The editosome also contains two sets of noncatalytic proteins, several of which have been shown to be important for the structural integrity of the editosome and RNA binding (Drozdz et al. 2002; Huang et al. 2002; O'Hearn et al. 2003; Wang et al. 2003; Salavati et al. 2006; Babbarwal et al. 2007; Tarun et al. 2008).
Recent studies have shown that there are three distinct types of editosomes, each with a common set of core proteins but which have different endonucleases and one or two other specific proteins (Fig. 1; Panigrahi et al. 2006; Carnes et al. 2008; C. Zelaya Soares, unpubl.) KREN1 editosomes contain KREPB8 and the KREX1 exoUase, KREN2 editosomes contain KREPB7, and KREN3 editosomes contain KREPB6. The common set of core proteins for each of these editosomes consists of two trimeric subcomplexes, KRET2/KREPA1/KREL2 and KREX2/KREPA2/KREL1, that catalyze the U addition/ligation and U removal/ligation steps of editing, respectively (Schnaufer et al. 2003). The core also contains KREPA3, A4, and A6 and KREPB4 and B5, which are essential for editosome integrity (Wang et al. 2003; Salavati et al. 2006; Babbarwal et al. 2007; Guo et al. 2008; Tarun et al. 2008), and KREPA5, which has not been studied yet. Three of these proteins, KREPA3, A4, and A6, are RNA binding proteins (Brecht et al. 2005; Salavati et al. 2006; Tarun et al. 2008), which is also likely to be true for KREPA1, KREPA2, and KREPA5. Thus, KREX1 is exclusively in KREN1 deletion endonuclease editosomes, and KREX2 is present in the deletion subcomplex within the core of all editosomes.
FIGURE 1.
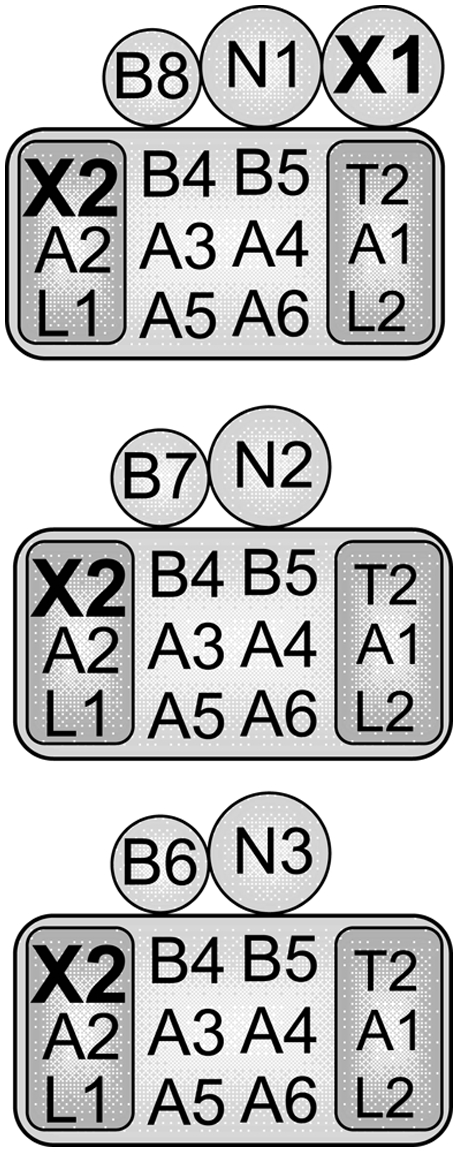
Composition of the three 20S editosomes. Each editosome contains a common set of core proteins (large rectangle) with a distinct endonuclease, KREN1, KREN2, or KREN3 (N1, N2, N3), and a specific KREPB protein (B6, B7, or B8). KREX1 (X1) is only in KREN1 editosomes. The core contains the KREPA1/KRET2/KREL2 (A1, T2, L2) insertion and the KREPA2/KREX2/KREL1 (A2, X2, L1) deletion subcomplexes (small rectangles). KREX2 is in the deletion subcomplex in all editosomes, but KREX1 is only in KREN1 editosomes.
KREX1 and KREX2 each have a C-terminal endonuclease/exonuclease/phosphatase (EEP) domain (Pfam 03372) that is present in a large family of proteins, including magnesium-dependent endonucleases such as AP endonucleases, DNase I, and phosphatases involved in cell-cycle regulation and cell signaling (Mol et al. 1995; Dlakic 2000; Worthey et al. 2003; Mian et al. 2006). The EEP domains in this diverse family of proteins have conserved catalytic amino acids and a similar four-layered α/β sandwich, indicating that these proteins share the same catalytic mechanism (Dlakic 2000). The KREX2 ortholog in Leishmania is a smaller protein, lacking the EEP domain and exonuclease activity (Rogers et al. 2007). RNAi knockdown studies have shown that KREX1, but not KREX2, is important for parasite growth (Kang et al. 2005; Rogers et al. 2007); however, these studies did not fully examine the consequences of knockdown of KREX1 or KREX2 on editing.
We show here that both KREX1 and KREX2 are exoUases and that knockdown of the latter did not affect cell growth but knockdown of the former or both did. Loss of KREX1 resulted in loss of KREN1, altered abundance of some edited and unedited RNAs in vivo, and loss of deletion endonuclease activity in vitro. This reflects the exclusive presence of KREX1 and KREN1 in one of the three types of editosomes. Loss of KREX2 did not alter the abundance of mt RNAs but did result in loss of KREPA2 and KREL1, consistent with the presence of these three proteins in the deletion subcomplex. Simultaneous knockdown of both KREX1 and KREX2 led to a more severe inhibition of deletion cleavage activity and cell growth. Thus, the slow growth phenotype observed upon knockdown of KREX1 or KREX1/X2 appears to be due to loss of deletion cleavage.
RESULTS
Trypanosoma brucei KREX1 and KREX2 are exoUases
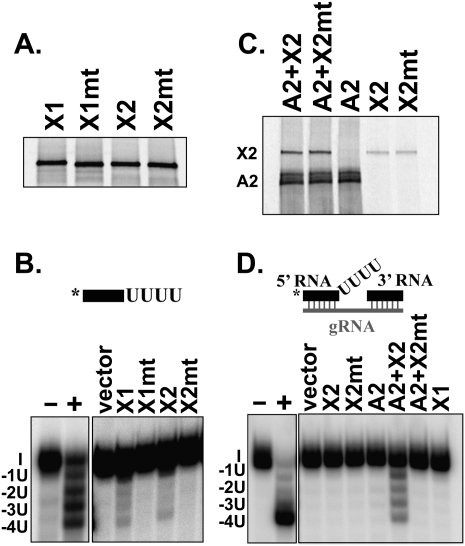
Recombinant Trypanosoma brucei KREX1 (TbKREX1) and T. brucei KREX2 (TbKREX2) proteins were shown to have U-specific exonuclease activity since mutation of conserved catalytic residues in the EEP domains of each protein abrogated this activity (Fig. 2). Labeled and unlabeled proteins were expressed by in vitro transcription/translation, immunoprecipitated with a monoclonal antibody (MAb) specific for the N-terminal Xpress tag on the proteins and analyzed by SDS-PAGE or activity assays, respectively. Two conserved amino acids shown to be important for catalysis and substrate binding in other EEP domain containing proteins (Mol et al. 1995; Dlakic 2000; Worthey et al. 2003) were mutated to generate KREX1 D789A/N791A and KREX2 D792A/N794A. Gel analysis of 35S-labeled proteins showed that the wild-type (wt) and mutant proteins were expressed to a similar level (Fig. 2A). The activity of the immunoprecipitated proteins was assayed using single-stranded (ss) U5-5′ RNA that ends in a stretch of four Us or U5-5′ RNA in combination with the 3′ fragment and gRNA that forms a double-stranded (ds) precleaved deletion editing substrate specifying the removal of four Us (Igo et al. 2002). Both wt KREX1 and KREX2 proteins removed the four Us from the ssRNA but did not remove the next G nucleotide (Fig. 2B). Substitution of an A for the second U resulted in the removal of only one U in the ssRNA (data not shown), indicating that both exonucleases are U specific, although removal of a terminal C was not tested here but was shown previously for KREX1 (Kang et al. 2005). The mutant KREX1 and KREX2 proteins were inactive and did not remove any nucleotides from the ssRNA, indicating that this domain is indeed catalytic.
