Abstract
MicroRNA (miRNA)-mediated silencing is a post-transcriptional mechanism that regulates translation of mRNAs primarily via their 3′-UTR. Ago2 binds miRNA directly and is the core component of miRNA-induced silencing complex. GW182 is another important factor in miRNA-mediated silencing, and its interaction with Ago2 is evolutionarily conserved. However, the GW182-Ago2 interaction in humans has not been characterized thoroughly, and the role of GW182 in the mammalian miRNA pathway remains unclear. In the current study, we generated a set of GST-, green fluorescence protein (GFP)-, or 3xFlag-tagged deletion constructs of GW182 and Ago2 to further analyze GW182-Ago2 interactions. The C-terminal half of Ago2 interacted with four nonoverlapping GW-rich regions of GW182, and this interaction recruited Ago2 to GWB. Furthermore, the interaction with GW182 was observed in all four human Ago proteins. Most interestingly, tethering the C-terminal half of Ago2 to the 3′-UTR of reporter mRNA recapitulated translational repression comparable to that of tethered Ago2, and this repression was greatly impaired upon GW182 knockdown. In comparison, the N-terminal half of Ago2 did not bind GW182 and did not retain the repression function of Ago2. Our data strongly support a model in which Ago2 recruits GW182 to the 3′-UTR of mRNA to mediate silencing, and suggest that GW182 may contribute to enhancement in translational repression by interacting with multiple Ago proteins from multiple miRNA target sites in the same or adjacent 3′UTR.
Keywords: Ago2, PIWI, GW182, GW-rich, microRNA, gene silencing
INTRODUCTION
MicroRNA (miRNA)-mediated gene silencing is an important post-transcriptional regulation which, in part, controls the half lives of mRNA targets. In this regulatory role, miRNA binds to the 3′-UTR of mRNA leading to translational inhibition, mRNA degradation, and mRNA sequestration (Nilsen 2007). MiRNAs are evolutionarily conserved in most of the species studied to date, and are estimated to regulate ∼30% of protein-encoding genes in humans (Lewis et al. 2005; Filipowicz et al. 2008).
There are four human Ago proteins that include Ago1–Ago4, which are the core components of silencing effecter complexes and are known to bind single-stranded miRNA. These Ago proteins share >80% identity and are primarily characterized by PAZ and PIWI domains. The PAZ domain contains a binding pocket for the 3′ overhanging nucleotides of miRNA. Interestingly, despite highly conserved sequences, only the PIWI domain of Ago2 harbors RNase H-type activity and, therefore, Ago2 also functions in small interfering RNA (siRNA)-mediated slicing of mRNA targets (Liu et al. 2004; Yuan et al. 2005). Tethering Ago proteins to the 3′-UTR of mRNA mimicked miRNA function and affected translational repression (Pillai et al. 2004).
GW182 is important for miRNA-mediated translational silencing, and interacts with Ago2 (Jakymiw et al. 2005; Liu et al. 2005a). GW182 contains several glycine/tryptophan-rich (GW-rich) regions, a glutamine/asparagine-rich (Q/N-rich) domain, and a C-terminal RNA recognition motif (RRM) (Eystathioy et al. 2002; Decker et al. 2007). GW182 localized to and was essential for the formation of GW bodies (GWB) (also known as mammalian P bodies) (Yang et al. 2004; Schneider et al. 2006), cytoplasmic structures closely linked to mRNA decay (Eystathioy et al. 2003; Sheth and Parker 2003), and the miRNA/siRNA pathway (Jakymiw et al. 2005; Lian et al. 2006, 2007; Pauley et al. 2006). Knockdown of GW182 greatly impaired miRNA-mediated gene silencing and subsequent mRNA degradation (Liu et al. 2005a; Rehwinkel et al. 2005). Interestingly, Ago proteins, miRNAs, and mRNAs targeted by miRNAs all colocalized with GW182 in GWB (Jakymiw et al. 2005; Liu et al. 2005b; Pillai et al. 2005; Sen and Blau 2005). Furthermore, GW182 interacted with Ago2, and this interaction was conserved from plants to humans (Ding et al. 2005; Jakymiw et al. 2005; Liu et al. 2005a; Behm-Ansmant et al. 2006; El-Shami et al. 2007; Till et al. 2007). However, the role of GW182 and the importance of GW182-Ago2 interactions in translational repression remain unclear. In the current study, we mapped the GW182-Ago2 interaction, and investigated the possible role of this interaction in miRNA-mediated silencing in human.
