Abstract
RNA interference (RNAi)-based gene silencing possesses great therapeutic potential for inhibiting replication of human viruses such as hepatitis C virus (HCV). However, one of the putative limitations for its use as a therapy is the rapid emergence of escape variants. These contain deletions or mutations within the viral genome sequences complementary to the small interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs) being used for treatment. As a potential solution to this problem, we constructed an expression system for duplex RNAs harboring two siRNA units using convergent H1 and U6 Pol III promoters. Here, the length and orientation of the transcript, tandem siRNA (tsiRNA), were optimized to be processed by the intracellular ribonuclease Dicer into functional siRNAs targeting different sequences. Assessment in transfected cells indicates that the length of the tsiRNA duplex (40–42 base pairs) is more critical for both siRNA-producing capacity and gene silencing activity than the orientation of each siRNA unit. In Huh7 cells replicating full-length HCV RNA, expression of length-optimized tsiRNA inhibited viral protein levels as efficiently as a single 21-nucleotide siRNA-expression construct, without affecting miRNA maturation or induction of an interferon response. We verified that the anti-viral activity of tsiRNA was achieved by precise cleavage of two target sites. A distinct advantage of this strategy is that each side of the optimized linear duplex RNA could enter into the Dicer-mediated processing machinery, thus likely providing more equal and efficient production of multiple siRNAs required for reducing the chance of viral escape.
Keywords: siRNA, HCV, full-length replicon, multiple gene silencing
INTRODUCTION
Hepatitis C virus (HCV) belongs to the Flaviviridae family and has an ∼9.6-kb positive strand RNA genome that encodes at least 10 viral structural and nonstructural proteins, which is flanked by 5′- and 3′-untranslated regions (UTRs) (Grakoui et al. 1993; Bartenschlager and Lohmann 2000). An estimated 170 million people worldwide are chronically infected with this virus, and it is recognized as a major causative agent for hepatitis, cirrhosis, and hepatocellular carcinoma (Alter 1997). Unfortunately, neither a prophylactic nor a therapeutic vaccine against HCV is available. Although combination therapy with interferon (IFN)-α and ribavirin has resulted in remarkable outcomes in clinical applications, only about half of the HCV-infected patients benefit from this treatment (Chander et al. 2002). Thus, there is an urgent need to develop alternative therapeutics to control HCV infection.
RNA interference (RNAi) is a post-transcriptional gene silencing process that is evolutionally conserved in plants, Caenorhabditis elegans, and mammals (Bosher and Labouesse 2000; Dykxhoorn et al. 2003). In mammalian cells, the RNase III-like cytoplasmic ribonuclease Dicer recognizes double-stranded RNA (dsRNA) and cleaves it into shorter duplex RNAs; these small interfering RNAs (siRNA) are between 21 and 25 nucleotides (nt) in length (Dorsett and Tuschl 2004). Previous studies have shown that RNAi-based technology has great promise, particularly for the treatment of RNA virus-derived diseases, including human immunodeficiency virus type 1 (HIV-1) and HCV (Capodici et al. 2002; Kapadia et al. 2003; Kim et al. 2006). However, the mutation rate of these viruses is high due to lack of proofreading activity of viral RNA polymerases, which allows variants to escape rapidly from RNAi inhibition (Das et al. 2004; Wilson and Richardson 2005). Several recent reports have suggested that this problem could be resolved by using a synthetic siRNA mixture containing different sequences, in vitro Dicer-generated siRNA products from long dsRNA, or cellularly expressed long hairpin RNA (lhRNA) (Wilson and Richardson 2005; Watanabe et al. 2006; Liu et al. 2007; Sano et al. 2008).