FIGURE 2.
ExoUase activity of recombinant TbKREX1 and TbKREX2. Recombinant proteins were expressed by in vitro transcription/translation and immunoprecipitated with monoclonal antibodies (MAbs) specific for the N-terminal Xpress tag on the KREX proteins or for KREPA2. (A) Expression of wt and mt (D789A/N791A) KREX1 and wt and mt (D792A/N794A) KREX2 proteins. 35S-labeled proteins were immunoprecipitated with the Xpress MAb and separated on polyacrylamide gels. (B) Activity of wt and mt KREX proteins immunoprecipitated with the Xpress MAb using U5-5′ ssRNA ending in four Us. (C) Coimmunoprecipitation of 35S-labeled KREX2 proteins with 35S-labeled KREPA2 using KREPA2 MAb. (D) Activity of individual KREX proteins or coprecipitates of KREX2 and KREPA2 with a dsRNA precleaved deletion substrate specifying the removal of four Us. Immunoprecipitations were performed with the Xpress MAb except KREPA2 alone was performed with the KREPA2 MAb. Diagrams of the RNA substrates are shown and the asterisk indicates the radiolabel. I (input), -1U, -2U, -3U, and -4U products are indicated. Positive control reactions (+) were performed using biochemically purified editosomes, which was omitted in negative control reactions (−). KREX1 and KREX2 have exoUase activity that is abrogated by mutations in the EEP domain.
Both wt and mutant KREX2 proteins were coimmunoprecipitated with KREPA2 using a MAb specific for KREPA2 (Fig. 2C). This confirms previous studies that showed their interaction in vivo and in vitro (Schnaufer et al. 2003; Kang et al. 2004), and shows that the mutations did not affect their interaction. A small amount of the KREX2 proteins was nonspecifically immunoprecipitated, i.e., in the absence of KREPA2. No interaction between KREX1 and KREPA2 was detected in parallel studies (data not shown). Individual coprecipitated KREPA2 and KREX2 proteins were assayed for U removal using a trimeric “precleaved” deletion RNA substrate that mimics the RNA after endonucleolytic cleavage and assesses the U removal and ligation steps of editing (Igo et al. 2002). The KREPA2/KREX2 coprecipitate efficiently removed Us from the precleaved substrate, although neither KREX1 nor KREX2 alone did so (Fig. 2D). KREX1 and KREX2 alone, when produced in insect cells and used in much larger amounts, can remove Us from dsRNA substrates (Kang et al. 2005; Rogers et al. 2007). Taken together, these results indicate that KREX1 and KREX2 are exoUases and that U removal by KREX2 from an editing-like substrate is enhanced by its binding partner KREPA2. The stimulation of KREX2 U removal activity by KREPA2 was also observed with ssRNA substrates (data not shown). This enhancement may reflect enhanced binding of the dsRNA substrate or an effect on catalysis.
RNAi knockdown of KREX1 but not KREX2 inhibits cell growth
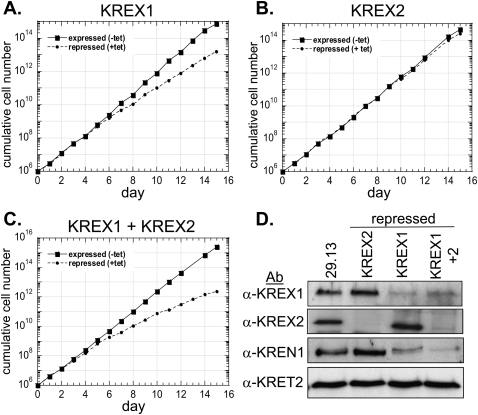
Repression of KREX1 expression by RNAi in procyclic form (PF) T. brucei resulted in a reduced growth rate, whereas repression of KREX2 had no effect, and simultaneous repression of both proteins had a greater effect on cell growth than KREX1 repression alone (Fig. 3). Three vectors were constructed that had either a ∼500-bp 5′ region of KREX1 or KREX2 or both of these regions between opposable tetracycline (tet)-regulated T7 promoters. PF 29.13 cells expressing T7 RNA polymerase and the tet repressor were transfected with the constructs, and dsRNA was induced by addition of tet. Growth of uninduced and induced cells was monitored over 15 d. Reduced cell growth was observed in KREX1 RNAi cells after 6 d of induction and continued throughout the time course and resulted in a 1.4-fold increase in the generation time compared with the uninduced cells (Fig. 3A), whereas growth of the induced KREX2 RNAi cells remained the same as the uninduced cells (Fig. 3B). Induction of dsRNA targeted to both KREX1 and KREX2 in the KREX1/KREX2 RNAi cell line resulted in a greater growth inhibition than observed with KREX1 repression alone, starting 6 d after induction and resulting in a twofold increase in the generation time compared with the uninduced cells (Fig. 3C).
FIGURE 3.
Effect of RNAi repression of KREX1 and KREX2 on cell growth. Growth of KREX1 RNAi (A), KREX2 RNAi (B), or KREX1/KREX2 RNAi (C) cell lines when the protein was expressed (squares, line) or repressed (circles, dashed line) in the absence or presence of RNAi induction, respectively. Growth was monitored over 15 d and the cumulative cell number calculated by multiplication of the cell densities by the dilution factor. (D) Western analysis of pooled ∼20S fractions (fractions 9–12) of glycerol gradient purified mitochondrial lysates from the parental 29.13 cell line and the RNAi cell lines repressed for 7 d (see Fig. 5) with polyclonal antibodies to KREX1, KREX2, KREN1, and KRET2. Cell growth was modestly or substantially reduced upon KREX1 or KREX1/KREX2 repression, respectively, whereas cell growth was unaffected by KREX2 loss.