RESULTS
C-terminal half of human Ago2 containing the PIWI domain was responsible for the interaction with GW182
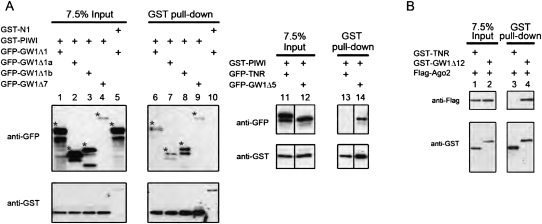
The interaction between human Ago2 and GW182 was first reported in 2005 (Jakymiw et al. 2005; Liu et al. 2005a). To further characterize this interaction, a series of deletion constructs covering different domains of GW182 and Ago2 were generated (Fig. 1A). The human GW182 gene identified as TNRC6A (GenBank) is currently predicted to have 2 isoforms: GW182 as previously described (Eystathioy et al. 2002), and a longer isoform TNGW1 (NM_014494.2) that contains an extra N-terminal 253 amino acid -polypeptide with a glutamine-repeat (Q-repeat) region encoded by CAG trinucleotide repeats (TNR) (Li et al. 2008). The deletion construct TNR (amino acids 1–204) included the Q-repeat region and did not overlap with GW182 (Fig. 1A). TNR (amino acids 1–204) and two other GW182 deletion constructs GW1Δ1 (amino acids 254–751) and GW1Δ10 (amino acids 566–1343), respectively, representing approximately the N-terminal 1/3 and middle 1/3 of GW182, were initially used to analyze the binding to Ago2 using the GST pull-down assay. GST-GW1Δ1 and GST-GW1Δ10 were demonstrated to interact with endogenous Ago2, whereas GST-TNR did not (Fig. 1B). To map the region of Ago2 responsible for this interaction, two deletion constructs corresponding to the N-terminal (PAZ, amino acids 1–480) and C-terminal (PIWI, amino acids 478–860) halves were constructed (Fig. 1A). By coexpression of GST- and Flag-tagged constructs and the GST pull-down assay, Ago2 consistently interacted with GW1Δ1 and GW1Δ10 but not with TNR (Fig. 1C,D). Importantly, PIWI but not PAZ was responsible for the association of Ago2 with GW1Δ1 and GW1Δ10 (Fig. 1C). Notably, there was less of the GST-GW1Δ10 in the soluble input when it was cotransfected with Flag-Ago2 (Fig. 1C, lane 1) or Flag-PIWI (Fig. 1C, lane 2) than when GST-GW1Δ10 was co-transfected with Flag-PAZ (Fig. 1C, lane 3). To investigate the reason for these observed differences, the expression of GST-GW1Δ10 was examined in both total cell lysates and the insoluble fractions. The expression of GST-GW1Δ10 in total cell lysates was relatively uniform no matter which Flag-tagged constructs it cotransfected with (Supplemental Fig. S1). Interestingly, higher levels of GST-GW1Δ10 were observed in the insoluble fraction when it was cotransfected with Flag-Ago2 (Fig. 1C, lane 4) and even higher when cotransfected with Flag-PIWI (Fig. 1C, lane 5), whereas GST-GW1Δ10 was barely detectable in the insoluble fraction when cotransfected with Flag-PAZ (Fig. 1C, lane 6). These data suggested that GW1Δ10 formed insoluble complexes with Ago2 or PIWI, which might explain why there was a lower level of GW1Δ10 in the soluble input for the GST pull-down assays. In summary, GW182 fragments GW1Δ1 and GW1Δ10 both bound to Ago2, and this binding was mediated by the C-terminal half of Ago2.
FIGURE 1.
Identifying the interaction of C-terminal half of Ago2 with GW182 fragments using GST pull-down assays. (A) Schematic of human GW182 and Ago2 deletion constructs used in this study. Amino acid residues of GW182 constructs are referenced to the TNGW1, the longer isoform of GW182 (GenBank Accession NM_014494.2). Q-repeat, glutamine repeat (box in white); Q/N-rich, glutamine/asparagine-rich region (box in magenta); RRM, RNA recognition motif (box in green); GW-rich, glycine/tryptophan-rich region (boxes in yellow); N-GW, N-terminal GW-rich region; M-GW, middle GW-rich region; C-GW, C-terminal GW-rich region. Human Ago2 contains two conserved domains: PAZ domain (box in blue) and PIWI domain (box in red). (B) Endogenous Ago2 coprecipitated with GW182 fragments GW1Δ1 (“aa254-751”) and GW1Δ10 (“aa566-1343”) but not with TNR (“aa1-204”). GST-tagged TNR (lanes 1,4), GW1Δ1 (lanes 2,5), or GW1Δ10 (lanes 3,6) was transfected into HeLa cells. Endogenous Ago2 was detected by using rabbit polyclonal anti-Ago2, which also contained anti-GST activity. In a longer exposure (lanes 7–9), Ago2 was detected more clearly being coprecipitated with GST-GW1Δ10 (lane 9) but still absent in GST-TNR precipitates (lane 7). (C) GW182 fragments GW1Δ1 and GW1Δ10 coprecipitated with Ago2 fragment PIWI (“aa478-860”) but not with PAZ (“aa1-480”). GST-GW1Δ10 (lanes 1–6) or GST-GW1Δ1 (lanes 7–10) was cotransfected with Flag-tagged Ago2, PIWI, or PAZ. Full-length Ago2 (lanes 4,9) and PIWI (lanes 5,10), but not PAZ (lane 6), coprecipitated with GST-GW1Δ10 or GST-GW1Δ1. (D) GW182 fragment TNR did not pull down Ago2. Flag-Ago2 was cotransfected with GST-tagged TNR (“aa1-204,” lanes 1,3) or with GW1Δ1 (lanes 2,4) as a positive control.
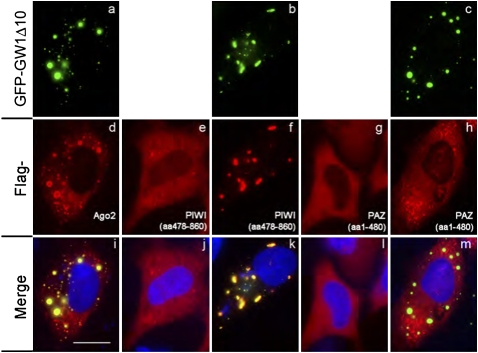
GW182-Ago2 interaction was important for the localization of Ago2 in cytoplasmic foci
Previous studies have shown that Ago2 colocalized with GW182 in cytoplasmic GWB and GW182 was essential for the formation of these foci (Yang et al. 2004; Jakymiw et al. 2005). However, the driving force for the localization of Ago2 to GWB remains unknown. Based on the above data that the GW182 fragment GW1Δ10 formed insoluble complexes with Ago2 and PIWI, but not with PAZ, we hypothesized that the GW182-Ago2 interaction is crucial for Ago2 to localize to GWB. To examine this hypothesis, Flag-Ago2, Flag-PIWI, or Flag-PAZ was coexpressed with GFP-GW1Δ10 in HeLa cells and Flag-PIWI or Flag-PAZ were expressed alone as controls (Fig. 2). Flag-Ago2 was shown to colocalize with GFP-GW1Δ10 in cytoplasmic foci, whereas singly expressed Flag-PIWI or Flag-PAZ were diffusely distributed in the cytoplasm (Fig. 2, panels e,g). Interestingly, coexpression of GFP-GW1Δ10 with Flag-PIWI dramatically changed the distribution of Flag-PIWI, which was recruited to cytoplasmic foci and colocalized with GFP-GW1Δ10 (Fig. 2, panels b,f,k). In contrast, coexpressing GFP-GW1Δ10 with Flag-PAZ did not recruit the diffusely distributed Flag-PAZ to cytoplasmic GFP-GW1Δ10-positive foci (Fig. 2, panels c,h,m). These data supported the contention that the interaction of GW182 with the C-terminal half of Ago2 mediated the localization of Ago2 to GWB.
