Abstract
A main unsolved problem in the RNA World scenario for the origin of life is how a template-dependent RNA polymerase ribozyme emerged from short RNA oligomers obtained by random polymerization on mineral surfaces. A number of computational studies have shown that the structural repertoire yielded by that process is dominated by topologically simple structures, notably hairpin-like ones. A fraction of these could display RNA ligase activity and catalyze the assembly of larger, eventually functional RNA molecules retaining their previous modular structure: molecular complexity increases but template replication is absent. This allows us to build up a stepwise model of ligation-based, modular evolution that could pave the way to the emergence of a ribozyme with RNA replicase activity, step at which information-driven Darwinian evolution would be triggered.
Keywords: RNA folding, structural motif, modular evolution, RNA ligation, hairpin ribozyme, RNA polymerase
INTRODUCTION
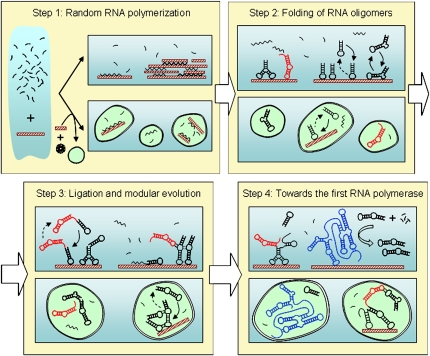
The RNA World model (Gilbert 1986), already suggested by Woese, Crick, and Orgel in the decade of the 1960s, envisages a plausible scenario for the origin and early evolution of life. It propounds that DNA-based life arose from an earlier world where RNA molecules stored genetic information and catalyzed biochemical reactions (reviewed in Joyce 2002; Joyce and Orgel 2006). The main support to this hypothesis comes from the discovery of a growing number of ribozymes in extant organisms, as well as from the possibility of selecting different ribozymes and aptamers by means of in vitro evolution experiments (Joyce 2004; Stoltenburg et al. 2007). Still, one of the relevant open questions for the establishment of an RNA World regards how the first template-dependent RNA polymerase ribozyme could have emerged. Orgel (2004) suggested that “one must postulate that a library of RNA strands with different sequences formed spontaneously on the primitive Earth and that this family of sequences included catalysts able to support self-replication of RNA,” and Joyce and Orgel (2006) argued that “The RNA-first view of the origin of life assumes […] that a catalyst existed to convert the activated nucleotides to an ensemble of random-sequence polynucleotides, a subset of which have the ability to replicate.” Nevertheless, up to now, no RNA polymerase activity has been found among the short RNA sequences that can be generated by nonenzymatic random polymerization. Indeed, a minimum size of about 165 nucleotides (nt) has been experimentally established for such a template-dependent RNA polymerase molecule (Johnston et al. 2001; Joyce 2004), a length three to four times that of the longest RNA oligomers obtained by random polymerization of activated ribonucleotides on clay mineral surfaces (Huang and Ferris 2003, 2006). Therefore, understanding how the RNA World could have arisen requires the investigation of how functional molecules may appear in the absence of template replication. Clues to this transition come from simple models where the degeneration between molecular genotype and phenotype is taken into account, since many different and unrelated sequences may fold into the same structure and eventually perform the same function. Among them, models using RNA secondary structure as a simplified representation of the phenotype are particularly relevant (Schuster et al. 1994; Hazen et al. 2007). This is the starting point of our stepwise model of ligation-based, modular evolution of RNA schematized in Figure 1, and developed in this contribution.
FIGURE 1.
Schematic representation of the stepwise process toward a template-dependent RNA polymerase. In every step we depict two possible and compatible scenarios: evolution on mineral surfaces (shown as brown rectangles) in bulk solution, as well as evolution inside vesicles that could also encapsulate mineral particles. Functional hairpin structures (with ligase activity) are shown in red. Solid and dotted arrows stand for the surface-bound to in-solution equilibria. The RNA polymerase emerging from this process is depicted in blue. Further details are given in the main text.