Knockdown of KREX1 and KREX2 expression was confirmed by Western analysis (Fig. 3D) and quantitative real-time PCR (qPCR) (Fig. 4). Pooled ∼20S fractions of glycerol gradient purified mt lysates from the 29.13 parental cells and the repressed cells were subjected to Western analysis using polyclonal antibodies (Pabs) to KREX1, KREX2, KREN1, and KRET2 (Fig. 3D). A significant reduction of KREX1 was observed in the KREX1 and KREX1/KREX2 repressed cells, although a small amount remained, indicating that the RNAi repression of KREX1 expression was substantial although incomplete. KREX2 expression was essentially undetectable in the KREX2 repressed cells and dramatically reduced in the KREX1/KREX2 repressed cells. No change was seen in the level of KREX1 in the KREX2 repressed cells or vice versa, indicating that the dsRNA was specific for the targeted gene. No KREX1 or KREX2 protein was detected outside of the 20S editosome upon their repression (data not shown). Additionally, no change was observed in the levels of the editing TUTase, KRET2 when KREX1, KREX2, or both were repressed. However, the deletion endonuclease, KREN1 was significantly reduced in the KREX1 repressed cells and even more so in the KREX1/KREX2 repressed cells but not in the KREX2 repressed cells. No KREN1 protein was detected outside of the 20S region of glycerol gradients upon KREX1 repression, indicating that no stable KREN1 subcomplexes were formed or that KREN1 is not stable outside of the 20S editosome (data not shown). KREX1 is present exclusively in KREN1 editosomes, and these results indicate that the presence of KREN1 depends on KREX1 and that these two proteins may interact directly. Based on these results, KREX1, but not KREX2, appears to be essential for growth of PFs. Nevertheless, repression of both exoUases results in a greater inhibition of cell growth than either alone suggesting that both proteins have roles in editing in PFs.
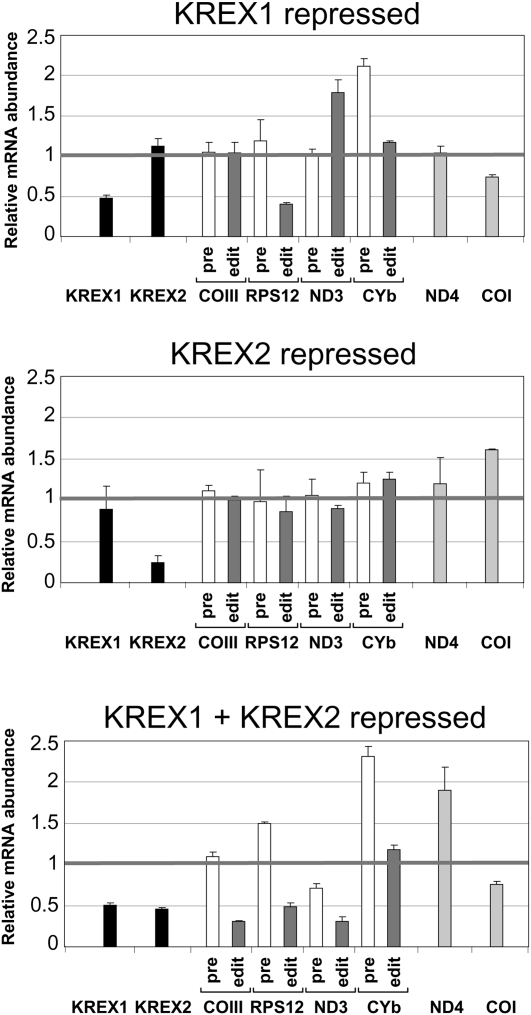
FIGURE 4.
Effect of KREX1 and KREX2 repression on editing in vivo. Quantitative real-time PCR analysis of total RNA isolated from the RNAi cell lines on day 7. The relative mRNA abundance is the ratio of RNA from the induced cells compared with the uninduced cells using 18S rRNA as an internal control. Analysis was performed in triplicate. The black line at 1 indicates no change in the mRNA levels. The black bars denote the KREX mRNAs, the white bars denote pre-edited mRNA, the dark gray bars denote edited mRNA, and the light gray bars denote never edited RNAs. Editing in vivo is not affected by KREX2 repression but is partially reduced upon KREX1 repression and significantly reduced upon repression of both KREX1 and KREX2. Error bars, SD from three replicates.
Simultaneous repression of KREX1 and KREX2 inhibits editing in vivo
Repression of both KREX1 and KREX2 expression resulted in reduction of edited RNAs in vivo, while KREX1 repression alone had only a partial effect and KREX2 repression alone had none (Fig. 4), consistent with the observed growth defects. The relative levels of the KREX1 and KREX2 mRNAs and several pre-edited, edited, and never edited mRNAs were determined using qPCR after 7 d of repression of the KREX proteins. Upon repression, the KREX1 and KREX2 mRNA levels were respectively reduced to ∼50% and ∼35% compared with nonrepressed cells, and both were reduced to ∼50% in the double knockdown. Repression of one did not affect the levels of the other mRNA. The reduction of KREX1, but not KREX2, mRNA mirrors the reduction observed at the protein level, and the greater effect at the protein level may reflect an effect on translation or protein stability (see Fig. 3). Edited RPS12 and pre-edited CYb mRNAs were primarily affected in KREX1 RNAi cells, and edited COIII and ND3 mRNAs were also affected in the double KREX1 and KREX2 RNAi cells. The never edited ND4 and COI mRNAs were variably affected in the different RNAi knockdowns. The reductions in editing in vivo also mirror the reductions in the growth rates of the cells. The reason for the preferential effects on edited and pre-edited RPS12 mRNA is unclear. The effect on pre-edited CYb mRNA with essentially no change in edited CYb mRNA upon knockdown of KREX1 may reflect the fact that this RNA is edited only by insertion and that KREX1 is only associated with editosomes that contain the KREN1 deletion endonuclease. The greater effect on editing in vivo with the double knockdown than that of KREX1 alone indicates that both KREX1 and KREX2 have roles in editing in PF T. brucei.
KREX2 repression leads to partial disruption of 20S editosomes
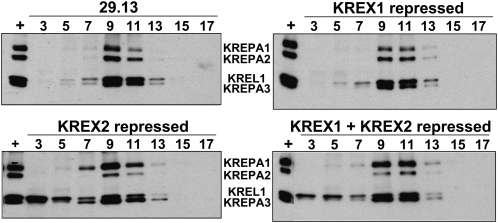
Repression of KREX1 did not affect the sedimentation of editosomes in glycerol gradients, whereas KREX2 repression resulted in partial disruption of the complexes and a shift of KREL1 higher in the gradient (Fig. 5). The overall integrity of ∼20S editosomes was assessed by glycerol gradient sedimentation of mt lysates of the 29.13 parental cells and KREX1 or KREX2 RNAi cells repressed for 7 d. The glycerol gradient fractions were analyzed by Western blot using a cocktail of MAbs for the KREPA1, KREPA2, KREL1, and KREPA3 editosome proteins (Panigrahi et al. 2001b). No change in sedimentation of editosomes was observed upon KREX1 repression compared with the parental 29.13 cells except for a slight decrease in KREPA1, with the peak of the proteins in fractions 9–11. Similarly, the peak of the editosome proteins was in fractions 9–11 when KREX2 was repressed alone or in combination with KREX1. However, less KREPA2 protein was detected upon KREX2 repression compared with KREPA1, and there was a marked shift of some of the KREL1 protein to the top of the gradient. Similar results were observed with glycerol gradient fractionated whole cell lysates from uninduced and induced cells (data not shown). Taken together, these results indicate that KREX2 repression results in partial disruption of the editosome and destabilization of the deletion subcomplex that contains KREX2, KREPA2, and KREL1. In addition, the loss of KREN1 upon KREX1 repression did not result in a change in editosome sedimentation.
