Abstract
We present a novel Phi29 DNA polymerase application in RCA-based target RNA detection and analysis. The 3′→5′ RNase activity of Phi29 DNA polymerase converts target RNA into a primer and the polymerase uses this newly generated primer for RCA initiation. Therefore, using target RNA-primed RCA, padlock probes may be targeted to inner RNA sequences and their peculiarities can be analyzed directly. We demonstrate that the exoribonucleolytic activity of Phi29 DNA polymerase can be successfully applied in vitro and in situ. These findings expand the potential for detection and analysis of RNA sequences distanced from 3′-end.
Keywords: exoribonuclease, GAPDH, Phi29 DNA polymerase, RCA, RNA analysis, RNase
INTRODUCTION
Rolling circle amplification (RCA) is used for amplification of DNA, including the amplification and analysis of complete genomes (Niel et al. 2005; Haible et al. 2006), and also for single or multiplex detection of all macromolecules already identified in the course of genome projects: DNA, RNA, and proteins (Landegren et al. 2004). In these methods a DNA template for RCA is created by circularization of oligonucleotide(s) on complementary nucleic acid target(s) (Nilsson et al. 1997; Baner et al. 1998; Lizardi et al. 1998). RCA has been proven as a useful method for detection of not only isolated or preformed targets in vitro, but also in situ, for analysis of individual molecules in single cells (Christian et al. 2001; Larsson et al. 2004).
Detection of RNA molecules by RCA is performed routinely using antibody-coupled RCA (immuno-RCA) (Zhou et al. 2001), DNA oligonucleotide circularization on RNA (Christian et al. 2001), or cDNA analysis with padlock probes (Ericsson et al. 2008). All these methods are based on the formation of circular DNA padlock probes, which are used as templates for RCA.
Direct detection of RNA molecules by RCA, using target RNA as a primer for RCA, was previously described for miRNA (Jonstrup et al. 2006). The method was upgraded using “Turtle Probe” formation on target RNA technique and it was successfully applied in situ on nonpolyadenylated RNA molecules (Stougaard et al. 2007). The target RNA–circular DNA probe duplexes described in these publications were perfectly matched at the 3′-end of RNA molecules. The RNA sequences distant from the 3′-end have not been examined. However, in certain cases this application is very important, e.g., in detection and analysis of eukaryotic mRNA molecules, which have long poly(A) sequences at the 3′-end (Mijatovic et al. 2000).
The observation that Phi29 DNA polymerase exhibits 3′→5′ exoribonucleolytic activity prompted us to use the enzyme for target RNA conversion into a primer for RCA (Lagunavicius et al. 2008). Previously we have shown that the enzyme's RNase activity degraded RNA up to the double stranded probe hybridization region transforming target RNA into a primer, from which synthesis of DNA, complementary to the DNA template, begins in the presence of dNTPs.
In this report we demonstrate that the exoribonucleolytic activity of Phi29 DNA polymerase can be successfully applied in RCA-based detection of RNA molecules and analysis of their sequence peculiarities in vitro and in situ.
RESULTS AND DISCUSSION
The observation that Phi29 DNA polymerase exhibits RNase activity in the presence of dNTPs prompted us to use the enzyme's polymerization activity following 3′→5′ exoribonucleolytic conversion of target RNA into a primer for RCA.
Detection of target RNA in RNA mixtures using preformed padlock probe or target RNA-templated circularization of linear oligonucleotide technique
Previously we have described that Phi29 DNA polymerase synthesized RCA product from the RNA target after its 3′→5′ exonucleolytical degradation (Lagunavicius et al. 2008). The 111-nucleotide (nt)-long RNA transcript RNA2 was hybridized to a specific circular DNA padlock probe PP2 in a region more than 50 nt inwards from the 3′-end of RNA2 and was successfully converted into a primer, which later was used in RCA.
We chose the same RNA2-PP2 hybrid as a model system testing if Phi29 DNA polymerase was suitable to detect inner RNA sequences (distant from the 3′-end of RNA molecule) of individual transcripts in RNA mixtures. Phi29 DNA polymerase catalyzed RCA reactions were carried out in two ways: using preformed circular padlock probe or after target RNA templated circularization of linear oligonucleotide.