FIGURE 2.
GW182 fragment GW1Δ10 (“aa566-1343”) recruited Ago2 to cytoplasmic foci by interacting with the C-terminal half of Ago2. GFP-GW1Δ10 (green, a–c) was cotransfected with Flag-Ago2 (d), PIWI (“aa478-860,” f) or PAZ (“aa1-480,” h) into HeLa cells. As controls, Flag-PIWI (e) or Flag-PAZ (g) was singly transfected. The cells were stained with anti-Flag antibody (red, d–h). Panels in the bottom row are the merged images (i–m). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar, 10 μM.
Ago2 bound to multiple nonoverlapping GW-rich regions of GW182
Since the GW182 fragments, GW1Δ1 and GW1Δ10, were both shown to bind Ago2 and these two fragments have an overlapping 186 amino acids, it is possible that the overlapping region of GW182 (amino acids 566–751) is the primary site for the GW182-Ago2 interaction. To examine this possibility, deletion constructs GW1Δ1a (amino acids 254–503) and GW1Δ1b (amino acids 502–751) were generated with the latter covering the overlapping region of GW1Δ1 and GW1Δ10 (Fig. 1A). In addition, other deletion constructs GW1Δ7 (amino acids 1034–1962) and GW1Δ5 (amino acids 1670–1962) were used to investigate whether regions of GW182 other than GW1Δ1 and GW1Δ10 bound Ago2. GFP-GW1Δ1a, GFP-GW1Δ1b, GFP-GW1Δ7, or GFP-GW1Δ5 was coexpressed with GST-tagged Ago2 fragment PIWI in HeLa cells and a GST pull-down assay was performed to examine the interaction. As a negative control, GFP-GW1Δ1 was coexpressed with GST-tagged fragment N1, the N-terminal amino acids 51–779 of a completely unrelated protein hZW10 (Famulski et al. 2008). Unexpectedly, GFP-GW1Δ1a, GFP-GW1Δ1b, GFP-GW1Δ7, and GFP-GW1Δ5 were all coprecipitated with GST-PIWI (Fig. 3A, lanes 7–9,14). Another GW182 truncated construct GW1Δ12, which contains the reported ortholog-conserved GW-rich region (amino acids 1074–1144) (Till et al. 2007), was also able to coprecipitate Ago2 (Fig. 3B). Interestingly, GW1Δ1a, GW1Δ1b, GW1Δ12, and GW1Δ5 are nonoverlapping fragments, and thus, these data showed that at least four separate regions of GW182 could bind Ago2. Moreover, the deletion constructs that bound Ago2 all contained a GW-rich region, whereas TNR, the only deletion construct that did not bind Ago2, lacked a GW-rich region. In summary, the GW182 deletion constructs containing GW-rich regions all bound to the C-terminal half of Ago2, indicating that multiple regions of GW182 mediated the interaction of GW182 with Ago2, and that GW repeats might be an key element for Ago2 binding.
FIGURE 3.
Ago2 bound to multiple nonoverlapping GW-rich regions of GW182. (A) GW182 fragments coprecipitated with C-terminal half of Ago2. GST-PIWI (“aa478-860”) was cotransfected with GFP-tagged GW1Δ1 (lane 1), GW1Δ1a (lane 2), GW1Δ1b (lane 3), GW1Δ7 (lane 4), TNR (lane 11), or GW1Δ5 (lane 12) into HeLa cells. Similar to positive control GW1Δ1 (lane 6), GW1Δ1a (lane 7), GW1Δ1b (lane 8), GW1Δ7 (lane 9), GFP-GW1Δ5 (lane 14), but no GFP-TNR (lane 13), were detected in GST-PIWI precipitates. GST-tagged N1, N-terminal fragment from an unrelated protein hZW10, was cotransfected with GFP-GW1Δ1 (lane 5) as a negative control and no interaction was detected (lane 10). Asterisks indicate the corresponding GFP-tagged constructs in Western blot. (B) GW182 fragment GW1Δ12 coprecipitated with Ago2. Flag-Ago2 was cotransfected with GST-tagged TNR (lanes 1,3) or with GW1Δ12 (lanes 2,4), which contains the conserved sequence for Ago2 interaction. Flag-Ago2 was coprecipitated with GST-tagged GW1Δ12 but not TNR.