A STEPWISE MODEL FOR THE ORIGIN OF THE RNA WORLD
Step 1: Abiotic polymerization of RNA oligomers
The catalytic role of minerals and metal ions in the abiotic synthesis of biopolymers has been thoroughly investigated during the last decades (Monnard 2005; Ferris 2006). In particular, much effort has been put into studying the outstanding catalytic properties of montmorillonite, an abundant phyllosilicate believed to have formed large deposits on the early Earth (Ferris 2006; Joshi et al. 2007). Montmorillonite catalyzes the formation of single-stranded (ss) RNA oligomers from ribonucleotides whose 5′-phosphate group is activated either by imidazole (Ferris and Ertem 1993; Ferris 2002) or by 1-methyladenine (Huang and Ferris 2003). The polymerization of up to 50 mers of poly(A) and poly(U) in “feeding reactions” has been described (Ferris 2002), as well as the formation of 30 to 40 mer RNA homopolymers in the absence of a primer (Huang and Ferris 2003). More recently, the montmorillonite-catalyzed reaction of methyladenine-activated monomers of the four ribonucleotides has been reported, yielding RNA oligomers of up to 20 nt long (Huang and Ferris 2006).
In parallel, montmorillonite can also catalyze the formation of vesicles from simple fatty acids supplied as micelles (Hanczyc et al. 2003). Interestingly, clay particles, even with RNA molecules adsorbed on them, can be encapsulated within vesicles, which provide compartmented environments for further biochemical reactions. Such vesicles can grow by incorporating fatty acid micelles, fuse to each other, and divide by extrusion through small pores. Hence, montmorillonite can help supplying two key components of life: nucleic acids, further useful as a genetic material, and a protocell-like compartment. Vesicle membranes composed of simple amphiphilic molecules show enough permeability to nucleotides to allow nucleic acid elongation inside the protocell (Mansy et al. 2008). For the purpose of the model presented here, the reactions leading to a modular evolution of RNA could have operated on mineral surfaces or interlayers in contact with a bulk aqueous medium, as well as inside vesicles.
What diversity of sequences and molecular structures could random polymerization supply? Little is actually known about the sequence composition and variability that could have been originated by RNA polymerization on montmorillonite, besides the observation that in short oligomers (2–5 mer) the relative sequence abundance is biased with respect to the values expected by random synthesis (Miyakawa and Ferris 2003; Ertem et al. 2007). More relevant is the repertoire of RNA secondary structures, the precursors of chemical function, originated in a pool of random sequences. Insights into the distribution of structural motifs can be obtained from theoretical studies (Fontana et al. 1993; Knight and Yarus 2003; Gevertz et al. 2005) and from analyses of the repertoire of secondary structures present in extant RNA molecules (Bourdeau et al. 1999; Hendrix et al. 2005).
Step 2: Folding of the RNA oligomers and ubiquity of hairpin structures
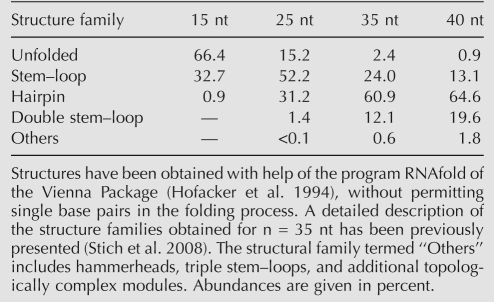
The folding process of RNA oligomers obtained by template-independent polymerization on mineral surfaces can be computationally studied. We have analyzed the structural properties of large pools of random ssRNA sequences of lengths 15, 25, 35, and 40 nt (Table 1). The minimum free energy structures obtained for every pool are ranked according to their frequency and classified into different structure families. Topologically simple RNA modules turn out to be the most abundant ones, especially stem–loops (composed of two basic structural motifs: one stem and one hairpin loop) and hairpin structures (being the simplest one formed by two stems joined by a bulge or an interior loop, and a terminal hairpin loop), as previously defined (Stich et al. 2008). Further, a strong correlation is found between nucleotide composition within each family and its structural complexity: abundant and topologically simple structures arise from sequences depleted in G, while rare and more complex structures (e.g., hammerheads) require enrichment in G. It has been experimentally proven that the longest RNA oligomers polymerized on montmorillonite correspond to poly-A, poly-U, and poly-(AU) chains (Huang and Ferris 2003, 2006), while the inclusion of G seems to produce shorter homo- or heteropolymers. Although indirectly, these results suggest that the first mineral-catalyzed RNA oligomers might have been depleted in G, and according to our computational results, the pool of secondary structures produced would have been further biased toward topologically simple modules, such as hairpin-like ones.
TABLE 1.