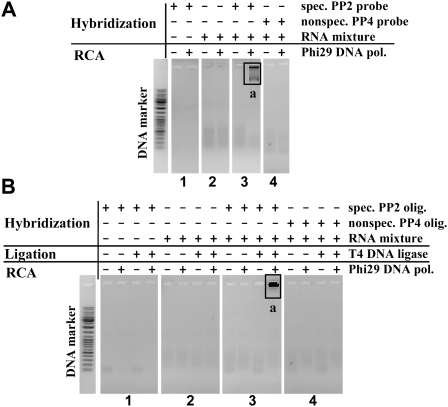
First, the initial RCA reactions with Phi29 DNA polymerase were performed using the preformed circular padlock probe PP2 and RNA Ladder, as RNA mixture, one fragment of which was RNA oligonucleotide RNA2. To verify reaction specificity, the control samples of free circular padlock probe PP2, free RNA mixture or RNA mixture with circular noncomplementary circular padlock probe PP4 were used. It was shown that the sample containing RNA mixture with specific padlock probe PP2 generated significant amounts of RCA product (Fig. 1A, panel 3, product a), while all control samples generated no RCA products (Fig. 1A, panels 1,2,4).
FIGURE 1.
Detection of target RNA molecules in RNA mixtures using preformed padlock probes (A) or circularization or target RNA-templated circularization of linear oligonucleotide technique (B). Reactions were carried out under the conditions described in Materials and Methods. Specific preformed PP2 probe (having complementary sequence to RNA2) or phosphorylated PP2 oligonucleotide (having complementary 3′- and 5′-ends to RNA2) mixtures with RNA LR Ladder (one fragment of which was RNA oligonucleotide RNA2) were used as testing samples (A,B, panels 3). Free preformed PP2 probe, phosphorylated PP2 oligonucleotide, RNA LR Ladder, or its mixture with nonspecific preformed PP4 probe (having no complementary sequence to RNA2) or nonspecific phosphorylated PP4 oligonucleotide (having no complementary sequence to RNA2) were used in other samples as controls of reaction specificity (A,B, panels 1,2,4). RCA reaction products were analyzed by electrophoresis through 1% agarose gel. The contexts of samples are shown above the gels' lines. a represents long RCA product.
Next, the detection of RNA transcripts could be also performed by RNA–templated DNA ligation technique using linear DNA probes and T4 DNA ligase. The literature data indicate that DNA ligases, such as T4 DNA ligase, in addition to conventional DNA substrates can catalyze the ligation of nicked DNA involved in complementary double stranded DNA–RNA structures (Nilsson et al. 2000; Christian et al. 2001; Nilsson et al. 2001). Such DNA padlock probes ligated on RNA targets could be used in analysis of RNA sequence peculiarities, including single nucleotide polymorphisms (SNPs), deletions, spliced transcripts, insertions, etc. Therefore, novel application of Phi29 DNA polymerase was also tested in ligation-based target RNA detection technique with the same RNA mixture–RNA LR Ladder. Linear specific padlock probe precursor, PP2 oligonucleotide, was used, 5′- and 3′-ends of which were designed to be complementary to RNA2 target. Our data revealed that the sample containing linear specific padlock probe PP2 precursor oligonucleotide and RNA mixture generated RCA product (Fig. 1B, panel 3, product a). All control samples, however, were negative (Fig. 1B, panels 1,2,4).
Detection of target RNA transcript in total cellular RNA
To test Phi29 DNA polymerase's ability to directly detect target transcripts within complex mixtures we chose to look for GAPDH transcripts in total RNA isolated from human HeLa cells. Control RT-PCR experiments confirmed that prepared total RNA contained GAPDH transcripts and was free of GAPDH encoding gene (data not shown).
GAPDH transcripts contain long poly(A) tails of about 70–220 adenylic residues (Mijatovic et al. 2000). Previously it was shown that Phi29 DNA polymerase has poor activity on RNA in double-stranded nucleic acid structures, but efficiently degrades poly(A) RNA substrate (Lagunavicius et al. 2008). Therefore, for RNA detection the target RNA sequence distant from 3′-end of RNA molecule was selected at the border of poly(A) tail.