In this study, as an another approach, we employed an RNA expression system using two convergent RNA polymerase III promoters, human H1 and U6, to produce linear duplex RNA that has similar secondary structure to extended synthetic siRNA molecules (Kaykas and Moon 2004; Zheng et al. 2004; Lin and Feng 2005; Shin et al. 2006). In comparison to a hairpin RNA-expression construct with inverted repeats or the use of different RNA Pol III promoters on one plasmid backbone, it was reported that this system has the benefits of increased success rates of sequencing reactions and genetic stability by reducing recombination probability of DNA (ter Brake et al. 2008). Importantly, the efficiency of siRNA processing could be improved by exposure of both sides of the stem bases of the duplex RNA molecules to Dicer. In brief, the major concept of this study is that the long duplex RNA, containing multiplexed siRNAs (named tandem siRNA or tsiRNA), can be processed via the cellular RNAi machinery into separate molecules that target their cognate genes. To evaluate this hypothesis, we designed a series of tsiRNAs against different reporters and optimized the length and orientation of each siRNA unit by monitoring siRNA-processing efficiency as well as RNAi activity. Based on the established rules for constructing multiple siRNAs, we further evaluated antiviral activity using constitutively expressed tsiRNAs targeting the HCV E2 and NS3-coding sequences in full-length HCV replicon-containing cells. In parallel, using a reporter and viral target sequence fusion system, the potential advantage of anti-HCV tsiRNA construct for preventing or covering the expression of escape variants was described. The results show that the most effective and sequence-specific gene silencing can be achieved when siRNA units are composed of 21 + 21-nt or 23 + 21-nt sequences, and thus the length of tsiRNA is 40 base pairs (bp) or 42 bp with 2-nt 3′-overhangs. It appears that the activity is not significantly affected by inverting the orientation of the siRNA sequences. This approach will provide a useful platform from which to design extended precursor siRNAs and to reduce the probability of the escape of viral mutants.
RESULTS
Gene silencing activity of double-stranded RNAs expressed from convergent RNA polymerase III promoters
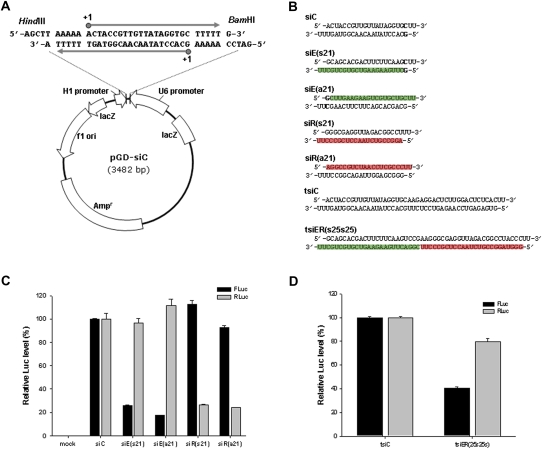
According to previous reports, we have constructed an siRNA expression vector, pGD-siC, which encodes sense and antisense RNA molecules from two convergent human H1 and U6 promoters, respectively, with (T)5-termination signals at their −5 to −1 positions (Fig. 1A; Kaykas and Moon 2004; Zheng et al. 2004; Lin and Feng 2005; Shin et al. 2006). And gene silencing by H1/U6-expressed siRNAs had been assessed by inserting several 21-nt exogenous viral sequences between the promoters (Kim et al. 2006; Shin et al. 2006). To test the ability of gene knockdown against other targets, we cloned fluorescent protein (EGFP)- and Renilla luciferase (RLuc)-specific 21-mer siRNA sequences into the HindIII and BamHI enzyme sites of pGD-siC, creating pGD-siE(s21) and pGD-siR(s21). We also prepared the corresponding inverted constructs pGD-siE(a21) and pGD-siR(a21) (Fig. 1B). Huh7 cells were co-transfected with pEGFPLuc (encoding the fused target EGFP and firefly luciferase [FLuc] gene) and phRL-CMV (encoding the target RLuc gene) together with the individual siRNA expression vectors. On day 2, the levels of luciferase reporters were determined by a dual-luciferase assay (Fig. 1C). All specific siRNA constructs caused significant gene knockdown (∼80% on average), independent of their sequence orientation. This result indicates that both RNA polymerase III promoters produce RNA molecules with comparable transcription efficiency.
FIGURE 1.