Abundance of structure families in pools of 108 (6.4 × 107 for n = 40 nt) random RNA oligomers of different lengths: 15, 25, 35, and 40 nt
Hairpin structures of different sizes are common in current viral and cellular RNAs, being involved in RNA–RNA and RNA–protein interactions that guide RNA folding, ribozyme function, RNP structure, and gene expression regulation (Svoboda and Di Cara 2006). Certain hairpin-like structures are endowed with ribozyme activity able to catalyze RNA cleavage/ligase reactions in a reversible way, as originally described in plant virus satellite RNAs (Buzayan et al. 1986; Hampel and Tritz 1989). The typical hairpin ribozyme consists of a 35-nt-long hairpin structure (as topologically defined in the previous paragraph), with a 5′ dangling end of 14 nt (the substrate-binding site) and one overhanging nucleotide in 3′ (reviewed in Puerta-Fernández et al. 2003). The rate constant for ligation in the hairpin ribozyme is 10-fold higher than that for cleavage of a cognate substrate (Nesbitt et al. 1999), and physical–chemical conditions that stabilize hairpin structure allegedly promote ligation (Fedor 1999). Interestingly, the 3′–5′-ligase activity is conserved even in truncated and fragmented derivatives of the hairpin ribozyme (Vlassov et al. 2004). These experimental observations support the robustness of the ligase function in the hairpin ribozyme and, relevant for our model, allow us to hypothesize that stable hairpin structures (such as most of those produced upon nonenzymatic polymerization) could have been endowed with a certain degree of primordial RNA ligase activity.
The number of stable and active hairpin structures with a fixed sequence at the catalytic site emerging as the result of random polymerization can be estimated as follows. Suppose that the environment contains a fraction f nt of each type of nucleotide, with nt standing for A, G, C, or U, and that an active hairpin requires Na precise positions occupied by A, Ng by G, and so on. Then, the product h(N) fANa fGNg fCNc fUNu yields the fraction of hairpin structures with the correct composition at the specified sites. The function h(N) stands for the fraction of hairpin structures found in randomly polymerized sequences of length N. According to the results summarized in Table 1, the value of h(N) is around 0.5 for the lengths of interest. As an example, an active hairpin of length 50 nt with 17 fixed positions (as it is the case in the consensus hairpin ribozyme described in Puerta-Fernández et al. 2003) in an environment with no nucleotide imbalance (fA = fG = fC = fU = 1/4), will be found in one out of [0.5 (1/4)17]−1 ≈ 3.4 × 1010 molecules. Therefore, the expected number of active hairpin ribozymes in 1 mL of 1 mM solution of random 50-mers of ssRNA is about 1.8 × 107.
In the pre-cellular scenario proposed here, we are devising a dynamic environment where some RNA modules could easily migrate from the mineral to the aqueous solution, since adsorption onto mineral surfaces or into phyllosilicate interlamellar channels mainly involves weak noncovalent van der Waals interactions. Their strength is in principle proportional to the length of the polymer, though it can be tuned by the RNA structure of the adsorbed molecule. A fraction of either the short original modules (e.g., some stem–loops) or the ligated products could have shown aptamer activities allowing a tighter binding to the mineral surface, and therefore increasing the ratio of surface-bound versus in-solution molecule. Although not experimentally observed yet, the likely existence of RNA aptamers with mineral-binding activity is supported by the possibility to select in vitro active RNA molecules with enhanced metal binding affinity (Gugliotti et al. 2004; Liu et al. 2006). Also, additional metal or mineral-binding RNAs could be discovered since a growing number of RNA aptamers, usually 12–60 nt long, are currently selected against many organic and inorganic ligands (Stoltenburg et al. 2007). Such an eventual aptamer–mineral interaction, though not a prerequisite for the feasibility of our model, could have represented a primordial selective pressure toward separating surface bound RNA modules from the dissolved ones. This could allow different groups of RNA molecules to be involved in subsequent ligation reactions in distinct structured micro- or nanoenvironments. An analogous repertoire of different RNA modules could have been provided by heterogeneous ensembles of RNA-containing vesicles. The availability of pools of different RNA modules for further reactions could have been influenced by several factors including fluctuations in temperature, freezing/melting cycles, variations in the ionic strength and metal concentration, evaporation processes, as well as watering cycles in tidal regions. In particular, drying and wetting cycles could have provided a favorable scenario since they promote the ligase activity of the hairpin ribozyme (Kazakov et al. 2006) and, in parallel, foster the capture of macromolecules by membrane-bounded vesicles (Deamer 1998).