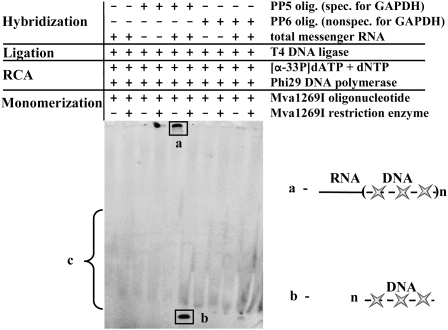
Linear specific padlock probe PP5 precursor oligonucleotide was used, 5′- and 3′-ends of which were designed to be complementary to GAPDH transcript. The RNA hybridization region overlapped with poly(A) region and comprised 12 A's of the tail and 18 nt upstream of them. After the padlock probe formation via ligation of hybridized precursor onto target RNA, the RCA reaction was performed and specific RCA product was detected by incorporation of labeled nucleotides into nascent RCA product. The specificity of the obtained RCA product was verified by hybridizing it with complementary DNA oligonucleotides (Mva1269I olig.) and forming double-stranded DNA duplexes containing Mva1269I restriction enzyme's recognition sites, which were later hydrolyzed by the same restriction endonuclease creating RCA product monomers.
Our data showed the presence of specific RCA products in the case of padlock probe PP5 precursor oligonucleotide (Fig. 2, products a,b). Except for reaction background (Fig. 2, product c), nonspecific oligonucleotide PP6 differing in few nucleotides in ligation junction region did not generate any other labeled RCA product. These results allowed us to conclude that in vitro direct detection of GAPDH transcript sequences distanced from 3′-end of RNA could be performed practicing both activities of Phi29 DNA polymerase: 3′–5′ single-strand RNA exoribonucleolytic and polymerization activities.
FIGURE 2.
Detection of GAPDH transcripts in total cellular RNA. Reactions were carried out under the conditions described in Materials and Methods. Phosphorylated PP5 oligonucleotide (specific for GAPDH) mixture with total mRNA was used as testing sample. Free PP5 oligonucleotide, free total mRNA, or its mixture with nonspecific oligonucleotide PP6 (differing from PP5 in few nucleotides in ligation junction region) were used in other samples as controls of reaction specificity. RCA reaction products were monomerized by cleavage with Mva1269I restriction enzyme and analyzed by electrophoresis through denaturing 8% polyacrylamide gel. The contexts of samples are shown above the gel lines. a and b represent labeled RCA products before and after monomerization with Mva1269I restriction enzyme, respectively, c is the nonspecific background of RCA reaction.
Detection of individual transcripts in situ
One of the main directions of cell biology is the development of methods allowing individual molecule detection in single cell. The novel activity of Phi29 DNA polymerase was tested for suitability to be used in situ.
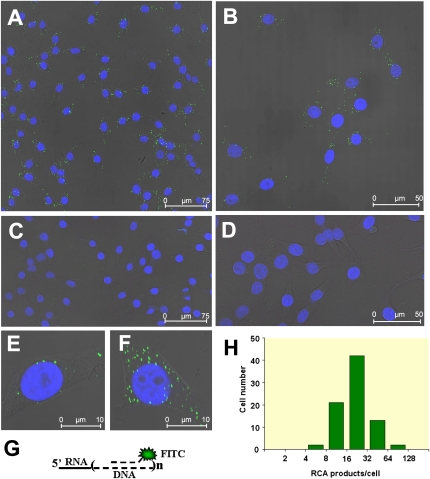
The obtained experimental data showed that RNA-primed RCA successfully detected GAPDH transcripts in single HeLa cells (Fig. 3). Using specific padlock probe PP5 precursor oligonucleotides for RCA and FITC labeled hybridization probes for RCA products, we have detected the accumulation of fluorescent label in HeLa cell cytoplasm (Fig. 3A,B,E,F), which indicated the location of specific reaction products. Control samples with nonspecific padlock probe PP6 precursor oligonucleotides having few mismatched nucleotides at 3′- and 5′-ends did not generate any labeled RCA product (Fig. 3C,D). The arithmetic mean of the number of visualized specific RCA products per cell was determined by summarizing the fluorescent signals in random nonoverlapping frames (one of them is presented in Fig. 3B) containing totally 80 cells and was 24 copies per cell (the standard deviation was 13). The analysis of individual cells revealed that the abundance of RCA products varied by one order in individual cells (see Fig. 3E,F, respectively) and had log-normal distribution (Fig. 3H), as previously described for other mRNA's in single cells (Bengtsson et al. 2005; Bengtsson et al. 2008). The geometric mean and geometric standard deviation of vizualized GAPDH-primed RCA products (these two statistical measurements are more informative than arithmetic mean and standard deviation for log-normally distributed data) corresponding to backtransformed mean and standard deviation of log transformed data were 21 and 1.7, respectively.