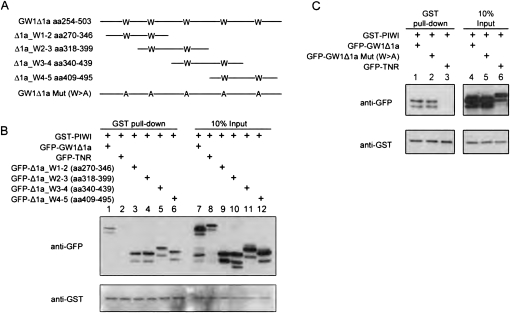
Tryptophan residues of GW1Δ1a were not required for interaction with Ago2
Previous studies from two groups have shown that short synthetic peptides containing one to two WG/GW were able to interact with Ago protein (El-Shami et al. 2007; Till et al. 2007). The interaction was significantly reduced when either one of the tryptophans was mutated to alanine (Till et al. 2007). We noted that GW1Δ1a, unlike GW1Δ1b and GW1Δ5, lacks sequence homology with the reported ortholog-conserved sequence (see Discussion). It is possible that the interaction of GW1Δ1a with Ago2 is different from that of the other GW182 truncated constructs. To examine if any of the five tryptophans in GW1Δ1a play important roles in Ago2-binding, four sequential truncated constructs were designed to span the sequence of GW1Δ1a and each containing two tryptophans (Fig. 4A). Interestingly, all these truncated constructs were coprecipitated by GST-PIWI (Fig. 4B). In addition, to examine whether tryptophan is essential for the GW1Δ1a-PIWI interaction, all of the five tryptophans in GW1Δ1a were mutated to alanine (W > A) (Fig. 4A). Surprisingly, the W > A mutation of GW1Δ1a did not abolish its ability to coprecipitate with the PIWI fragment (Fig. 4C). In summary, the GW1Δ1a fragments containing two tryptophans all bound to the C-terminal half of Ago2. However, loss of tryptophan in GW1Δ1a did not disrupt the GW1Δ1a-PIWI association, implying that tryptophan might not be an essential feature for mediating the interaction of GW182 with Ago.
FIGURE 4.
Four subregions of GW1Δ1a were capable of binding Ago2, and the interaction between GW1Δ1a and PIWI was not dependent on the five tryptophan residues. (A) Schematics of the four overlapping fragments of GW1Δ1a each containing two tryptophan residues and the mutant designed with all five tryptophans substituted with alanine. (B) Truncated constructs of GW1Δ1a containing any two continuous tryptophans were able to coprecipitate with PIWI. GST-PIWI was transfected with GFP-tagged TNR, GW1Δ1a, and its truncated constructs: Δ1a_W1-2 (“aa270-346,” contains the first and second tryptophans of GW1Δ1a), W2-3 (“aa318-339,” contains the second and third trytophans), W3-4 (“aa340-439,” contains the third and fourth tryptophan), and W4-5 (“aa409-495,” contains the fourth and fifth tryptophans). Compared with GFP-TNR (lane 2), all truncated constructs of GW1Δ1a (lanes 3–6) coprecipitated with GST-PIWI as full-length GW1Δ1a did (lane 1). (C) GW1Δ1a mutant without tryptophan still coprecipitated with PIWI. A GW1Δ1a mutant with all tryptophan (W) mutated to alanine (A) (GW1Δ1a Mut (W > A), lane 2) was cotransfected with GST-PIWI into HeLa cells. Compared with wild-type GW1Δ1a (lane 1), GW1Δ1a mutant was coprecipitated with PIWI at the comparable level. GFP-TNR (lane 3) served as a negative control in the GST pull-down assay.
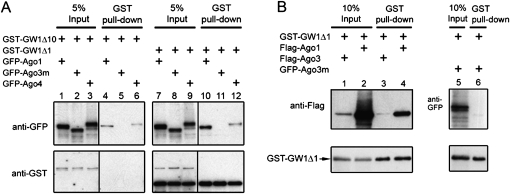
The interaction of Ago2 with GW182 was conserved in other human Ago proteins
There are four Ago proteins in human cells that share a high degree of sequence similarity. To examine whether Ago1, Ago3, and Ago4 also interact with GW182, GFP-Ago1, GFP-Ago3, GFP-Ago3m, or GFP-Ago4 was coexpressed with GST-tagged GW182 fragments GW1Δ1 or GW1Δ10 in HeLa cells and a GST pull-down assay was performed. Ago3m is a splicing variant of Ago3 and is missing amino acids 757–823, the C-terminal 66 amino acids of the PIWI domain. Interestingly, human Ago1, Ago3, and Ago4 bound GW1Δ1 and GW1Δ10 (Fig. 5A,B). Ago3m did not bind GW1Δ1 or GW1Δ10, indicating that the C terminus of that cognate PIWI domain was required for the binding to GW182 (Fig. 5A,B). Notably, both GFP-Ago1 (Fig. 5A, lane 4) and Flag-Ago1 (Fig. 5B, lane 4) bound GST-GW1Δ1, demonstrating that different N-terminal fusion tags did not affect the binding of Ago1 with GW182. In summary, the interaction of human GW182 with Ago2 was observed with other human Ago proteins and the C-terminal region of PIWI domain was critical for the interaction of GW182 with Ago3.
FIGURE 5.
Both GW182 fragments GW1Δ1 and GW1Δ10 coprecipitated with other human Ago proteins. (A) Ago 1 and Ago4, but not Ago3 mutant, coprecipitated with GW182 fragments GW1Δ1 and GW1Δ10. GFP-tagged Ago1, Ago3m (Ago3 mutant), or Ago4 was cotransfected with GST-GW1Δ10 (lanes 1–6) or GST-GW1Δ1 (lanes 7–12). Ago3m is missing an exon (“aa757-823”), the C-terminal 66 amino acids of the PIWI domain compared with the reference sequence (NM_024852.2). Both Ago1 (lanes 4,10) and Ago4 (lanes 6,12) were pulled down by GST-GW1Δ10 or GST-GW1Δ1. In comparison, Ago3m was absent from either pull-down (lanes 5,11). (B) Ago3 coprecipitated with GW182 fragment GW1Δ1. Flag-Ago3 (lanes 1,3) or Flag-Ago1 (lanes 2,4) was cotransfected with GST-GW1Δ1 (lanes 1–4). GFP–Ago3m (lanes 5,6) was cotransfected with GST-GW1Δ1 as a negative control.