Step 3: Ligation-based modular evolution of RNA
Pools of random RNA sequences are abundant in hairpin structures, and a fraction of them may display RNA ligase activity. Hairpin ubiquity and activity set the basis for a progressive structural and functional complexation of RNA molecules. Computer simulations indicated that upon hairpin-catalyzed ligation of two preformed RNA modules, up to 1% of the assembled molecules retain the previous modular structure of their building blocks (Manrubia and Briones 2007). This RNA thermodynamics-driven observation suggests that hairpin ribozymes, both as individual modules and in combined structures, could have catalyzed the synthesis of progressively longer RNA molecules from short and structurally simple modules. The ligase ability of hairpin ribozymes linked to other RNA molecules has been experimentally demonstrated by combining two ribozyme units into one functional twin molecule (Ivanov et al. 2005). In any case, our in silico RNA folding study indicated that most ligation events yield molecules whose minimum free energy structure does not retain that of the two appended RNA modules. The alternative structures originated by the progressively longer RNA molecules increase the repertoire of newly accessible structural variants (such as large hammerheads, tRNA-like structures or pseudoknot-promoting conformations), on hand to undergo further ligation events.
Additional processes could have increased the diversity of the pool of available RNA modules. Among them, nonenzymatic, metal–ion-catalyzed RNA polymerization could have occurred both in template-independent and template-directed reactions, as experimentally shown (reviewed in Joyce and Orgel 2006; Ferris 2006). In particular, some of the abundant hairpin and stem–loop structures could have been elongated in the presence of metal–ion catalysts, using their 5′-overhangs as the templating sequence, through a process similar to that described in the eutectic phase in water–ice (Monnard and Szostak 2008). This mechanism could have led to the subsequent metal–ion-catalyzed polymerization of certain modules into their continuous fully complementary strands, again increasing the repertoire of RNA sequences and structures available.
During this stage, the abundant stem–loop and hairpin modules could have promoted different kinds of intra- and intermolecular tertiary interactions through loop–loop (“kissing”) contacts (Horiya et al. 2003). Kissing-loop interactions, as well as other kinds of long-range tertiary interactions or pseudoknots, could have increased even more the repertoire of modules noncovalently joined to the nascent multimodular RNA structure, some of which could have been covalently ligated to it in further stages of the stepwise process. The experimental structural analysis of random, 85 nt-long RNA polymers has shown that unevolved sequences frequently acquire compact folds characterized by specific secondary structures and, in general, one or a few stable tertiary interactions (Schultes et al. 2005).
Ligation of RNA molecules eventually occurred in contemporary microenvironments, causing a shift from a previous situation driven by the competition among modules to a new framework where the assembled modules constituted selectable molecules in subsequent evolutionary steps.
Step 4: Toward a template-dependent RNA polymerase
Simple hairpin ligases enriched with additional RNA modules could have led to more evolved ligase ribozymes. The possibility of evolving complex and highly active RNA ligases has been documented in vitro, using as the starting material pools of random RNA sequences (Ekland et al. 1995), natural ribozymes such as group I introns (Jaeger et al. 1999), or previously evolved RNA ligases (Voytek and Joyce 2007). Among the first group, the L1 ligase ribozyme constitutes a relevant example of the feasibility of our model: a relatively small ligase ribozyme can be linked to an aptameric domain that responds to small organic effectors, in such a way that the resulting aptazyme retains the two functional modules and acquires the potential for allosteric regulation (Robertson and Scott 2007). Additionally, other ligases could have emerged and eventually established self-replicating RNA ligase networks. An example is provided by in vitro evolved derivatives of the 61-nt-long R3C ligase, a ribozyme able to ligate two substrate molecules to synthesize an exact copy of itself (Paul and Joyce 2002). It has been recently shown that improved, cross-replicating R3C derivatives can originate self-sustained amplification systems that grow exponentially, can compete for a common pool of substrates, and eventually select recombinant variants fitter than the parental ribozymes (Lincoln and Joyce 2009). Thus, different kinds of ligation-based genetic systems could have arisen, coexisted, and competed—in terms of catalytic activity, growth rate, and ability to withstand product inhibition—at this stage of the RNA World previous to the appearance of the first RNA polymerase.
In parallel, ligation-based modular evolution could have originated RNA molecules endowed with novel functionalities. Indeed, in vitro selection experiments have shown that the ligation and exchange of RNA structural domains through recombination can be used to engineer new functional RNAs (Burke and Willis 1998; Joyce 2004). It is of particular interest how the evolution of the class I ligase ribozyme (Bartel and Szostak 1993), appended with a track of 76 random nucleotides at its 3′ end, allowed the selection of a 189-nt-long template-dependent RNA polymerase ribozyme (Johnston et al. 2001). The efficiency of this ribozyme has been improved by further in vitro evolution (Zaher and Unrau 2007), as well as by appending it to micelle-forming hydrophobic molecules (Müller and Bartel 2008).