FIGURE 5.
Editosome sedimentation after KREX1 and KREX2 repression. Western analysis of fractions of glycerol gradient purified mitochondrial lysates from parental 29.13 cells and RNAi cells in which the KREX proteins were repressed for 7 d as indicated. Western analysis of fractions 3–17 (top to bottom) used a mixture of four MAbs specific for the editosome proteins indicated. A fraction that contains ∼20S editosomes was used as a positive control (+). Editosome sedimentation is unchanged by KREX1 repression but KREX2 repression results in a reduction of KREPA2 and KREL1 in 20S editosomes and a shift in KREL1 to higher fractions.
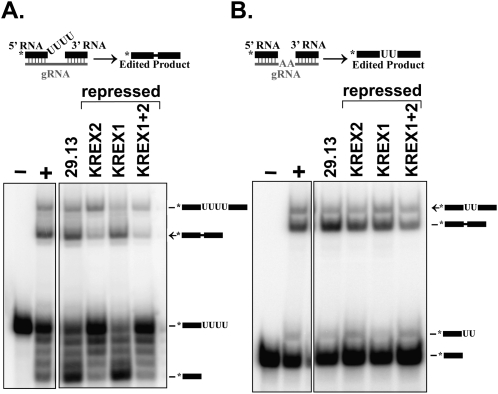
KREX2 repression results in a significant reduction of U removal in vitro
To evaluate the effects of KREX1 and KREX2 repression on editing activities in vitro, precleaved insertion and deletion assays were performed using pooled ∼20S glycerol gradient fractions from the parental and repressed cells shown in Figure 5. These assays use trimeric RNA substrates that mimic the pre-edited RNA after endonuclease cleavage and assess the U removal, U addition, and ligation activities of editing (Igo et al. 2002). Robust amounts of edited RNA and input RNA with four Us removed were detected in ∼20S fractions from the 29.13 parental cells. A dramatic reduction in edited RNA and input RNA with four Us removed was observed after repression of KREX2 alone or both KREX1 and KREX2 (Fig. 6A), along with an increase in products with zero, one, or two Us removed. No change in U removal or edited products was detected in ∼20S fractions from KREX1 repressed cells compared with the 29.13 cells. No significant change in U addition or ligation was observed with fractions from any of the repressed cells compared with the parental cells in precleaved insertion assays. Similar amounts of RNA with two Us added, ligated RNA with no added Us, and edited RNA were detected except for a slight increase of RNA with two added Us added in KREX2 and KREX1/KREX2 repressed cells, possibly as a consequence of less U removal in these fractions. The specific loss of U removal with KREX2 repression is consistent with its known activity; however, the lack of an effect on U removal with KREX1 repression is surprising.
FIGURE 6.
ExoUase activity is reduced after KREX2 repression. Precleaved deletion (A) and insertion (B) editing assays with ∼20S fractions from glycerol gradient purified mitochondrial lysates from parental 29.13 cells and RNAi cells in which KREX proteins were repressed for 7 d as indicated. RNA substrates and products are diagrammed, and the asterisk denotes the radiolabel. Positive control reactions (+) were performed using a ∼20S fraction containing peak editing activity, which was omitted in negative control reactions (−). U removal is strongly reduced upon KREX2 but not KREX1 repression.
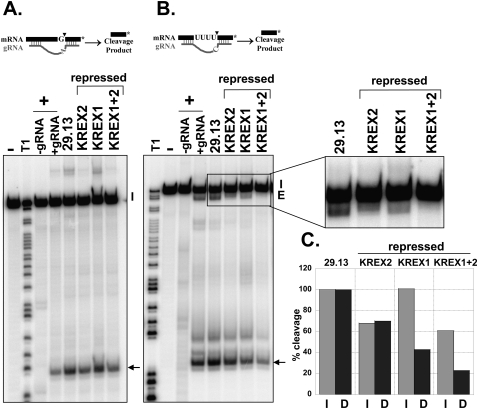
Deletion endonuclease activity is significantly reduced after KREX1 repression
Repression of KREX1 led to a reduction in deletion but not insertion endonuclease cleavage activity, whereas repression of KREX2 resulted in a smaller reduction of both activities and repression of both proteins had an additive effect. Endonucleolytic cleavage activities in pooled ∼20S glycerol gradient fractions were examined using full round insertion and deletion assays with A6 pre-mRNA substrates that specify the insertion of two Us or the removal of four Us, respectively (Kable et al. 1996; Carnes et al. 2005). Insertion cleavage activity was unaffected upon KREX1 repression compared with the parental cells but was reduced between 35% and 40% in fractions from the KREX2 and KREX1/KREX2 repressed cells, respectively (Fig. 7A,C). Deletion cleavage activity was reduced to the same extent as insertion cleavage after KREX2 repression (Fig. 7B,C). KREX2 repression also resulted in a significant reduction in the amount of edited RNA with four Us removed in deletion editing assays; however, ligated products with fewer Us removed accumulated (Fig. 7B), mirroring the reduction in U removal observed in precleaved deletion assays (Fig. 6A). Deletion cleavage was more substantially reduced in fractions from KREX1 repressed cells, and although less edited RNA was produced, all four Us were removed. Deletion cleavage was dramatically reduced in fractions from the KREX1/KREX2 repressed cells, and edited RNA was virtually undetectable, probably because of the low amount of cleavage. Thus, deletion cleavage activity was preferentially reduced after KREX1 repression, which is not surprising given the concomitant reduction of the KREN1 deletion endonuclease in these cells (Fig. 3D). Repression of KREX2 led to a modest reduction of both insertion and deletion cleavage activities, likely as a consequence of the observed partial disruption of the editosome in these cells (Fig. 5). The level of inhibition of insertion and deletion cleavage after repression of both KREX1 and 2 appears to be a combination of the reductions of these activities when either protein was repressed alone.
FIGURE 7.
Deletion cleavage activity is preferentially reduced upon KREX1 repression. Full round insertion (A) and deletion (B) editing assays with ∼20S fractions from glycerol gradient purified mitochondrial lysates from parental 29.13 cells and RNAi cells in which KREX proteins were repressed for 7 d as indicated. Diagrams of the RNA substrates and products are shown. The 3′ cleavage products are indicated by arrows. Input RNAs (I) and edited products (Ed) are indicated. Positive control reactions (+) were performed in the presence or absence of gRNA demonstrating gRNA dependence, and a ∼20S fraction that contains peak editing activity which was omitted in negative control reactions (−). T1 digested input RNA (T1) was used as a size marker. (C) Quantification of insertion (I, light gray) and deletion (D, dark gray) cleavage products. The percent of cleavage product is the amount of cleavage and edited products divided by the total input relative to the 29.13 parental control.