Tethering C-terminal half of Ago2 to the 3′-UTR of mRNA recapitulated Ago2-mediated silencing which required the presence of GW182
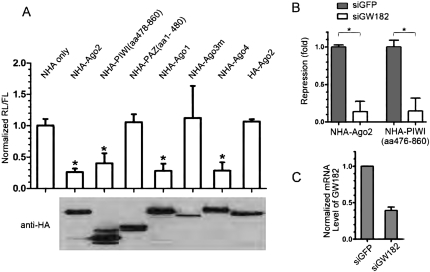
It was reported that tethering Ago2 to the 3′-UTR of mRNA causes repression of protein synthesis (Pillai et al. 2004). Because the C-terminal half of Ago2 bound GW182, whereas the N-terminal half of Ago2 did not, we examined whether the C-terminal half of Ago2 was able to mediate silencing when tethered to the 3′-UTR of mRNA. The dual luciferase and tethering assay was used as described previously (Pillai et al. 2004). In this assay, the reporter Renilla luciferase (RL) contains five 19-nucleotide (nt) BoxB hairpin structures in the 3′-UTR of its mRNA (RL-5BoxB). The λN peptide, which is derived from λ phage and binds to BoxB structures with high affinity (Legault et al. 1998), was fused to the N terminus of HA-tagged Ago2, PIWI, or PAZ. In this way, Ago2, PIWI, or PAZ was brought directly to the 3′-UTR of mRNA, bypassing the requirement for miRNA. Interestingly, tethered PIWI was attended by almost as much repression as tethered full-length Ago2 (Fig. 6A). In contrast, tethered PAZ was totally devoid of the repression function of Ago2 (Fig. 6A). These data indicated that the functional domain mediating silencing lay within the C-terminal half of Ago2. The repression effect of other human Ago proteins was also examined. Both Ago1 and Ago4 exerted a similar repression effect on the reporter as Ago2 and PIWI (Fig. 6A). Interestingly, Ago3m, which lost the interaction with GW182, was not able to induce repression. These data supported the conclusion that the repression effect mediated by the tethered construct might be associated with its interaction with GW182. To examine whether GW182 is required for Ago2- or PIWI-mediated repression, siRNA was used to knockdown GW182 before Ago2, PIWI, or PAZ was tethered to the reporter mRNA. Very interestingly, both Ago2- and PIWI-mediated repression was significantly reduced upon GW182 knockdown (Fig. 6B). The GW182 knockdown was confirmed by quantitative real-time polymerase chain reaction (PCR) (Fig. 6C). In summary, tethering the C-terminal half of Ago2 to the 3′-UTR of mRNA recapitulated the repression function of Ago2, and this repression required GW182.
FIGURE 6.
Gene silencing mediated by tethered C-terminal half of Ago2 required GW182. (A) Tethered PIWI (“aa478-860”) down-regulated protein synthesis to the same extends of other Ago proteins. HeLa cells were transfected with constructs expressing the RL-5BoxB reporter, control FL reporter, and indicated NHA-tagged proteins. Bar graphs represent normalized mean values of RL/FL activities with standard errors. The RL/FL values in cells with tethered NHA-tagged Ago2, PIWI, Ago1, and Ago4 were significantly reduced compared with the value in NHA only group, which was normalized as 1. The NHA tagged PAZ, Ago3m, or HA-Ago2 did not show repression effect on the reporters. The expression of fusion proteins were determined by Western Blot using anti-HA mAb and are indicated below the bar graphs. The assay was repeated a minimum of three times. Asterisks indicate groups have significant difference compared with the NHA only group (unpaired t-test, P < 0.0001). No significant difference was shown between any two groups with asterisk (unpaired t-test, P > 0.05). (B) Translational repression mediated by tethered Ago2 or PIWI (“aa478-860”) was greatly impaired upon GW182 knockdown. HeLa cells were transfected with siRNA for either GW182 (siGW182) or GFP (siGFP). Thirty hours later, cells were transfected again with constructs expressing reporter RL-5BoxB, control FL reporter, and the indicated NHA-tagged proteins. Bar graphs represent the reduction of RL/FL in cells with tethered NHA-Ago2 or NHA-PIWI compared with those in cells with tethered NHA. The reduced values of RL/FL in cell transfected with siGFP were set as 1. Error bars indicate standard errors. The assay was performed in triplicate and was repeated two times. *Significant difference (unpaired t-test, P < 0.01). (C) GW182 knockdown by siRNA was confirmed by quantitative real-time PCR. The bar graphs represent normalized mRNA level of GW182 with standard errors. The mRNA level of GW182 in cells transfected with GFP-siRNA was set as 1. The experiment was performed in triplicate.
DISCUSSION
Formation of GW182 and Ago protein complexes
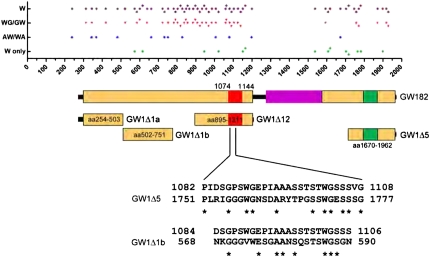
Two studies have identified that one GW-rich region capable of binding Ago2 is conserved in the plant and yeast orthologs of GW182, and that the GW repeat within this region is critical for GW182-Ago2 interaction (El-Shami et al. 2007; Till et al. 2007). Consistent with these studies, our data showed that multiple human GW-rich regions were able to bind Ago2 (Fig. 7). GW182 fragments GW1Δ10 (amino acids 566–1343), GW1Δ7 (amino acids 1034–1962), and GW1Δ12 (amino acids 895–1211) containing the ortholog-conserved GW-rich region (amino acids 1074–1144) (Till et al. 2007) were shown to bind Ago2 (Fig. 7). In addition, our data showed that at least four nonoverlapping regions of GW182 could independently bind Ago2 and, interestingly, three of these Ago2-binding fragments are outside of the ortholog-conserved GW-rich region (Fig. 7). Sequence alignment analysis showed that 27 amino acid and 23 amino acid residues of the ortholog-conserved GW-rich region shared 40.7% and 34.8% identity with the GW1Δ5 and GW1Δ1b, respectively (Fig. 7). However, there was no significant sequence identity between the ortholog-conserved region and GW1Δ1a. Further analysis of GW1Δ1a showed the four subregions, two of which did not overlap, were all capable of binding Ago2 and, surprisingly, the five Trp residues within GW1Δ1a were not apparently important for the interaction with Ago2. Our data implied that different GW-rich regions were capable of binding Ago proteins but that the requirement for tryptophan residues varies. Future studies of the crystal structure of the Ago2-GW182 complex should enhance our understanding of how these two molecules interact with each other.