Therefore, the experimental evidence accumulated on the outstanding catalytic capabilities of in vitro evolved RNA molecules supports one of the main assumptions of the model presented here: a stepwise transition could have occurred from a simple hairpin ligase ribozyme (highly abundant upon folding of short, random monomers synthesized on mineral surfaces), through the progressive ligation and evolution of different RNA modules, to an RNA-dependent RNA polymerase that could be the substrate—and catalyst—of Darwinian evolution.
Once the first RNA polymerase was assembled, its template-dependent copying activity could trigger the replication of the pools of modules and multimodular molecules so far originated and maintained. This likely resulted in fast amplifications of the surviving and available molecules, representing the first “genotypes” whose sequence information could originate phylogenetically related lines of descent. Modular evolution confers an additional advantage once replication appears, namely, it substantially decreases the evolutionary time required to select small modules with respect to the time needed to obtain a combined, larger structure. Doubling the length of a sequence under selection requires a time about 10-fold larger for a given secondary structure to be fixed in the population (Manrubia and Briones 2007). As noticed, the primordial RNA-catalyzed RNA replication might have been associated to an error rate of about 1.1 × 10−2 (Johnston et al. 2001), promoting a thorough exploration of the available sequence space. This would foster the selection of advantageous mutations in different replicating functional molecules, including the RNA replicase itself. Thanks to that, the fidelity of the template-dependent polymerase ribozyme could have progressively increased, in an optimization process that also benefited from the concomitant ligation of modules selectable as regulatory domains. Progressively smaller mutation rates would permit the emergence of a complex ecological organization of quasispecies of replicators (Takeuchi and Hogeweg 2008) as well as the coevolution of populations of different functional RNA molecules. Additionally, the overall length of the evolving molecules would steadily increase, since genome length is strongly dependent on the accuracy in replication (Eigen 1971).
During this process, the fixation of point mutations in specific positions of functional hairpin ribozymes maintained in the multidomain RNAs could modulate their ligase versus RNase ratio (Gaur et al. 2008) and promote cleavage and domain shuffling in the protogenome-like organization of suitable evolutionary lines. Additionally, modular evolution could have produced bifunctional enzymes endowed with RNA ligase and cleavage functionalities in separate structural modules, as experimentally reported (Kumar and Joyce 2003). This could allow the integration of the ribozyme into a target RNA substrate following an intron-like behavior. Indeed, the heterologous recombination of preformed, short modular elements is a relevant mechanism for the evolution of RNA, as observed in current functional RNA molecules (Ancel and Fontana 2000; Pasquali et al. 2005), viroids (Flores et al. 2004), and RNA virus genomes (Cheng et al. 2005). Together, mutation and recombination constituted the basic evolutionary mechanisms allowing the appearance of RNA variants further selectable and replicable in a progressively successful RNA World.
CONCLUSIONS
Here we present a plausible model for the origin of an RNA World based on the random polymerization of short RNA oligomers that, by means of hairpin ligase-mediated modular evolution, could have led to the emergence of a ribozyme with template-dependent RNA polymerase activity. Once replication-driven evolution was initiated, the fixation of advantageous mutations could fine tune the evolutionary process, thus shaping the functional properties of the information-bearing RNA molecules.
It is unlikely that a single mechanism can account for the processes that eventually triggered an RNA-dominated period in the precellular history of life. Also, different evolutionary pathways were likely explored within different environments available in the early Earth. Nevertheless, our evolutionary model shows three main advantages with respect to previous hypotheses put forward for the origin of the RNA World, in particular, to the unrealistic spontaneous emergence of an RNA polymerase activity from a pool of short, random RNA oligomers. First, short RNA modules resulting from template-independent polymerization on surfaces might suffice to produce the first functional RNAs, being template replication not needed at this stage. Second, the abundance of hairpin structures in any random pool of short RNA oligomers enables the appearance of ligation activity to build progressively longer and functionally more complex RNA molecules. Third, modular evolution shortens adaptation times and allows attaining complex structures that could not be otherwise directly selected. Therefore, ligation-based modular evolution might have bridged the gap between the first random RNA oligomers and a template-dependent RNA polymerase ribozyme at the dawn of the RNA World.