Transcription of siRNAs from convergent RNA Pol III promoters and their RNAi activity in cultured cells. (A) Construction of the duplex siRNA-expression plasmid, pGD-siC. Two complementary RNA strands with poly(U) at their 3′-ends are synthesized from the human H1 and U6 promoters. [(T)5] Transcription termination signals; (+1) the transcription initiation site. The restriction enzyme sites HindIII and BamHI were used for cloning oligonucleotides encoding siRNAs. (B) Sequence of H1/U6 promoter-driven siRNAs and tsiRNAs. (siC) Control, (siE) EGFP-specific, and (siR) Renilla luciferase-specific siRNAs. (s21) The 21-nt sense-strand transcription from the H1 promoter; (a21) the 21-nt antisense-strand transcription from the H1 promoter. tsiC and tsiER(s25s25) are longer duplex RNAs, fusing two different control siRNAs or the extended 25 nt siE(s25) and 25 nt siR(s25). Colored boxes show the position of the antisense sequences for (green) siE and (red) siR within the duplex RNAs. (Bold) The extra nucleotides required for efficient initiation of Pol III promoter-driven transcription from a purine sequence. The two U's at the 3′-end of each strand are from the transcription termination signal. (C) RNAi activity of the siE(s21)-, siE(a21)-, siR(s21)-, and siR(a21)-expression plasmids. Luciferase activity from cells transfected with the control siRNA construct (siC) was set at 100%. Mock-transfected cells were used as a negative control. Values represent means (+SD) from three replicate transfections. (D) RNAi activity of a tsiRNA, tsiER(s25s25), targeting EGFP and RLuc genes. Luciferase activity from cells transfected with the irrelevant tsiC-encoding plasmid was set at 100%. Transfections and measurements were performed as described above.
As a preliminary test of tandem siRNA, we attempted to combine the two siRNA sequences (25 + 25 nt in length) to silence both EGFP-FLuc and RLuc expression, based on the most recently published report on the length requirement for lhRNA (Sano et al. 2008). The tsiRNA-encoding plasmid pGD-tsiER(s25s25) was prepared by directly fusing two 25-mer sequences, siE(s25) and siR(s25), which contain extensions of their original 21-mer to 3′. We expected that the resulting 48-bp linear RNA duplex with 2-nt U sequences at both 3′-ends could be processed into functional siRNA units by the cellular RNAi machinery to cleave EGFP and RLuc mRNAs (Fig. 1B). Interestingly, expression of the EGFP-FLuc fusion was efficiently inhibited by 59%, whereas another target RLuc expression was marginally suppressed by 20% in the same reaction tube (Fig. 1D). These data suggest that further molecular modification is required to determine the optimal length or orientation of siRNA component. Such a refined siRNA stack would enhance not only the production rate of functional siRNA molecules, but also incorporation efficiency of the antisense guide strand from processed siRNA into the RNA-induced silencing complex (RISC) and thus result in more improved inhibition of distinct target genes.
RNAi activity of H1/U6 promoter-expressed tandem siRNAs with successively combined target sequences
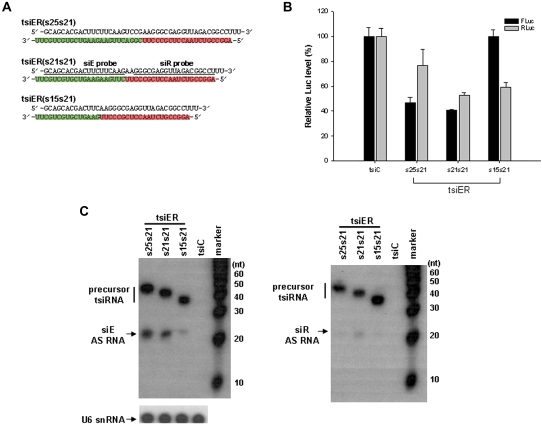