FIGURE 3.
Detection of individual GAPDH transcripts in situ. Reactions in situ were carried out under the conditions described in Materials and Methods. The phosphorylated oligonucleotides PP5 (specific for GAPDH) (A,B,E,F) were used as specific tools for GAPDH transcript detection in situ. Nonspecific phosphorylated oligonucleotides PP6 (differing from PP5 in few nucleotides in ligation junction region) (C,D) were used as controls of reaction specificity. The RCA reaction products were hybridized with FITC containing oligonucleotides and slides were analyzed using confocal microscope with 40× (A,C) and 63× (B,D–F) objectives. For best resolution of individual cells fivefold stronger magnification was used (E,F). The images are presented as an overlay of DAPI and FITC fluorescence projections and single bright field image. The green FITC label spots in cell cytoplasm surrounding blue-colored nuclei indicate the locations of specifically labeled RCA products. The specific RCA product hybridized with FITC-labeled oligonucleotides is schematically presented in (G). The log-normal distribution of vizualized RNA-primed RCA products per cell are presented in (H). The histogram shows the number of individual cells in each subset having specific RCA products within indicated bin ranges.
Our data correlate well with the recent data of GAPDH gene expression analysis in murine blood progenitors by “digital RT-PCR” technique (Warren et al. 2006), which concluded that GAPDH transcript levels can routinely fluctuate over full one-log range in the same cell line and the log-normal model was clearly preferred. The arithmetic and geometric means of GAPDH gene expression were quite similar in five tested cell lines. The arithmetic means varied from 37 to 72 (coefficient variations were 102% and 58%, respectively) per cell, the geometric means varied from 22 to 57 (geometric standard deviations were 3.1 and 2.3, respectively) per cell when measured as GAPDH cDNA. In summary, the absolute values of arithmetic and geometric means in both single-cell techniques either using “digital RT-PCR” or target RNA-primed RCA are close.
The relative detection efficiency of target GAPDH transcript-primed RCA was estimated by comparing the in situ data with RT-qPCR results (data not shown). RT-qPCR from two independent experiments showed that the arithmetic mean of GAPDH cDNA in HeLa cells was ∼105 cDNA molecules/1 ng total RNA and ∼2.6 × 103 cDNA molecules per cell. Therefore obtained detection efficiency of target GAPDH transcript-primed RCA in situ was ∼1% relative to the RT-qPCR outcome. Similar to target DNA-primed RCA (overall detection efficiency of the mitochondrial or chromosomal DNA targets' in situ was 1%–10%) (Larsson et al. 2004; Lohmann et al. 2007) current detection efficiency of the target RNA-primed RCA technique in situ precludes the application of this method for single-copy target examination in single cell. However, it still can be used for the detection and analysis of high-copy RNA transcripts as shown for GAPDH transcript and needs additional improvements and efforts rendering the method suitable for low-copy RNA transcript analysis.
CONCLUSIONS
We present “proof of principle” of novel Phi29 DNA polymerase application in RCA-based target RNA detection and analysis. The conversion of the target RNA into a primer by Phi29 DNA polymerase and the following padlock probe sequence amplification were successfully applied in vitro and in situ. We show that the padlock probes may be targeted to inner RNA sequences, and RNA molecules or their sequence peculiarities can be detected and analyzed without the requirement of additional DNA primers. Besides, since individual RNA targets are visualized as discrete spots in situ, preliminary information can be obtained about the distribution at single-cell level and the cellular localization of the transcripts.
However, in every experimental setting the method needs individual adjustments, such as the selection of target RNA and padlock probe sequences, the optimization of sample preparation, hybridization, and reaction conditions. Moreover, in some cases additional efforts may be required related to strong intramolecular RNA–RNA structures, which interfere with single-stranded 3′→5′ RNase activity of Phi29 DNA polymerase and encumber padlock probe formation on RNA targets.