Other limitations to the RNA World hypothesis remain to be solved and are out of the scope of our model, among them the prebiotic synthesis of RNA monomers and the limited stability of RNA molecules in solution (Joyce 2002; Ferris 2006), as well as the appealing possibility that certain nucleic acid analogs, such as peptide nucleic acids, could have been the first genetic molecules in a pre-RNA World (Joyce and Orgel 2006). As exemplified in this work, the interplay between in silico simulation and in vitro experimentation can provide valuable clues to unveil some of the riddles regarding the origin and early evolution of life.
ACKNOWLEDGMENTS
This work was supported by Ministerio de Ciencia e Innovación (grants BIO2007-67523 and FIS2008-05273), INTA, INSA, EU, and CAM.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1488609.
REFERENCES
- Ancel L.W., Fontana W. Plasticity, evolvability, and modularity in RNA. J. Exp. Zool. 2000;288:242–283. doi: 10.1002/1097-010x(20001015)288:3<242::aid-jez5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Bartel D.P., Szostak J.W. Isolation of new ribozymes of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- Bourdeau V., Ferbeyre G., Pageau M., Paquin B., Cedergren R. The distribution of RNA motifs in natural sequences. Nucleic Acids Res. 1999;27:4457–4467. doi: 10.1093/nar/27.22.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D.H., Willis J.H. Recombination, RNA evolution, and bifunctional RNA molecules isolated through chimeric SELEX. RNA. 1998;4:1165–1175. doi: 10.1017/s1355838298980542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzayan J.M., Gerlach W.L., Bruening G. Nonenzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature. 1986;323:349–353. [Google Scholar]
- Cheng C.P., Panavas T., Luo G., Nagy P.D. Heterologous RNA replication enhancer stimulates in vitro RNA synthesis and template-switching by the carmovirus, but not by the tombusvirus, RNA-dependent RNA polymerase: implication for modular evolution of RNA viruses. Virology. 2005;341:107–121. doi: 10.1016/j.virol.2005.06.042. [DOI] [PubMed] [Google Scholar]
- Deamer D.W. Membrane compartments in prebiotic evolution. In: Brack A., editor. The molecular origins of life. Cambridge University Press; Cambridge, UK: 1998. pp. 189–205. [Google Scholar]
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- Ekland E.H., Szostak J.W., Bartel D.P. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science. 1995;269:364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- Ertem G., Hazen R.M., Dworkin J.P. Sequence analysis of trimer isomers formed by montmorillonite catalysis in the reaction of binary monomer mixtures. Astrobiology. 2007;7:715–722. doi: 10.1089/ast.2007.0138. [DOI] [PubMed] [Google Scholar]
- Fedor M.J. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry. 1999;38:11040–11050. doi: 10.1021/bi991069q. [DOI] [PubMed] [Google Scholar]
- Ferris J.P. Montmorillonite catalysis of 30–50 mer oligonucleotides: laboratory demonstration of potential steps in the origin of the RNA world. Orig. Life Evol. Biosph. 2002;32:311–332. doi: 10.1023/a:1020543312109. [DOI] [PubMed] [Google Scholar]
- Ferris J.P. Montmorillonite-catalysed formation of RNA oligomers: The possible role of catalysis in the origins of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:1777–1786. doi: 10.1098/rstb.2006.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris J.P., Ertem G. Montmorillonite catalysis of RNA oligomer formation in aqueous solution. A model for the prebiotic formation of RNA. J. Am. Chem. Soc. 1993;115:12270–12275. doi: 10.1021/ja00079a006. [DOI] [PubMed] [Google Scholar]
- Flores R., Delgado S., Gas M.E., Carbonell A., Molina D., Gago S., De la Peña M. Viroids: The minimal noncoding RNAs with autonomous replication. FEBS Lett. 2004;567:42–48. doi: 10.1016/j.febslet.2004.03.118. [DOI] [PubMed] [Google Scholar]
- Fontana W., Konings D.A.M., Stadler P.F., Schuster P. Statistics of RNA secondary structures. Biopolymers. 1993;33:1389–1404. doi: 10.1002/bip.360330909. [DOI] [PubMed] [Google Scholar]