In the work described by Elbashir et al. (2001) using a Drosophila in vitro system, dsRNA molecules longer than 38 bp in length were processed into 21- to 22-nt siRNA fragments, subsequently causing 20–23-nt spacing cleavage of sense and antisense target RNAs. This finding provided evidence that each terminus of linear duplex RNA is equally exposed to Dicer attack in the initiation steps of RNAi. It is intriguing to speculate that stacking functional siRNA may be a promising approach to make multi-targeted siRNA precursors. However, in our preliminary study, stacked tsiER(s25s25) RNA exhibited poor activity, especially against the RLuc gene (Fig. 1D). In an attempt to learn the rule of combining different siRNA sequences in a single expression cassette, the less potent 25-mer RLuc siRNA sequences were reduced to 21-mer, which generated pGD-tsiER(s25s21) (Fig. 2A). Following co-transfection into cells as described above, we measured dual gene knockdown on day 2. Unfortunately, there was no significant improvement in multiple silencing compared to the previously used pGD-tsiER(s25s25) construct (Fig. 2B). EGFP-Luc gene expression was reduced by 53%, while inhibition efficiency of RLuc expression was still marginal (23% knockdown). Thus, the 25-nt siE RNA sequence was reduced further to 21 and 15 nt, generating pGD-tsiER(s21s21) and pGD-tsiER(s15s21), respectively (Fig. 2A). Notably, in cells treated with pGD-tsiER(s21s21), FLuc and RLuc expression was silenced, most efficiently, by 59% and 47%, although it was not as efficient as separately transfected pGD-siE or pGD-siR (Figs. 2B, 1C). This finding reflects the fact that tsiRNA might be as potent as single siRNA treatment in each target RNA cleavage. However, it is certain that tsiRNA structure has advantages of multiple gene silencing over a single version. And when pGD-tsiER(s15s21) was treated, gene silencing of FLuc was completed abolished, whereas considerable RNAi activity against the RLuc gene was observed (Fig. 2B).
FIGURE 2.
Dual gene silencing activity of linear tsiRNAs, tsiER(s25s21), tsiER(s21s21), and tsiER(s15s21) against EGFP and RLuc genes. (A) The predicted structure of tsiER RNAs transcribed from RNA Pol III promoters. (Boxes) Sites complementary to (green) EGFP and (red) RLuc target genes. In the pGD constructs, all colored guide/antisense strand sequences are located downstream from the U6 promoter. (Underlined) Probe sequences for detection of siE and siR antisense strands. (B) Luciferase assay of cellularly expressed tsiRNAs with sequential siE and siR components using the same direction of transcription. Transfection into Huh7 cells was performed as described in Figure 1C. The FLuc and RLuc expression levels of cells transfected with plasmid encoding control tsiRNA (tsiC) were set at 100%. Relative means (+SD) from three independent experiments are shown. (C) Northern blot analysis for detection of intracellularly processed siE and siR siRNAs from precursor tsiRNAs. Detection of cellular U6 snRNA was used as a loading control. (Arrows) Processed 21–23-nt antisense strand (AS) siRNA products. Probe sequences and positions are described in A. (Right) Marker sizes (in nucleotides, nt).
Next, we examined the processing efficiency of these tsiRNAs into siRNA products. The Northern blot data were obtained using the siE and siR sense strands as probes, which can hybridize to their antisense-strand RNAs. As shown in Figure 2C, both 21–23-nt siE and siR antisense products appeared in pGD-tsiER(s21s21)-transfected cells. In contrast, partial or relatively inefficient siRNA production was detected from transcription of tsiER(s25s21) and tsiER(s15s21). These results correlate with the luciferase assay (Fig. 2B). They suggest that with respect to RNAi activity and siRNA maturation, ∼21 nt is the optimal length of successively connected siRNA units.