MATERIALS AND METHODS
Plasmids, proteins, buffers, and reagents
The following products purchased from Fermentas UAB were used: BSA, DNaseI, DEPC-treated water, Maxima SYBR Green qPCR Master Mix (2×), Mva1269I restriction enzyme, polymerases (Phi29 DNA polymerase, Taq DNA polymerase), RevertAid H Minus First Strand cDNA Synthesis Kit, Ribolock RNase inhibitor, T4 polynucleotide kinase, T4 DNA ligase, Orange loading dye solution (6×), PBS buffer (10×), SSC buffer (20×), TAE buffer (50×), Tango buffer (10×), TBE buffer (10×), and RNA Loading Dye Solution (2×). Fetal bovine serum was purchased from Biochrom AG; creatinphosphate, Dulbecco's Modified Eagle's medium without L-glutamine, L-glutamine, gentamycine sulfate were purchased from Sigma; and QIAamp RNA Blood Mini Kit was purchased from Qiagen.
Nucleotides and oligonucleotides
[α33P]dATP (∼3000 Ci/mmol) was purchased from Amersham Biosciences. Unlabeled dNTPs and RNA LR Ladder were products of Fermentas UAB. The origin and sequences of oligonucleotides PP2, PP4, PL2, and PL4, have been presented previously (Lagunavicius et al. 2008). The following DNA oligonucleotides were obtained from Metabion:
PP5 (5′-TGAGCACAGGGTACTATGCTGCTGCTGTACTACGAGCGGTCTCCAGGAATGCGCATTTTTTTTTTTTGGT);
PP6 (5′-AGAGCACAGGGTACTATGCTGCTGCTGTACTACGAGCGGTCTCCAGGAATGCGCATTTTTTTTTTTTGGA);
Mva1269I olig. (5′-CCAGGAATGCGCATTTATGT);
EM1 (5′-GGTTGAGCACAGGGTACTTTATTGATGG); and
DP-FITC (5′-FITC-AATGCTGCTGCTGTACTACGG).
Target RNA detection in RNA mixture using preformed padlock probes
Circular single stranded DNA probes (PP2, PP4) were prepared as previously described (Lagunavicius et al. 2008). The hybridization reaction was performed in 225 μL of hybridization buffer (50 mM Tris-HCl at pH 8.0; 0.1 mM EDTA, 80 mM NaCl) containing 5 μg of RNA LR Ladder and 20 nM preformed specific padlock probe PP2 (having complementary sequence to one of the RNA size standards, RNA2) by heating for 10 min at 65°C and cooling to room temperature for 10 min. Free circular PP2 oligonucleotide, RNA LR Ladder or its mixture with nonspecific circular PP4 oligonucleotide (having no complementary sequence to RNA1) was used in other samples as controls of reaction specificity. Reactions with Phi29 DNA polymerase were performed for 2 h at 37°C in 50 μL reaction mixture (1 × Tango, 1 mM DTT, 1 mM dNTP, 50 U Ribolock, 0.6 μg RNA LR Ladder, 10 nM circular PP2, 7.5 U Phi29 DNA polymerase). After incubation 10 μL of 6 × orange loading dye solution were added to stop the reaction, samples were heated for 10 min at 65°C and cooled on ice, then loaded onto agarose gel (1% agarose, 0.4 μg/mL ethidium bromide, 1 × TAE). The gels were analyzed by an Ultra Lum Electronic U.V. Transilluminator (Ultra-Lum).
Ligation-based target RNA detection in RNA mixtures
RNA–DNA hybridization reaction was performed in 50 μL of ligation buffer (10 mM Tris-HCl at pH 7.5; 10 mM MgCl2, 20 μM ATP) containing 400 nM phosphorylated PP2 oligonucleotide and 0.9 μg RNA LR Ladder (one of the RNA size standards was the target RNA transcript, RNA2) by heating for 3 min at 65°C and cooling to room temperature for 10 min. After hybridization 50 U Ribolock and 25 U T4 DNA ligase were added. Reaction mixtures were incubated for 2 h at 37°C following 10 min incubation at 70°C and placed on ice. Different mixtures lacking essential additives (nucleic acids or enzymes) were used as controls of reaction efficiency and specificity. Reactions with Phi29 DNA polymerase were performed for 2 h at 37°C in 50 μL of reaction mixture (1 × Tango, 1 mM DTT, 1 mM dNTP, 50 U Ribolock, 12.5 μL of ligation probe, 7.5 U Phi29 DNA polymerase). After incubation 10 μL of 6 × orange loading dye solution were added to stop the reaction. Samples were heated for 10 min at 65°C and placed on ice before loading onto agarose gel.