- Gaur S., Heckman J.E., Burke J.M. Mutational inhibition of ligation in the hairpin ribozyme: Substitutions of conserved nucleobases A9 and A10 destabilize tertiary structure and selectively promote cleavage. RNA. 2008;14:55–65. doi: 10.1261/rna.716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevertz J., Gan H.H., Schlick T. In vitro RNA random pools are not structurally diverse: A computational analysis. RNA. 2005;11:853–863. doi: 10.1261/rna.7271405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Origin of life—The RNA world. Nature. 1986;319:618. doi: 10.1038/319618a0. [DOI] [Google Scholar]
- Gugliotti L.A., Feldheim D.L., Eaton B.E. RNA-mediated metal-metal bond formation in the synthesis of hexagonal palladium nanoparticles. Science. 2004;304:850–852. doi: 10.1126/science.1095678. [DOI] [PubMed] [Google Scholar]
- Hampel A., Tritz R. RNA catalytic properties of the minimum (−)sTRSV sequence. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- Hanczyc M.M., Fujikawa S.M., Szostak J.W. Experimental models of primitive cellular compartments: Encapsulation, growth, and division. Science. 2003;302:618–622. doi: 10.1126/science.1089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen R.M., Griffin P.L., Carothers J.M., Szostak J.W. Functional information and the emergence of biocomplexity. Proc. Natl. Acad. Sci. 2007;104:8574–8581. doi: 10.1073/pnas.0701744104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix D.K., Brenner S.E., Holbrook S.R. RNA structural motifs: Building blocks of a modular biomolecule. Q. Rev. Biophys. 2005;38:221–243. doi: 10.1017/S0033583506004215. [DOI] [PubMed] [Google Scholar]
- Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer L.S., Tacker M., Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188. [Google Scholar]
- Horiya S., Li X., Kawai G., Saito R., Katoh A., Kobayashi K., Harada K. RNA LEGO: Magnesium-dependent formation of specific RNA assemblies through kissing interactions. Chem. Biol. 2003;10:645–654. doi: 10.1016/s1074-5521(03)00146-7. [DOI] [PubMed] [Google Scholar]
- Huang W., Ferris J.P. Synthesis of 35–40 mers of RNA oligomers from unblocked monomers. A simple approach to the RNA world. Chem. Commun. (Camb.) 2003;12:1458–1459. doi: 10.1039/b303134a. [DOI] [PubMed] [Google Scholar]
- Huang W., Ferris J.P. One-step, regioselective synthesis of up to 50-mers of RNA oligomers by montmorillonite catalysis. J. Am. Chem. Soc. 2006;128:8914–8919. doi: 10.1021/ja061782k. [DOI] [PubMed] [Google Scholar]
- Ivanov S.A., Vauléon S., Müller S. Efficient RNA ligation by reverse-joined hairpin ribozymes and engineering of twin ribozymes consisting of conventional and reverse-joined hairpin ribozyme units. FEBS J. 2005;272:4464–4474. doi: 10.1111/j.1742-4658.2005.04865.x. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Wright M.C., Joyce G.F. A complex ligase ribozyme evolved in vitro from a group I ribozyme domain. Proc. Natl. Acad. Sci. 1999;96:14712–14717. doi: 10.1073/pnas.96.26.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston W.K., Unrau P.J., Lawrence M.S., Glasner M.E., Bartel D.P. RNA-catalyzed RNA polymerization: Acurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- Joshi P.C., Pitsch S., Ferris J.P. Selectivity of montmorillonite catalyzed prebiotic reactions of D, L-nucleotides. Orig. Life Evol. Biosph. 2007;37:3–26. doi: 10.1007/s11084-006-9013-x. [DOI] [PubMed] [Google Scholar]
- Joyce G.F. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- Joyce G.F. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- Joyce G.F., Orgel L.E. Progress toward understanding the origin of the RNA world. In: Gesteland R.F., et al., editors. The RNA world. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 23–56. [Google Scholar]
- Kazakov S.A., Balatskaya S.V., Johnston B.H. Ligation of the hairpin ribozyme in cis induced by freezing and dehydration. RNA. 2006;12:446–456. doi: 10.1261/rna.2123506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R., Yarus M. Finding specific RNA motifs: Function in a zeptomole world? RNA. 2003;9:218–230. doi: 10.1261/rna.2138803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R.M., Joyce G.F. A modular, bifunctional RNA that integrates itself into a target RNA. Proc. Natl. Acad. Sci. 2003;100:9738–9743. doi: 10.1073/pnas.1334190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln T.A., Joyce G.F. Self-sustained replication of an RNA enzyme. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Gugliotti L.A., Wu T., Dolska M., Tkachenko A.G., Shipton M.K., Eaton B.E., Feldheim D.L. RNA-mediated synthesis of palladium nanoparticles on Au surfaces. Langmuir. 2006;22:5862–5866. doi: 10.1021/la060426c. [DOI] [PubMed] [Google Scholar]