RNAi activity of H1/U6 promoter-expressed tandem siRNAs with convergently combined target sequences
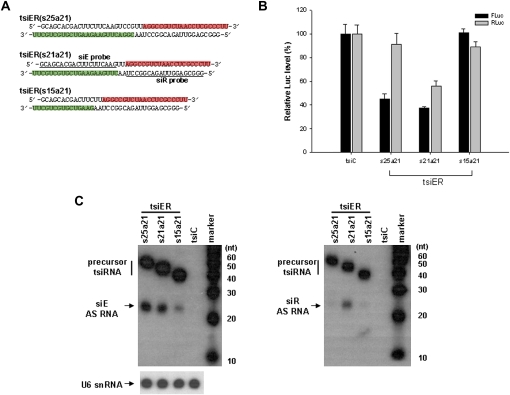
Most shRNA design programs for RNA Pol III-derived expression cassettes prefer the use of a purine sequence (adenosine or guanosine) as an initiation nucleotide for efficient transcription. However, when the two siRNAs are linked consecutively, the first nucleotide of the latter siRNA's antisense sequence can be occasionally occupied with a pyrimidine (uridine or cytidine), possibly lowering transcription yield of that strand relative to its complementary one. This may be a disadvantageous factor for preparing extensive tsiRNA expression vectors. Indeed, this problem has been simply solved by an addition of an extra purine sequence at the deficient transcription initiation site. However, it can threaten to affect siRNA's sequence identity and thus reduce the specificity of RNAi-mediated gene knockdown. Thereby, we used an alternative tsiRNA expression approach by connecting siRNA unit sequences convergently, in which purine sequences could be readily located at both first positions from the promoters upon shRNA design programs (Fig. 3A). We assessed the inhibition of EGFP-FLuc and RLuc expression by tsiER(s25a21), tsiER(s21a21), and tsiER(s25a21). Notably, transfection of pGD-tsiER(s21a21) achieved simultaneous gene silencing against FLuc (63% inhibition) and RLuc (46% inhibition), while only partial or no activity resulted from pGD-tsiER(s25a21) and pGD-tsiER(s15a21) (Fig. 3B). Consistent with this observation, pGD-tsiER(s21a21)-transfected cells, but not cells transfected with the other constructs, produced prominent guide strands of the expected size (21–23 nt) for both mature siE and siR sequences in Northern blot analysis (Fig. 3C). These data indicate that the tsiRNA expression construct with two convergent siRNAs of 21 nt functions as efficiently as the consecutively 21 + 21-linked one.
FIGURE 3.
Dual gene silencing activity of linear tsiRNAs, tsiER(s25a21), tsiER(s21a21), and tsiER(s15a21), against the EGFP and RLuc genes. (A) The predicted structure of tsiER RNAs. (Boxes) Antisense strands of (green) siE and (red) siR genes. In these pGD vector constructs, the green antisense strands of siE duplexes are expressed from the U6 promoter, whereas those of (red) siR are from the H1 promoter (see also Fig. 1A). Probe sequences for detection of siE and siR antisense strands are underlined. (B) Luciferase assay following transfection of three cellularly expressed tsiRNA constructs, in which the siE and siR components are combined in opposing transcriptional directions. FLuc and RLuc expression levels of cells transfected with the plasmid encoding control tsiRNA (tsiC) were set at 100%. Relative means (+SD) from three independent experiments are shown. (C) Northern blot analysis for detection of antisense strands of siE and siR siRNAs. Detection of cellular snU6 RNA was used as a loading control. (Arrows) Processed 21–23-nt antisense strand (AS) siRNA products (siE and siR). Probe sequences and positions are described in A. (Right) Marker sizes (in nucleotides, nt).
Additionally, we also measured intracellular concentration of these tsiRNA-derived sense-strand molecules by Northern blot (Supplemental Fig. 1). Interestingly, mature siLuc sense strand was detected efficiently in tsiER(s21s21)- and tsiER(s21a21)-encoding DNA-treated cells. However, siE sense strand molecules were barely detected in the same samples, consistent with the work of Lee et al. (2008). Given the predicted thermodynamic internal stability profiles, antisense strands of all these siRNAs gain higher scores for RISC selection (data not shown). This suggests that additional factors, such as nuclease resistance or structure, besides thermodynamic properties or biased strand retention by RISC, might be involved in determining the intracellular kinetics of siRNA single strands.