DISCUSSION
This study shows that both KREX1 and KREX2 proteins are important for editosome stability but that knockdown of these proteins in PF T. brucei results in differential effects on the editing catalytic activities and parasite viability. Repression of KREX1 expression leads to a concomitant reduction of KREN1 in ∼20S editosomes, whereas KREX2 repression results in reductions of KREPA2 and KREL1 in ∼20S editosomes. Thus, both of these catalysts are important for retention of other editosome proteins. Knockdown of KREX1 results in reduced cell viability, reduction of some edited RNA in vivo, and a significant reduction in deletion but not insertion endonuclease activity in vitro. In contrast, KREX2 knockdown does not affect cell growth or editing in vivo but results in modest reductions of both insertion and deletion endonuclease activities and a significant reduction of U removal in vitro. Simultaneous knockdown of both proteins leads to a more severe inhibition of cell growth and editing in vivo and an additive effect on endonuclease cleavage in vitro. Taken together, these results suggest that the reduction in cell viability upon KREX1 knockdown is likely due to the loss of the essential deletion endonuclease KREN1. Furthermore, although KREX2 appears dispensable for cell growth, the increased inhibition of editing and parasite viability upon knockdown of both KREX1 and KREX2 together suggests that both proteins have roles in editing.
The experiments presented here and elsewhere (Kang et al. 2005; Rogers et al. 2007) show that the KREX1 and KREX2 editosome proteins are exoUases. Both specifically remove Us from the 3′ end of ssRNA but do not remove As or Gs (Fig. 2; data not shown) or Cs (Kang et al. 2005; Rogers et al. 2007). The abrogation of activity upon mutation of two amino acids conserved in the EEP domains of both KREX1 and KREX2 confirms that this domain is essential for catalysis as previously seen with other mutations of this domain (Rogers et al. 2007). Recombinant proteins of both enzymes produced by in vitro transcription/translation remove Us from the 3′ end of a ssRNA substrate but not the 3′ unpaired Us from a partial dsRNA substrate that mimics the pre-mRNA after endonucleolytic cleavage. However, both recombinant exoUases produced in insect cells remove Us from both ssRNA and dsRNA substrates (Kang et al. 2005; Rogers et al. 2007). The difference between these results probably reflects the larger amounts of the enzymes used in the previous studies than with the in vitro transcribed/translated proteins described here, although differences in the dsRNA substrate and/or assay conditions cannot be excluded. Nevertheless, interaction of KREX2 with its binding partner KREPA2 (Schnaufer et al. 2003; Kang et al. 2004) resulted in efficient removal of Us from both ssRNA and the dsRNA (Fig. 2; data not shown). Thus, interaction with KREPA2 enhances KREX2 activity. Similar studies showed that interaction of KREPA1 with its binding partners, KRET2 TUTase and KREL2 ligase, enhanced the activities of the enzymes (Ernst et al. 2003; Schnaufer et al. 2003). The increased enzymatic activities might be the result of enhanced substrate binding via the RNA binding domains of KREPA1 and KREPA2 and/or protein folding effects. Indeed, KREPA1 and KREPA2 have been proposed to aid substrate recognition and coordination of the KRET2 and KREL2, and KREX2 and KREL1 catalytic activities within the insertion and deletion editosome subcomplexes, respectively (Schnaufer et al. 2003; Law et al. 2005).
Repression of KREX1 or KREX2 alone or together did not result in gross sedimentation changes in editosomes despite the loss of some of the other editosome proteins. The loss of KREN1 from 20S editosomes upon KREX1 repression reflects the mutual presence of both of these proteins only in KREN1 editosomes. It also indicates that these two proteins interact directly or via interactions involving other proteins and that KREX1 is essential for association of KREN1 with the editosome. Although the glycerol gradients may lack sufficient resolution to detect minor sedimentation changes, the absence of detectable subcomplexes implies that entire KREN1 editosomes might be lost upon KREX1 knockdown. Similar results, namely, the absence of detectable subcomplexes, were obtained upon knockdown of the KREN1, KREN2, or KREN3 endonucleases (Carnes et al. 2005, 2008; Trotter et al. 2005). The knockdown of the endonuclease proteins was more complete in these studies than in the current study and importantly resulted in an overall reduction of editosomes. Thus, the loss of each endonuclease might have resulted in destabilization and loss of that type of editosome. Whether loss of KREX1, or loss of the endonucleases, results in complete loss of each type of editosome requires further study. KREX2 is a component of all three ∼20S editosomes, unlike KREX1. The substantial reduction of KREPA2 and KREL1 in ∼20S editosomes upon KREX2 repression is consistent with the interactions among these three proteins that comprise the deletion subcomplex (Fig. 1; Schnaufer et al. 2003). The accumulation of KREL1 but not KREPA2 near the top of the gradient may reflect the greater stability of the former compared with the latter when free of the editosome. The reductions of KREL1 and KREPA2 proteins upon KREX2 knockdown may be insufficient to disrupt editing and inhibit cell growth, which is the consequence of direct knockdown of these proteins (Schnaufer et al. 2001; Huang et al. 2002; Gao and Simpson 2003). The sedimentation profile and composition of editosomes after knockdown of both KREX1 and 2 appears to be a combination of the effects observed upon repression of both proteins individually (Figs. 3, 5), with reductions in KREN1, KREPA2, and KREL1 and a shift of KREL1 to the top of the gradient.
Although KREX1 repression inhibited growth, it did not affect in vitro U removal activity of 20S editosomes, while a significant decrease in U removal activity resulted from KREX2 repression, which did not affect growth (Figs. 6, 7). This may reflect the presence of KREX2 in all three types of editosomes while KREX1 is only in one type (Panigrahi et al. 2006), or it may reflect differences in substrate preferences and activities between the two exoUases. KREX1 repression has been reported to reduce exoUase activity in studies in which the KREX knockdown appears more complete based on growth inhibition than here, but these studies also used different substrate and assay conditions, and the reduction in U removal was primarily observed with editosomes collected at much later time points after growth inhibition (Kang et al. 2005). Our studies used time points just after the onset of the growth defect to reduce the possibility of secondary effects arising from the growth inhibition. Knockdown of both exoUases did not increase the effect on in vitro exoUase activity, and thus, we could not attribute any effect on exoUase to KREX1 activity. Similar effects on exoUase activity that result from KREX1 and/or KREX2 knockdown were observed using full round substrates, except for the differential effects on endonucleolytic cleavage activities that are discussed below. Namely, there is a reduction in U removal following cleavage upon KREX2 knockdown and a reduction in cleavage of deletion sites upon KREX1 knockdown. The former resulted in less edited product with fewer Us removed due to the effect on U removal, while the latter resulted in a reduction in deletion editing product due the effect on deletion cleavage. Overall, KREX2 is active in cells, and there may be sufficient residual enzyme in the knockdowns for cell survival or it may be dispensable for cell viability. KREX2 is truncated in Leishmania, lacking the catalytic domain, but is an integral part of the editosome and thus exoUase activity from KREX1 may be sufficient for cell survival (Simpson et al. 2004; Rogers et al. 2007).