Ligation-based GAPDH gene transcript detection in total cellular RNA
Total RNA was isolated from human HeLa cells using acid phenol-guanidinium thiocyanate-chloroform extraction and following DNaseI treatment (Sambrook and Russel 2001). The presence of GAPDH transcripts and the absence of GAPDH encoding gene were confirmed by RT-PCR experiments (data not shown). Hybridization reaction was performed in 9 μL of reaction mixture (1 × Tango, 11 nM phosphorylated PP5 oligonucleotide [specific for GAPDH], 0.25 μg total RNA) by heating for 5 min at 65°C and cooling to room temperature for 10 min. Free PP5 oligonucleotide, free total mRNA, or its mixture with nonspecific PP6 oligonucleotide (differing from PP5 in few nucleotides in ligation junction region) were used in other samples as controls of reaction specificity. After hybridization 10 U Ribolock 20 μM ATP and 5 U T4 DNA ligase were added to each sample and incubated for 2 h at 37°C. The ligase was inactivated for 10 min at 70°C. RCA reactions were initiated by adding 1 mM dNTP (10 μCi [α33-P]) and 2.5 U Phi29 DNA polymerase to 10 μL of each sample. The samples were incubated for 3 h at 37°C and reactions were terminated for 10 min at 70°C. Further, RCA products specificity was tested. Complementary oligonucleotides, containing Mva1269I restriction endonuclease recognition site, were annealed to RCA product in 2 × Tango buffer in 10 μL volume at 10 μM final concentration. Annealing step was performed: 5 min at 95°C, 2 min at 80°C, 2 min at 70°C, 2 min at 60°C, 2 min at 50°C, 2 min at 40°C, 2 min at 30°C, 2 min at 20°C, and 2 min at 10°C. Then each sample was divided into two portions and one of them was supplemented with 5 U Mva1269I restriction endonuclease. Monomerization reaction was performed for 1 h at 37°C. Before electrophoretic analysis all samples were treated with proteinase K (200 μg/mL) in the presence of SDS (0.4%) for 30 min at 37°C. Following incubation equal volume of 2 × RNA loading dye solution was added to stop the reaction. The samples were heated for 10 min at 95°C and placed on ice, then loaded onto the 8% (w/v) denaturing polyacrylamide gel (29:1 [w/w] acrylamide/bisacrylamide; 7 M urea, 1 × TBE buffer). The gels were analyzed by Cyclone Storage Phosphor System and OptiQuant Image Analysis Software (Packard Instruments).
Individual transcript detection in situ
The HeLa cells were cultured in 10-well glass slide (Electron Microscopy Sciences) at a density of 3 × 103 cells/well in Dulbecco's Modified Eagle's medium (without L-glutamine), supplemented with 10% fetal bovine serum, 0.3 mg/mL L-glutamine, 50 μg/mL gentamycine sulfate for 24 h at 37°C in CO2 incubator. The cells on the slide were washed with 1 × PBS buffer for 3 min and fixed with 4% paraformaldehyde in 1 × PBS for 20 min at room emperature. After fixation the cells were washed with 1 × PBS buffer for 3 min, dehydrated with 70% (5 min), 85% (3 min), 99.9% (3 min) ethanol and air-dried at room temperature. The GAPDH transcript and specific PP5 oligonucleotide hybridization reaction was performed in 30 μL/well of RNA hybridization buffer (475 mM Tris-HCl at pH 8; 0.95 mM EDTA, 760 mM NaCl) containing 100 nM phosphorylated PP5 for 2–3 h at 37°C in a humidity chamber. The glass slide was washed at 37°C in preheated washing buffer (100 mM Tris at pH 7.5; 150 mM NaCl, 0.05% Tween20) for 5 min, dehydrated through serial washes in increasing concentration of an ethanol (70%, 85%, and 99.9%, for 3 min each) and air dried. The ligation of PP5 oligonucleotide on GAPDH transcripts was performed with a 3 U T4 DNA ligase in 30 μL/well of 1 × T4 DNA ligase buffer supplemented with BSA (0.1 mg/mL) and a 60 U Ribolock Ribonuclease inhibitor in a humidity chamber for 1 h at 37°C. The glass slide was washed at 37°C in preheated washing buffer for 3 min, dehydrated, and dried as described above. Nonspecific PP6 oligonucleotide (differing from PP5 in few nucleotides in ligation junction region) was used in other sample as control of reaction specificity. RCA was performed in 30 μL of 1 × Phi29 DNA polymerase buffer containing BSA (0.2 mg/mL), 0.25 mM dNTP, 30 U Ribolock, 30 U Phi29 DNA polymerase, for 16 h at 37°C in a humidity chamber. The glass slide was washed, dehydrated, and air-dried as described above. The RCA product was detected by hybridization with DP-FITC oligonucleotide in 30 μL of a hybridization buffer (2 × SSC buffer, 20% formamide, 5% glycerol) containing 250 nM DP-FITC oligonucleotide for 2 h at 37°C in a humidity chamber. The glass slide was washed at 37°C in preheated washing buffer twice for 5 min, dehydrated and air-dried. Slides were mounted in Vectashield mounting medium for fluorescence with 1.5 μg/mL of 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) and viewed with TSC SP5 Confocal Microscope (Leica).