Inhibition of HCV replication by tandem siRNAs
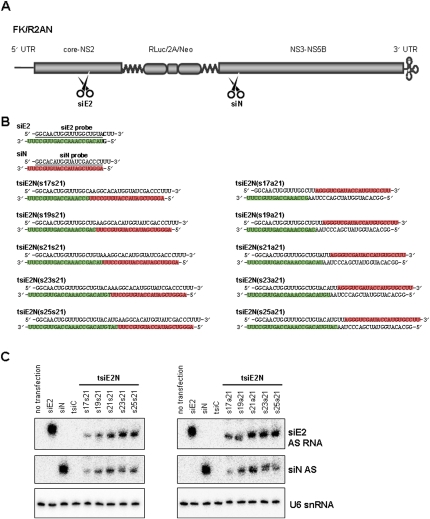
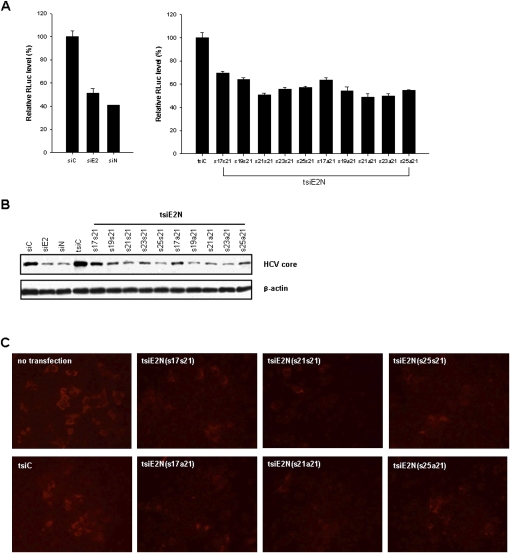
Previous work from our laboratory evaluated the anti-viral activity of several HCV-specific siRNAs using dual promoter-based siRNA expression vectors and full-length HCV replicon (Kim et al. 2006). Among those functional siRNAs, siE2 and siN-expressing plasmids pGD-siE2 and pGD-siN, which harbor 22-nt and 21-nt sequences from E2 and NS3 genes and exhibit thermodynamic antisense strand bias for RISC incorporation, were prepared (Fig. 4A,B; Khvorova et al. 2003). And to determine the optimal tsiRNAs for RNAi trigger, we constructed a series of vectors by combining siE2 and siN with various lengths (i.e., 17 + 21, 19 + 21, 21 + 21, 23 + 21, and 25 + 21) and different alignments (i.e., sense + sense and sense + antisense) (Fig. 4B). Their siRNA production efficiency in replicon cells was monitored by Northern blot analysis using radio isotope-labeled sense-strand probes (Fig. 4B,C). Highest amounts of mature siRNAs which potentially target E2 and NS3 genes were accumulated especially in 21 + 21 and 23 + 21 (sense + sense or sense + antisense) tsiRNA-encoding plasmid-treated cells (Fig. 4C). We next measured the RNAi activity of all constructs by luciferase assay and anti-viral protein immunoblot analysis following transfection of FK/R2AN cells (Fig. 5). The data confirm that single siRNAs, siE2 and siN, are effective for silencing of Renilla luciferase expression levels by 49% and 59% relative to irrelevant siC (Fig. 5A, left). Here, the luciferase reporter intensity reflects relative viral replication or expression levels. When tsiRNAs were tested, the luciferase assay showed that the 21 + 21 and 23 + 21 constructs, such as tsiE2N(s21s21), tsiE2N(s23s21), tsiE2N(s21a21), and tsiE2N(s23a21), inhibited HCV replication, most significantly, by 52% on average, whereas the tsiE2N(s17s21) and tsiE2N(s17a21) were the least potent (Fig. 5A, right). In Western blot analysis, viral core protein was also suppressed by the single siRNA-encoding plasmids used as positive controls (Fig. 5B). Significant knockdown of viral core protein was detected in all dual constructs, except for 17 + 21 tsiRNAs. Notably, the reproducible and the most noticeable RNAi activity was induced in 21 + 21 and 23 + 21 tsiRNA-transfected cells independently of siRNA orientation (Fig. 5B). Consistently, significant knockdown of another HCV protein NS5A by 21 + 21 tsiRNA appeared through immunostaining of replicon cells (Fig. 5C). Taken together, these findings suggest that between 21 + 21-mer and 23 + 21-mer is the optimal length to create multi-functional duplex RNAs.
FIGURE 4.