The differential effects on endonucleolytic cleavage upon knockdown of KREX1 or KREX2 are likely due to differential effects on editosome composition (Fig. 6). The specific reduction of deletion cleavage upon KREX1 repression is consistent with the loss of the KREN1 deletion endonuclease (Fig. 3), while the modest reduction of both insertion and deletion cleavage upon KREX2 repression is consistent with the reduced level of ∼20S editosomes with their full complement of proteins (Fig. 5). The lack of an effect on cell viability and in vivo editing as a result of the reduced endonuclease cleavage activities associated with KREX2 repression indicates that the reductions were insufficient to become limiting in vivo. The reduction of deletion cleavage activity upon KREX1 repression, however, was sufficient to affect both in vivo editing and cell growth. However, the basis for the preferential effect on edited RPS12 mRNA compared with COIII and ND3 mRNAs in vivo is unclear since all three require deletion editing. The effect on a wider variety of edited RNAs upon repression of both KREX1 and KREX2 is likely to be due to the combination of the reduction of KREN1 editosomes as well as editosomes with a full complement of proteins. The lack of an effect on edited CYb mRNA may be due to the fact that it has a relatively small 5′ edited region that only entails insertion editing and perhaps preferential recognition by the remaining functional editosomes. The increased pre-edited CYb mRNA in KREX1 knockdowns might reflect the preferential association of this RNA with the remaining editosomes that are specific for insertion editing sites. Thus reduced in vivo editing and cell growth appear to result from effects on the endonuclease step of editing rather than from the exoUase step.
These results raise the question of the role of KREX2 in editing in vivo. It is important for editosome integrity, including the retention of KREPA2 and KREL1 in ∼20S editosomes. However, its exoUase activity may not be essential, at least in laboratory culture since it does not seem to be required in Leishmania. It is possible that the exoUase activities of KREX1 and KREX2 are redundant and can compensate for each other's loss or reduction. However, the increased inhibition of deletion endonucleolytic activity, in vivo editing, and cell growth upon knockdown of both KREX1 and KREX2 compared with KREX1 alone suggests that both proteins are important for editing. Hence, the two exoUases may have complementary roles such as KREX1 removing Us from deletion sites and KREX2 removing excess Us added at insertion sites, or they may function differently in different life-cycle stages.
MATERIALS AND METHODS
Cell free protein expression and immunoprecipitation
Constructs for the expression of recombinant KREX1 and KREX2 were made by PCR amplification of the ORFs from T. brucei genomic DNA (strain 427). pRSET-KREX1 was made by amplifying the KREX1 ORF minus the first 32 amino acids using primers 4429 (5′-ATACTCGAGAATTGAATGTGACTACG-3′) and 4724 (5′-ATAAAGCTTTTACGGTAACTTCAATG-3′). The restriction sites are italicized. The PCR product was digested with XhoI and HindIII and ligated with similarly digested pRSET-B (Invitrogen). Mutation of amino acids D794 and N791 to alanines in this construct was performed using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) with primers 5340 (5′-CGGGGATGTGGTAACTATGGGAGCGTTCGCGGACTGGCCTACAAATGAATTT-3′) and 5341 (5′-AAATTCATTTGTAGGCCAGTCCGCGAACGCTCCCATAGTTACCACATCCCCG-3′) according to the manufacturer's instructions. pRSETA-KREX2 was made by amplifying the entire KREX2 ORF using primers 3275 (5′-ATAGGATCCATGTTGCGCCGCAGTCGC-3′) and 3309 (5′-GCGGATCCGAGCTCTAAACCACCTGAAACTC-3′). The PCR product was digested with BamHI and SstI and ligated with similarly digested pRSET-A (Invitrogen). Mutation of amino acids D792 and N794 to alanines was performed as above using primers 4901 (5′-AAGCAGGATTAATTGTTATGGGTGCGTTTGCGGACTGCGCGAAGAATTACTTCAC-3′) and 4902 (5′-GTGAAGTAATTCTTCGCGCAGTCCGCAAACGCACCCATAACAATTAATCCTGCTT-3′). The construct for expression of KREPA2, pSG-TbMP63, was previously described (Panigrahi et al. 2001b).
Cell-free protein expression and coimmunoprecipitation were performed as previously described (Ernst et al. 2003). Briefly, proteins were expressed individually using the TNT T7 Quick Coupled Transcription/Translation System (Promega) in the presence of unlabeled or 35S-labeled methionine according to the manufacturer's instructions. Mixtures of the proteins were pre-incubated for 5 min at room temperature. KREX1 and KREX2 proteins were immunoprecipitated with anti-Xpress (Invitrogen) and KREPA2 (TbMP63) with anti-KREPA2 (P1H3) monoclonal antibodies (Panigrahi et al. 2001b). Goat anti-mouse IgG-coated immunomagnetic beads (2 × 107 beads) (Dynabeads M-450; Dynal) were coupled with 2.5 μL anti-Xpress antibody in IP buffer (10 mM Tris-HCl at pH 7.2, 10 mM MgCl2, 200 mM KCl, and 0.1% Triton X-100) or 1 mL P1H3 tissue culture supernatant in the presence of 1% BSA for 90 min at 4°C with mixing. The beads were washed three times with 800 μL IP buffer. The antibody bound beads were incubated with either individual proteins or mixtures of proteins in IP buffer containing 1% BSA for 90 min at 4°C with mixing. The beads were washed four times with 800 μL IP buffer. 35S-labeled proteins were run out on 10% SDS–polyacrylamide gels, dried, and visualized by phosphorimaging, and unlabeled immunoprecipitated proteins were assayed for U removal activity.
RNAi constructs
Constructs for expressing tet-inducible dsRNA (RNAi) for KREX1 or KREX2 were generated by inserting a 505-bp or 508-bp fragment of the respective gene into pZJM (Wang et al. 2000). The fragments were PCR amplified from T. brucei genomic DNA (strain 427) using primers 4434 (5′-ATACTCGAGTTCCTTTGACTGGGTAGCTG-3′) and 4425 (5′-ATAAAGCTTTTCTGCTCGACACACCCTTG-3′) or primers 3637 (5′-GATACTCGAGATGTTGCGCCGCAGTCGC-3′) and 3638 (5′-GATAAAGCTTCCGCTCCATCGGGAAGTG-3′) to create pZJM-KREX1 and pZJM-KREX2, respectively. To create pZJM-KREX1/KREX2, the same region of KREX1 as in pZJM-KREX1 was amplified using primer 4434 above and primer 4426 (5′-ATACTCGAGTTCTGCTCGACACACCCTTG-3′), digested with XhoI, and inserted into pZJM-KREX2 that was digested with XhoI.