FIGURE 7.
At least three nonoverlapping GW-rich regions that are different from the ortholog-conserved GW-rich region can independently bind Ago2. The dot graph on top indicates the distribution of tryptophan in GW182. W, every tryptophan (magenta diamond); WG/GW, a glycine right adjacent to tryptophan (red triangle); AW/WA, an alanine right adjacent to tryptophan (blue diamond); W only, no glycine or alanine right adjacent to tryptophan (green diamond). The majority of the tryptophans are adjacent to either a glycine or alanine. The schematic of GW182 is indicated below the dot graph. GW-rich region, box in yellow; ortholog-conserved GW-rich region (“aa1074-1144”) (Till et al. 2007), box in red. Compared with GW1Δ12, which contains the ortholog-conserved region, GW1Δ1a, GW1Δ1b, and GW1Δ5 are the three nonoverlapping regions identified in the current study that binds Ago2. The amino acid sequence alignment between GW1Δ5/GW1Δ1b and ortholog-conserved GW-rich region was performed using ExPASy website tool. *Represent sequence identity.
Since our data also indicated that GW182 can bind multiple Ago proteins, it is possible that the different Ago proteins incorporate into the same complex and contribute to the formation of functional translational silencing complexes. It is yet to be determined if the function of the silencing complex depends on which Ago proteins it contains. In support of our speculation that GW182 helps to stabilize the binding of multiple Ago-miRNA complexes to the 3′-UTR of target mRNA for more efficient translational repression, it was reported that more closely spaced miRNA binding sites in the 3′-UTR of target mRNA led to more efficient miRNA-mediated translational repression (Grimson et al. 2007). It is also possible that GW182 simultaneously binds to Ago-miRNA complexes on several different mRNAs and this GW182-Ago interaction may be the driving force for the assembly of submicroscopic and microscopic GWB. This hypothesis is supported by current observations that the GW1Δ1 GW182 fragment or the PIWI Ago2 fragment could mediate GW182-Ago2 interaction and by our previous data that overexpression of either of these two constructs disassembled GWB, possibly due to disruption of GW182-Ago2 interaction by a dominant-negative effect (Jakymiw et al. 2005).
C-terminal half of Ago2 preserved the silencing function of Ago2 probably because it maintained the interaction with GW182
Our GW182-Ago2 interaction mapping showed that the C-terminal half of Ago2 (amino acids 478–860) was sufficient for the binding with GW182, whereas the N-terminal half (amino acids 1–480) was not required. Interestingly, only the C-terminal half of Ago2 preserved the silencing function of Ago2 when directly brought to the 3′-UTR of target mRNA. The silencing function mediated by Ago2 or the C-terminal half of Ago2 was abolished upon GW182 knockdown. Our data strongly suggested that interaction of Ago2 with GW182 is critical for the silencing process mediated by Ago2 at the 3′-UTR of target mRNA. This hypothesis is also supported by two recent studies where overexpressing the Ago-binding fragment of yeast or Drosophila ortholog of GW182 greatly disrupted GW182-Ago interaction and significantly impaired miRNA-mediated silencing in vitro and in vivo (Eulalio et al. 2008; Till et al. 2007).
Furthermore, our recent study showed that the repression effect caused by tethering GW182 was independent of Ago2 (Li et al. 2008) and our data herein again suggested that Ago2 is not the “final repressor” because its silencing function relied greatly on GW182. Interestingly, the PIWI domain of Ago2 was reported to be responsible for the interaction with Dicer (Tahbaz et al. 2004). It would be interesting to determine if Dicer and GW182 compete for the binding to Ago2 through the PIWI domain.
Based on the data from current study, we propose a model for miRNA-mediated gene silencing in which, after miRNA guides the Ago-miRNA complex to the 3′-UTR of target mRNA, Ago protein recruits GW182 to stabilize Ago-miRNA-mRNA binding and represses translation. In addition, GW182-Ago2 interaction recruits Ago2 to GWB, which accumulate many Ago2-miRNA-mRNA complexes and become centers for miRNA-mediated silencing. We speculate that the ability of GW182 to potentially bind multiple Ago proteins may contribute to aggregation and formation of the cytoplasmic foci GW/P bodies, which have been implicated to be critical components of miRNA activity.