RNA isolation and RT-qPCR
Total RNA was isolated from 0.5 × 106 HeLa cells using QIAamp RNA Blood Mini Kit following manufacturer's recommendations. Five dilutions of total RNA preparation (ranging from 100 ng to 10 pg) were used for cDNA synthesis with RevertAid H Minus First Strand cDNA Synthesis Kit according to manufacturer's instructions. The GAPDH specific primer EM1 was used to prime reverse transcription in a reaction of 20 μL. Quantitative RT-PCR's were performed in duplicates for each sample using 2 μL of the cDNA reaction supplemented with 0.3 μM of forward and reverse primers specific for GAPDH (control primers in RevertAid H Minus First Strand cDNA Synthesis Kit) and Maxima SYBR Green qPCR Master Mix in a reaction of 25 μL, ampified in 7000 Real-Time PCR System (Applied Biosystems) instrument. A standard curve was prepared from cDNA solutions corresponding to the serially diluted solutions (range of 102–107 copies) of human GAPDH mRNR (control RNA in RevertAid H Minus First Strand cDNA Synthesis Kit). The volumes and components of RT and qPCR reaction mixtures were the same as those for the test samples. Obtained results from five dilutions of the test sample were compared to the standard curve, averaged, and presented as the copies of DNA/ng or the copies of DNA per single cell. The experiment was repeated twice and the average was calculated.
ACKNOWLEDGMENTS
This work was partially funded by EU FP6 Integrated Project “MolTools” award. We are grateful to Dr. Jorn Koch (University of Aarhus) and Dr. Lolita Zaliauskiene (Fermentas UAB) for the expert advice on in situ methods, Professor Vida Kirveliene (University of Vilnius) for lending the confocal microscopy, Jurgita Kupciunaite and Birute Gagiliene (Fermentas UAB) for assistance in RT-qPCR experiments, and Dr. Laurynas Riauba (Fermentas UAB) for help in statistical analysis of in situ data. Also, we express our appreciation to Dr. Ramune Leipuviene (Fermentas UAB) for critical reading of the manuscript and linguistic help.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1279909.