Trypanosome growth, transfection, and RNAi induction
T. brucei 29-13 procyclic cells (Wirtz et al. 1999) were grown in SDM-79 medium containing 15% FBS with 15 μg/mL G418 and 25 μg/mL hygromycin. RNAi cell lines were generated in 29.13 cells by transfection with 15 μg of NotI-linearized pZJM-KREX1, pZJM-KREX2, or pZJM-KREX1/KREX2 as described previously (Schnaufer et al. 2001). The stable cell lines were designated KREX1 RNAi, KREX2 RNAi, and KREX1/KREX2 RNAi and grown in medium with 15 μg/mL G418, 25 μg/mL hygromycin, and 2.5 μg/mL phleomycin. dsRNA was induced with 1 μg/mL tet, and the uninduced and induced cells were counted daily using a Coulter Counter. Cells were maintained between 1 × 106 and 2 × 107cells/mL.
Mitochondria isolation and glycerol gradient sedimentation
Mitochondria were isolated from 1.25 × 1010 29.13 parental cells or the RNAi cell lines induced for 7 d essentially as described previously (Harris et al. 1992). Mitochondria were lysed and the cleared lysates loaded onto 10%–30% glycerol gradients and centrifuged at 38,000 RPM for 9 h at 4°C (SW40 rotor; Beckman). Subsequently, twenty-four 500-μL fractions were collected from the top, flash-frozen in liquid nitrogen, and stored at −80°C.
Real-time PCR
RNA was isolated from the uninduced and induced RNAi cell lines using Trizol reagent as described by the manufacturer (GIBCO BRL). Real-time PCR was used to assess the levels of KREX1, KREX2, pre-edited, edited, and never edited mRNAs as previously described (Carnes et al. 2005). Briefly, 10 μg of RNA was DNase I treated using the DNA-free Kit (Ambion) as described by the manufacturer, and the integrity of the DNase-treated RNA was confirmed using a BioAnalyzer (Agilent Technologies). Half of the RNA was used to generate cDNA using random hexamers with TaqMan Reverse Transcription Reagents (Applied Biosystems) as described by the manufacturer. The other half of the RNA was used for control reactions without reverse transcriptase to confirm the absence of contaminating genomic DNA. cDNA reactions were diluted between seven- and 10-fold depending on how many targets were to be analyzed and amplified in 25 μL reactions containing 2.5 μL of each cDNA, 5 μL each of 1.5 μM forward and reverse primers, and 12.5 μL SYBR Green PCR Master Mix (Applied Biosystems) in 96-well plates using the ABI Prism 7000 Sequence Detection System (Applied Biosystems). The cDNA for the 18S control reactions was further diluted 1:50 to ensure that the Ct value for this more abundant RNA was in the same range as the other targets. Amplification conditions were 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. Primers used for real-time PCR of KREX1 were CCAACTGCTTCAGCCAACCT and TGTGTTGCCCGGACTATCCT; for KREX2, CTGATGTCGGGAGCACTGAAC and TGTCAGCGCAAATGTCCAAT. Primers for the pre-edited, edited, and never edited RNAs were previously described (Carnes et al. 2005). Each mRNA species was analyzed in triplicate. Analysis of the relative amounts of RNA was carried out using the ΔΔCt method (Ingham et al. 2001). Relative changes for each target amplicon were determined after normalization to β-tubulin mRNA and 18S rRNA and expressed as relative mRNA abundance from the respective control cells.
Western blots
Glycerol gradient fractions were resolved by SDS-PAGE, transferred to PVDF membranes and probed either with a cocktail of monoclonal antibodies (Mabs) specific for KREPA1, KREPA2, KREL1, and KREPA3 (Panigrahi et al. 2001b) or with individual Pabs specific for KREX1, KREX2, KREN1, or KRET2 that were prepared as described below.
Rabbit Pabs were produced to purified recombinant KREX1, KREX2, KREN1, and KRET2 by Pocono Research Farm and Laboratory, Inc. The plasmids used to express the proteins were as follows. Each ORF was amplified from T. brucei genomic DNA (strain 427) with the indicated primers. The restriction sites are italicized. The N terminal half of KREX2 (amino acids 8–451) was amplified from using primers 3537 (5′-ACAGGATCCCACCTGCTAGCGGACTAC-3′) and 3538 (5′-CACGAGCTCGATCATTTCTGCCTTGGGGG-3′). The PCR product was digested with BamHI and SstI and ligated with similarly digested pRSET-A (Invitrogen) to create pRSETA-KREX2 N-term. The KREN1 ORF minus the first 11 amino acids was amplified with primers 5410 (5′-GGAATTCTCCAACTGGTGCCACATGCG-3′) and 4246 (5′-TAAGCTTTCACGCACCAACCGAGATG-3′), digested with EcoRI and HindIII, and ligated with similarly digested pRSET-B (Invitrogen) to create pRSETB-KREN1. pRSETB-KREX1 was described above. The His-tagged proteins were induced with 0.4 mM IPTG for 3 h. The proteins were largely insoluble and purified under denaturing conditions as per the manufacturer's protocol. The proteins were refolded by stepwise dialysis to remove the urea. The KREX2 antiserum was affinity purified by absorption to recombinant protein immobilized on PVDF membrane as previously described (Ernst et al. 2003).
RNA editing activity assays
Precleaved deletion and insertion activities were assayed as previously described using 5′-labeled U5-5′, U5-3′, and the gA6[14]PC-del RNAs (Igo et al. 2002) and 5′-labeled 5′CL18, 3′CL13pp, and gPCA6-2A RNAs (Igo et al. 2000), respectively. In vitro full-round deletion and insertion editing assays based on ATPase synthetase subunit 6 (A6) were performed as described previously (Kable et al. 1996; Seiwert et al. 1996) with modifications to enhance accumulation of the endonucleolytic cleavage product (Carnes et al. 2005). Insertion editing reactions used A6-eES1 pre-mRNA and gA6[14] gRNA in the absence of ATP. Deletion editing assays were performed with A6short/TAG.1 pre-mRNA and D34′ gRNA (5′-GGAUAUACUAUAACUCCACCCUCACAACUUUCUU) (Trotter et al. 2005) in the presence of 1 mM ADP. U removal from ssU5-5′ RNA was performed using the same conditions as for precleaved deletion editing assays. Each experiment was performed at least twice.
ACKNOWLEDGMENTS
We thank G. Cross and E. Wirtz for T. brucei strain 29.13, P. Englund for the pZJM vector, and the members of the Stuart laboratory for helpful suggestions and discussions. This work was supported by NIH grant AI14102 to K.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1373009.