- Manrubia S.C., Briones C. Modular evolution and increase of functional complexity in replicating RNA molecules. RNA. 2007;13:97–107. doi: 10.1261/rna.203006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansy S.S., Schrum J.P., Krishnamurthy M., Tobé S., Treco D.A., Szostak J.W. Template-directed synthesis of a genetic polymer in a model protocell. Nature. 2008;454:122–125. doi: 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa S., Ferris J.P. Sequence- and regioselectivity in the montmorillonite-catalyzed synthesis of RNA. J. Am. Chem. Soc. 2003;125:8202–8208. doi: 10.1021/ja034328e. [DOI] [PubMed] [Google Scholar]
- Monnard P.A. Catalysis in abiotic structured media: An approach to selective synthesis of biopolymers. Cell. Mol. Life Sci. 2005;62:520–534. doi: 10.1007/s00018-004-4342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnard P.A., Szostak J.W. Metal-ion catalyzed polymerization in the eutectic phase in water-ice: A possible approach to template-directed RNA polymerization. J. Inorg. Biochem. 2008;102:1104–1111. doi: 10.1016/j.jinorgbio.2008.01.026. [DOI] [PubMed] [Google Scholar]
- Müller U.F., Bartel D.P. Improved polymerase ribozyme efficiency on hydrophobic assemblies. RNA. 2008;14:552–562. doi: 10.1261/rna.494508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt S.M., Erlacher H.A., Fedor M.J. The internal equilibrium of the hairpin ribozyme: Temperature, ion, and pH effects. J. Mol. Biol. 1999;286:1009–1024. doi: 10.1006/jmbi.1999.2543. [DOI] [PubMed] [Google Scholar]
- Orgel L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004;39:99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- Pasquali S., Gan H.H., Schlick T. Modular RNA architecture revealed by computational analysis of existing pseudoknots and ribosomal RNAs. Nucleic Acids Res. 2005;33:1384–1398. doi: 10.1093/nar/gki267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul N., Joyce G.F. A self-replicating ligase ribozyme. Proc. Natl. Acad. Sci. 2002;99:12733–12740. doi: 10.1073/pnas.202471099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerta-Fernández E., Romero-López C., Barroso-delJesús A., Berzal-Herranz A. Ribozymes: Recent advances in the development of RNA tools. FEMS Microbiol. Rev. 2003;27:75–97. doi: 10.1016/S0168-6445(03)00020-2. [DOI] [PubMed] [Google Scholar]
- Robertson M.P., Scott W.G. The structural basis of ribozyme-catalyzed RNA assembly. Science. 2007;315:1549–1553. doi: 10.1126/science.1136231. [DOI] [PubMed] [Google Scholar]
- Schultes E.A., Spasic A., Mohanty U., Bartel D.P. Compact and ordered collapse of randomly generated RNA sequences. Nat. Struct. Mol. Biol. 2005;12:1130–1136. doi: 10.1038/nsmb1014. [DOI] [PubMed] [Google Scholar]
- Schuster P., Fontana W., Stadler P.F., Hofacker I. From sequences to shapes and back: A case study in RNA secondary structures. Proc. R. Soc. Lond. B. Biol. Sci. 1994;255:279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- Stich M., Briones C., Manrubia S.C. On the structural repertoire of pools of short, random RNA sequences. J. Theor. Biol. 2008;252:750–763. doi: 10.1016/j.jtbi.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Stoltenburg R., Reinemann C., Strehlitz B. SELEX-a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Svoboda P., Di Cara A. Hairpin RNA: A secondary structure of primary importance. Cell. Mol. Life Sci. 2006;63:901–918. doi: 10.1007/s00018-005-5558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi N., Hogeweg P. Evolution of complexity in RNA-like replicator systems. Biol. Direct. 2008;3:11. doi: 10.1186/1745-6150-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov A.V., Johnston B.H., Landweber L.F., Kazakov S.A. Ligation activity of fragmented ribozymes in frozen solution: Implications for the RNA world. Nucleic Acids Res. 2004;32:2966–2974. doi: 10.1093/nar/gkh601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytek S.B., Joyce G.F. Emergence of a fast-reacting ribozyme that is capable of undergoing continuous evolution. Proc. Natl. Acad. Sci. 2007;104:15288–15293. doi: 10.1073/pnas.0707490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher H.S., Unrau P.J. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA. 2007;13:1017–1026. doi: 10.1261/rna.548807. [DOI] [PMC free article] [PubMed] [Google Scholar]