REFERENCES
- Baner J., Nilsson M., Mendel-Hartvig M., Landegren U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 1998;26:5073–5078. doi: 10.1093/nar/26.22.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson M., Ståhlberg A., Rorsman P., Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson M., Hemberg M., Rorsman P., Ståhlberg A. Quantification of mRNA in single cells and modeling of RT-qPCR induced noise. BMC Mol. Biol. 2008;9:63. doi: 10.1186/1471-2199-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian A.T., Pattee M.S., Attix C.M., Reed B.E., Sorensen K.J., Tucker J.D. Detection of DNA point mutations and mRNA expression levels by rolling circle amplification in individual cells. Proc. Natl. Acad. Sci. 2001;98:14238–14243. doi: 10.1073/pnas.251383598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson O., Jarvius J., Schallmeiner E., Howell M., Nong R.Y., Reuter H., Hahn M., Stenberg J., Nilsson M., Landegren U. A dual-tag microarray platform for high-performance nucleic acid and protein analyses. Nucleic Acids Res. 2008;36:e45. doi: 10.1093/nar/gkn106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haible D., Kober S., Jeske H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods. 2006;135:9–16. doi: 10.1016/j.jviromet.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Jonstrup S.P., Koch J., Kjems J. A microRNA detection system based on padlock probes and rolling circle amplification. RNA. 2006;12:1747–1752. doi: 10.1261/rna.110706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunavicius A., Kiveryte Z., Zimbaite-Ruskuliene V., Radzvilavicius T., Janulaitis A. Duality of polynucleotide substrates for Phi29 DNA polymerase: 3′→5′RNase activity of the enzyme. RNA. 2008;14:503–513. doi: 10.1261/rna.622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landegren U., Schallmeiner E., Nilsson M., Fredriksson S., Banér J., Gullberg M., Jarvius J., Gustafsdottir S., Dahl F., Söderberg O., et al. Molecular tools for a molecular medicine: Analyzing genes, transcripts and proteins using padlock and proximity probes. J. Mol. Recognit. 2004;17:194–197. doi: 10.1002/jmr.664. [DOI] [PubMed] [Google Scholar]
- Larsson C., Koch J., Nygren A., Janssen G., Raap A.K., Landegren U., Nilsson M. In situ genotyping individual DNA molecules by target-primed rolling-circle amplification of padlock probes. Nat. Methods. 2004;1:227–232. doi: 10.1038/nmeth723. [DOI] [PubMed] [Google Scholar]
- Lizardi P.M., Huang X., Zhu Z., Bray-Ward P., Thomas D.C., Ward D.C. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Lohmann J.S., Stougaard M., Koch J. Detection of short repeated genomic sequences on metaphase chromosomes using padlock probes and target primed rolling circle DNA synthesis. BMC Mol. Biol. 2007;8:103. doi: 10.1186/1471-2199-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijatovic T., Houzet L., Defrance P., Droogmans L., Huez G., Kruys V. Tumor necrosis factor-α mRNA remains unstable and hypoadenylated upon stimulation of macrophages by lipopolysaccharides. Eur. J. Biochem. 2000;267:6004–6012. doi: 10.1046/j.1432-1327.2000.01676.x. [DOI] [PubMed] [Google Scholar]
- Niel C., Diniz-Mendes L., Devalle S. Rolling-circle amplification of Torque teno virus (TTV) complete genomes from human and swine sera and identification of a novel swine TTV genogroup. J. Gen. Virol. 2005;86:1343–1347. doi: 10.1099/vir.0.80794-0. [DOI] [PubMed] [Google Scholar]
- Nilsson M., Krejci K., Koch J., Kwiatkowski M., Gustavsson P., Landegren U. Padlock probes reveal single-nucleotide differences, parent of origin and in situ distribution of centromeric sequences in human chromosomes 13 and 21. Nat. Genet. 1997;16:252–255. doi: 10.1038/ng0797-252. [DOI] [PubMed] [Google Scholar]
- Nilsson M., Barbany G., Antson D.O., Gertow K., Landegren U. Enhanced detection and distinction of RNA by enzymatic probe ligation. Nat. Biotechnol. 2000;18:791–793. doi: 10.1038/77367. [DOI] [PubMed] [Google Scholar]
- Nilsson M., Antson D.O., Barbany G., Landegren U. RNA-templated DNA ligation for transcript analysis. Nucleic Acids Res. 2001;29:578–581. doi: 10.1093/nar/29.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russel D.V. Molecular cloning: A laboratory manual. 3rd ed. Vol. 1. Cold Spring Harbor Laboratory Press; Cold Spring Harbor. NY: 2001. pp. 7.4–7.12. [Google Scholar]
- Stougaard M., Lohmann J.S., Zajac M., Hamilton-Dutoit S., Koch J. In situ detection of nonpolyadenylated RNA molecules using Turtle Probes and target primed rolling circle PRINS. BMC Biotechnol. 2007;7:69. doi: 10.1186/1472-6750-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L., Bryder D., Weissman I.L., Quake S.R. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc. Natl. Acad. Sci. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Calciano M., Hamann S., Leamon J.H., Strugnell T., Christian M.W., Lizardi P.M. In situ detection of messenger RNA using digoxigenin-labeled oligonucleotides and rolling circle amplification. Exp. Mol. Pathol. 2001;70:281–288. doi: 10.1006/exmp.2001.2365. [DOI] [PubMed] [Google Scholar]