REFERENCES
- Aphasizhev R., Sbicego S., Peris M., Jang S.H., Aphasizheva I., Simpson A.M., Rivlin A., Simpson L. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R., Aphasizheva I., Nelson R.E., Gao G., Simpson A.M., Kang X., Falick A.M., Sbicego S., Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003a;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R., Aphasizheva I., Simpson L. A tale of two TUTases. Proc. Natl. Acad. Sci. 2003b;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbarwal V.K., Fleck M., Ernst N.L., Schnaufer A., Stuart K.D. An essential role of KREPB4 in RNA editing and structural integrity of the editosome in Trypanosoma brucei . RNA. 2007;2007:737–744. doi: 10.1261/rna.327707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht M., Niemann M., Schlüter E., Müller U.F., Stuart K., Göringer H.U. TbMP42, a protein component of the RNA editing complex in African trypanosomes has endo-exoribonuclease activity. Mol. Cell. 2005;17:621–630. doi: 10.1016/j.molcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Carnes J., Trotter J.R., Ernst N.L., Steinberg A.G., Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei . Proc. Natl. Acad. Sci. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J., Trotter J.R., Peltan A., Fleck M., Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell. Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M. Functionally unrelated signaling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem. Sci. 2000;25:272–273. doi: 10.1016/s0968-0004(00)01582-6. [DOI] [PubMed] [Google Scholar]
- Drozdz M., Palazzo S.S., Salavati R., O'Rear J., Clayton C., Stuart K. TbMP81 is required for RNA editing in Trypanosoma brucei . EMBO J. 2002;21:1791–1799. doi: 10.1093/emboj/21.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst N.L., Panicucci B., Igo R.P., Jr, Panigrahi A.K., Salavati R., Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Gao G., Simpson L. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J. Biol. Chem. 2003;278:27570–27574. doi: 10.1074/jbc.M303317200. [DOI] [PubMed] [Google Scholar]
- Guo X., Ernst N.L., Stuart K.D. The KREPA3 zinc finger motifs and OB-fold domain are essential for RNA editing and survival of Trypanosoma brucei . Mol. Cell. Biol. 2008;28:6939–6953. doi: 10.1128/MCB.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M., Decker C., Sollner-Webb B., Hajduk S. Specific cleavage of pre-edited mRNAs in trypanosome mitochondrial extracts. Mol. Cell. Biol. 1992;12:2591–2598. doi: 10.1128/mcb.12.6.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.E., O'Hearn S.F., Sollner-Webb B. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 2002;22:3194–3203. doi: 10.1128/MCB.22.9.3194-3203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo R.P., Jr, Weston D.S., Ernst N.L., Panigrahi A.K., Salavati R., Stuart K. Role of uridylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Eukaryot. Cell. 2002;1:112–118. doi: 10.1128/EC.1.1.112-118.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham D.J., Beer S., Money S., Hansen G. Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques. 2001;31:132–140. doi: 10.2144/01311rr04. [DOI] [PubMed] [Google Scholar]
- Kable M.L., Seiwert S.D., Heidmann S., Stuart K. RNA editing: A mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- Kang X., Falick A.M., Nelson R.E., Gao G., Rogers K., Aphasizhev R., Simpson L. Disruption of the zinc finger motifs in the Leishmania tarentolae LC-4 (=TbMP63) L-complex editing protein affects the stability of the L-complex. J. Biol. Chem. 2004;279:3893–3899. doi: 10.1074/jbc.M310185200. [DOI] [PubMed] [Google Scholar]
- Kang X., Rogers K., Gao G., Falick A.M., Zhou S., Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl. Acad. Sci. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Gao G., Rogers K., Falick A.M., Zhou S., Simpson L. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc. Natl. Acad. Sci. 2006;103:13944–13949. doi: 10.1073/pnas.0604476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Huang C.E., O'Hearn S.F., Sollner-Webb B. In Trypanosoma brucei RNA editing, band II enables recognition specifically at each step of the U insertion cycle. Mol. Cell. Biol. 2005;25:2785–2794. doi: 10.1128/MCB.25.7.2785-2794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison-Antenucci S., Hajduk S. RNA editing-associated protein 1 is an RNA binding protein with specificity for preedited mRNA. Mol. Cell. 2001;7:879–886. doi: 10.1016/s1097-2765(01)00231-3. [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci S., Grams J., Hajduk S.L. Editing machines: The complexities of trypanosome RNA editing. Cell. 2002;108:435–438. doi: 10.1016/s0092-8674(02)00653-0. [DOI] [PubMed] [Google Scholar]
- McManus M.T., Shimamura M., Grams J., Hajduk S.L. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei . RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian I.S., Worthey E.A., Salavati R. Taking U out with two nucleases? BMC Bioinformatics. 2006;7:1–10. doi: 10.1186/1471-2105-7-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol C.D., Kuo C.F., Thayer M.M., Cunningham R.P., Tainer J.A. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- O'Hearn S., Huang C.E., Hemann M., Zhelonkina A., Sollner-Webb B. Trypanosoma brucei RNA editing complex: Band II is structurally critical and maintains band V ligase, which is nonessential. Mol. Cell. Biol. 2003;23:7909–7919. doi: 10.1128/MCB.23.21.7909-7919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Gygi S., Ernst N., Igo R.P., Jr, Palazzo S.S., Schnaufer A., Weston D., Carmean N., Salavati R., Aebersold R., et al. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001a;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Schnaufer A., Carmean N., Igo R.P., Jr, Gygi S.P., Ernst N.L., Palazzo S.S., Weston D.S., Aebersold R., Salavati R., et al. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001b;21:6833–6840. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Schnaufer A., Ernst N.L., Wang B., Carmean N., Salavati R., Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Ernst N.L., Domingo G.J., Fleck M., Salavati R., Stuart K.D. Compositionally and functionally distinct editosomes in Trypanosoma brucei . RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K., Gao G., Simpson L. Uridylate-specific 3′-5′ exoribonucleases involved in uridylate-deletion RNA editing in Trypanosomatid mitochondria. J. Biol. Chem. 2007;282:29073–29080. doi: 10.1074/jbc.M704551200. [DOI] [PubMed] [Google Scholar]
- Rusche L.N., Cruz-Reyes J., Piller K.J., Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusché L.N., Huang C.E., Piller K.J., Hemann M., Wirtz E., Sollner-Webb B. The two RNA ligases of the Trypanosoma brucei RNA editing complex: Cloning the essential Band IV gene and identifying the Band V gene. Mol. Cell. Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salavati R., Ernst N.L., O'Rear J., Gilliam T., Tarun S., Jr, Stuart K. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei . RNA. 2006;12:819–831. doi: 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A., Panigrahi A.K., Panicucci B., Igo R.P., Jr, Salavati R., Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei . Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- Schnaufer A., Ernst N., O'Rear J., Salavati R., Stuart K. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- Seiwert S.D., Heidmann S., Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- Simpson L., Sbicego S., Aphasizhev R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L., Aphasizhev R., Gao G., Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K., Allen T.E., Heidmann S., Seiwert S.D. RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev. 1997;61:105–120. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K.D., Schnaufer A., Ernst N.L., Panigrahi A.K. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Tarun S.Z., Jr, Schnaufer A., Ernst N.L., Proff R., Deng J., Hol W., Stuart K. KREPA6 is an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei . RNA. 2008;14:347–358. doi: 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter J.R., Ernst N.L., Carnes J., Panicucci B., Stuart K. A deletion site editing endonuclease in Trypanosoma brucei . Mol. Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Wang Z., Morris J.C., Drew M.E., Englund P.T. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Wang B., Ernst N.L., Palazzo S.S., Panigrahi A.K., Salavati R., Stuart K. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei . Eukaryot. Cell. 2003;2:578–587. doi: 10.1128/EC.2.3.578-587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C., Cross G.A.M. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei . Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Worthey E.A., Schnaufer A., Mian I.S., Stuart K., Salavati R. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 2003;31:6392–6408. doi: 10.1093/nar/gkg870. [DOI] [PMC free article] [PubMed] [Google Scholar]