MATERIALS AND METHODS
Construction of deletion constructs of GW182 and Ago2
The details of constructing GW1Δ1 (amino acids 254–751, formerly known as GW182Δ1), TNR (amino acids 1–204), Ago2 (amino acids 1–860), and PIWI (amino acids 478–860) were described previously (Jakymiw et al. 2005; Li et al. 2008). GW1Δ1a (amino acids 254–503), GW1Δ1b (amino acids 502–751), GW1Δ7 (amino acids 1034–1962), GW1Δ12 (amino acids 895–1211), and GW1Δ1a truncated constructs (W1-2, W2-3, W3-4, W4-5) were cloned from cDNA of full-length GW182 or its deletion constructs by PCR (see Supplemental Materials and Methods for information of primers and conditions). The human Ago3 mutant (Ago3m) in the pCMV-SPORT6 vector was obtained from Invitrogen (Clone number: CS0DB008YP10). Ago3m sequence was cloned by PCR (see Supplemental Materials and Methods for primers and condition) to adapt to Gateway cloning system (Invitrogen). The products from the above PCR reactions were then cloned into pDONR207 (Invitrogen) using the Gateway BP recombination reaction as per the manufacturer's instructions (Invitrogen). To construct pENTR-GW1Δ5 (amino acids 1670–1962), pENTR-GW182 was digested with SalI (5′ end linker) and SpeI (nt5008) to release a 4.2-kb vector fragment containing the C terminus of GW182. The overhangs of this fragment were filled in and then ligated. To construct pENTR-GW1Δ10 (amino acids 566–1343), phrGFP-KIAA1460 was digested with XhoI (5′ end linker) and SmaI to release a 2.3-kb fragment, which was then subcloned into the SalI and EcoRV sites of pENTR2B (Invitrogen). The cDNA of GW1Δ1a mutant (tryptophan > alanine, W > A) was directly synthesized and subcloned in pUC57 vector by GenScript using the sequence as shown in the Supplemental Materials and Methods. The mutated gene sequence was moved to pDONR207 vector (Invitrogen) using Gateway BP recombination for further applications. To construct pENTR-PAZ (amino acids 1–480), pENTR-Ago2 was digested with XhoI (nt1467) and XhoI (3′ end linker) to generate the vector fragment containing the N-terminal half of Ago2, which was purified and then ligated. To construct pENTR-Ago1, EST clone pBluescript hAgo1 was first digested with BamHI (3′ end linker) and the overhang was filled in to generate a blunt 3′ end. Then the digested product was cut by KpnI (5′ end linker) to generate a 4.0-kb fragment which was subcloned into the KpnI and EcoRV sites of pENTR1A (Invitrogen). To construct pENTR-Ago4, EST clone pBluescript hAgo4 was digested with SmaI (5′ end linker) and ScaI (3′ end linker) to generate a 3.5-kb fragment which was then subcloned into the DraI and EcoRV sites of pENTR1A (Invitrogen). All of the variants used in current study were subcloned into Gateway compatible GST, GFP, 3xFlag, or NHA (Li et al. 2008) vectors by using Gateway LR recombination reaction (Invitrogen). pIreSneo-Flag/HA Ago3 was obtained from Thomas Tuschl (Meister et al. 2004) through Addgene. The tethering assay plasmids including the pClneo-NHA vector, NHA-Ago2, Renilla luciferase RL-5BoxB, and firefly luciferase (FL) were kind gifts from Dr. Witold Filipowicz (Friedrich Miescher Institute for Biomedical Research) (Pillai et al. 2004). All DNA constructs used in this study were confirmed by direct DNA sequencing.
Antibodies
Rabbit anti-Ago2 and rabbit anti-GST were gifts from Dr. Tom Hobman (University of Alberta) and Dr. Peter Sayeski (University of Florida), respectively. Mouse monoclonal anti-HA was purchased from Covance. Mouse monoclonal anti-Flag M2 and antitubulin were purchased from Sigma-Aldrich. Rabbit polyclonal anti-GFP was purchased from Invitrogen.
Plasmid transfection, GST pull-down, and Western blot analysis
HeLa cells were cultured in DMEM containing 10% fetal bovine serum in a 37°C incubator with 5% CO2. HeLa cells were grown to 90%–100% in six-well plates at the day of transfection. GST-tagged construct was singly transfected or cotransfected with other tagged constructs into HeLa cells using Lipofectamine 2000 (Invitrogen) as per the manufacturer's instructions for 24 h. To detect expression of GST-tagged proteins in whole cell lysate, the cells were lysed in Laemmli sample buffer directly. For most of the GST pull-down assay, the cells were lysed with NET/0.3% NP40 buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris at pH 7.4, 3% NP40) with Complete Protease Cocktail Inhibitor (Roche Diagnostics) and then sonicated at 20% amplitude for 10 sec three times on ice. The GST pull-down assay in Figure 5 was performed under conditions that followed a published protocol (Till et al. 2007). Afterward, the lysates were centrifuged at 13,200 rpm for 5 min. The pellets (insoluble fractions) were lysed in Laemmli sample buffer directly. The soluble fractions were incubated with Glutathione Sepharose 4B (GE Healthcare) and mixed at 4°C for 2 h for GST pull-down. After the incubation, the beads were washed with NET/0.3% NP40 buffer for four times and the samples eluted in Laemmli sample buffer. The soluble fraction of cell lysates (input), GST pull-down samples, whole cell lysates, and insoluble fractions were separated on 10% polyacrylamide gel and transferred to nitrocellulose. Western blotting was performed as described previously (Lian et al. 2007). The dilutions of primary antibodies were: 1:1000 for anti-GST, 1:400 for anti-Flag, 1:1000 for anti-GFP, 1:500 for anti-Ago2, and 1:1000 for anti-HA.
Indirect immunofluorescence
Cell were fixed and permeabilized as described previously (Jakymiw et al. 2005; Lian et al. 2007). The dilution of the anti-Flag antibody was 1:1000.
Tethering assay using a dual luciferase system
HeLa cells were grown to about 90%–100% confluence in 24-well plates on the day of transfection. To determine the effect of tethering PIWI (amino acids 478–860) and PAZ (amino acids 1–480), the cells were transfected with 0.1 ng of constructs expressing reporter Renilla luciferase (RL-5BoxB), 100 ng of control firefly luciferase (FL), plus 700 ng of NHA tag, NHA-Ago2, NHA-PIWI, or NHA-PAZ using Lipofectamine 2000 (Invitrogen) for 48 h as per the manufacturer's instructions. Cells were harvested 48 h after transfection, and the FL and RL activities were measured using Dual-Luciferase Reporter Assay System (Promega), following the manufacturer's protocol. Relative luciferase activities (ratio of targeted luciferase activities over control luciferase activities) were calculated as described previously (Pillai et al. 2004). Briefly, FL/RL activity in NHA vector transfected (control) group was regarded as 0% translational repression. The repression levels of other experimental groups were calculated by the percentage reduction of relative luciferase activities compared with that in the NHA control group. The assay was performed in triplicate and repeated for 2–3 times. To detect the expressions of tethered NHA-tagged proteins, the above cell lysates were mixed at a 1:1 ratio with Laemmli sample buffer, and Western blot was performed as described in “Western blot analysis” of the Materials and Methods.
RNAi and Quantitative Real-Time PCR
The sequence of siRNA for GW182 or for GFP was described previously (Lian et al. 2007). To determine how GW182 knockdown affects the Ago2- or PIWI-mediated suppression, cells were grown to 30%–50% confluence and were transfected with 100 nM of siRNA for GW182, or siRNA for GFP as a negative control using Lipofectamine 2000 (Invitrogen). Thirty hours later, these cells were transfected again with constructs expressing reporter RL-5BoxB, control FL, and NHA-tagged constructs as described above. Forty-eight hours after the second transfection, total RNA was extracted from HeLa cells using RNeasy Mini Kit (Qiagen). RNase-Free DNase Set (Qiagen) was applied to eliminate potential DNA contamination. The relative mRNA level of GW182 was measured in duplicate using the ΔΔCt method (Livak and Schmittgen 2001) and TaqMan Fast Universal Master Mix (Applied Biosystems) with the corresponding TaqMan Gene Expression Assay (TNRC6A, Hs00379422_m1, Applied Biosystems). The level of 18S rRNA was measured as internal control (18S rRNA, 4310893E, Applied Biosystems). The melting curve in each individual measurement was monitored to guard against nonspecific amplification.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. Witold Filipowicz (Friedrich Miescher Institute for Biomedical Research) for providing tethering assay plasmids, Dr. Gordon K.T. Chan (University of Alberta) for providing expression constructs and GST protein controls, Dr. Tom Hobman (University of Alberta) for providing rabbit antibody to hAgo2, and Dr. Peter Sayeski (University of Florida) for the rabbit anti-GST antibody. This work was supported in part by Canadian Institutes for Health Research Grant MOP-38034, NIH Grant AI47859, and the Andrew J. Semesco Foundation. M.J.F. holds the Arthritis Society Chair. S.L. and S.L.L. are supported by NIDCR Oral Biology Training Grant T32 DE007200.
Footnotes
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; FL, firefly luciferase; GFP, green fluorescence protein; GWB, GW/P bodies; IIF, indirect immunofluorescence; NHA, hemagglutinin tag with a 22-amino acid-long N peptide specifically recognizing the BoxB hairpin; PCR, polymerase chain reaction; RL, Renilla luciferase; miRNA, microRNA; nt, nucleotide; siRNA, small interfering.RNA.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1229409.
REFERENCES
- Behm-Ansmant I., Rehwinkel J., Doerks T., Stark A., Bork P., Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Teixeira D., Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Spencer A., Morita K., Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M.A., Jacobsen S.E., Cooke R., Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes & Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Huntzinger E., Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Eystathioy T., Chan E.K.L., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T., Jakymiw A., Chan E.K.L., Seraphin B., Cougot N., Fritzler M.J. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulski J.K., Vos L., Sun X., Chan G. Stable hZW10 kinetochore residency, mediated by hZwint-1 interaction, is essential for the mitotic checkpoint. J. Cell Biol. 2008;180:507–520. doi: 10.1083/jcb.200708021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K.L. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Legault P., Li J., Mogridge J., Kay L.E., Greenblatt J. NMR structure of the bacteriophage λ N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li S., Lian S.L., Moser J.J., Fritzler M.L., Fritzler M.J., Satoh M., Chan E.K. Identification of GW182 and its novel isoform TNGW1 as translational repressors in Ago2-mediated silencing. J. Cell Sci. 2008;121:4134–4144. doi: 10.1242/jcs.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian S., Jakymiw A., Eystathioy T., Hamel J.C., Fritzler M.J., Chan E.K.L. GW bodies, microRNAs, and the cell cycle. Cell Cycle. 2006;5:242–245. doi: 10.4161/cc.5.3.2410. [DOI] [PubMed] [Google Scholar]
- Lian S., Fritzler M.J., Katz J., Hamazaki T., Terada N., Satoh M., Chan E.K.L. Small interfering RNA-mediated silencing induces target-dependent assembly of GW/P bodies. Mol. Biol. Cell. 2007;18:3375–3387. doi: 10.1091/mbc.E07-01-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Liu J., Rivas F.V., Wohlschlegel J., Yates J.R., Parker R., Hannon G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pauley K.M., Eystathioy T., Jakymiw A., Hamel J.C., Fritzler M.J., Chan E.K.L. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Artus C.G., Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Behm-Ansmant I., Gatfield D., Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M.D., Najand N., Chaker S., Pare J.M., Haskins J., Hughes S.C., Hobman T.C., Locke J., Simmonds A.J. Gawky is a component of cytoplasmic mRNA processing bodies required for early Drosophila development. J. Cell Biol. 2006;174:349–358. doi: 10.1083/jcb.200512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen G.L., Blau H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahbaz N., Kolb F.A., Zhang H., Jaronczyk K., Filipowicz W., Hobman T.C. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till S., Lejeune E., Thermann R., Bortfeld M., Hothorn M., Enderle D., Heinrich C., Hentze M.W., Ladurner A.G. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- Yang Z., Jakymiw A., Wood M.R., Eystathioy T., Rubin R.L., Fritzler M.J., Chan E.K.L. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- Yuan Y.R., Pei Y., Ma J.B., Kuryavyi V., Zhadina M., Meister G., Chen H.Y., Dauter Z., Tuschl T., Patel D.J. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]