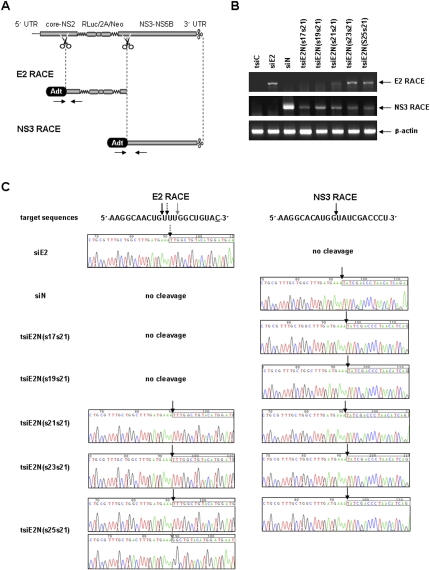
Construction of tsiRNA-expression vectors targeting HCV genome. (A) Schematic representation of the HCV replicon FK-R2AN RNA and anti-HCV siRNA target sites against E2 (siE2) and NS3 (siN) coding genes. (5′ UTR) 5′-Untranslated region; (core-NS2) coding region for viral core, E1, E2, p7 to NS2; (RLuc) Renilla luciferase; (2A) foot-and-mouth disease virus (FMDV) 2A; (Neo) neomycin resistance gene; (NS3-NS5B) coding regions for viral proteins NS3, NS4A, NS4B, NS5A to NS5B; and (3′ UTR) 3′-untranslated region. (B) Sequence composition of HCV E2- (siE2) and NS3-specific (siN) siRNAs, as well as tsiE2N RNAs with sense + sense (s17s21, s19s21, s21s21, s23s21, and s25s21) and sense + antisense (s17a21, s19a21, s21a21, s23a21, and s25a21) alignments. In the pGD RNA-expression vector, the upper and lower strand RNAs are transcribed from the H1 and U6 promoters, respectively. (Green boxes) With E2 sequences and (red boxes) with NS3 sequences show the positions of antisense sequences in each duplex RNA. (Underlined) Probe sequences for Northern blot. (C) Northern blot analysis for detection of (AS) antisense strands of intracellularly processed siE2 and siN from tsiRNAs. Detection of cellular U6 snRNA was used as a loading control.
FIGURE 5.
Antiviral activity of Pol III promoter-driven siRNAs and tsiRNAs in HCV replicon cells, FK-R2AN. (A) tsiRNA-mediated inhibition of HCV replication in FK/R2AN cells. HCV replicon cells expressing the Renilla luciferase reporter were transfected with 1 μg of the siRNA- or tsiRNA-expression pGD plasmids indicated. On day 2, cell lysates were harvested for the Renilla luciferase assay. Reporter protein expression levels of cells transfected with (left) the control siRNA (siC) or (right) tsiRNA (tsiC) construct were set at 100% in each graph. Assays were performed in triplicate. (B) tsiRNA-mediated inhibition of HCV core gene expression. Immunoblotting was performed using a core-specific antibody and an antibody against β-actin (an internal loading control). (C) tsiRNA-mediated inhibition of HCV NS5A gene expression in FK/R2AN cells. Cells transfected with representative tsiE2N RNA-expressing pGD vectors indicated were immunostained with rabbit anti-NS5A polyclonal antibody and goat anti-rabbit IgG-Texas Red (red) on day 2.
Identification of cleavage sites within the HCV RNA genome
To determine RNAi cleavage sites within the viral RNA, 5′ rapid amplification of cDNA ends (RACE) was performed with total RNA from FK/R2AN cells as described in the Materials and Methods (Fig. 6A). As shown by gel electrophoresis, transfection of single siRNA siE2- and siN-encoding plasmids resulted in their specific amplification products (Fig. 6B). No PCR bands were detected in control tsiC-expression plasmid-treated cells. It is important to note that both E2 and NS3 RACE products were amplified from tsiE2N(s21s21), tsiE2N(s23s21), and tsiE2N(s25s21) RNA-expressing cells. In contrast, only NS3 RNA cleavage was achieved by tsiE2N(s17s21) and tsiE2N(s19a21), indicating that these tsiRNAs are processed into functional siN but not siE2 (Fig. 6B).
FIGURE 6.
Identification of the 5′-end cleavage sites of tsiRNA using 5′ rapid amplification of cDNA ends (RACE) technology. (A) Schematic illustration of the RACE strategy. An adaptor RNA (Adt) was ligated to total RNA from FK/R2AN cells transfected with siRNA- or tsiRNA-encoding constructs. The RNA pool was reverse-transcribed with random oligomers and then amplified with adaptor-specific primers and either E2-specific (for E2 RACE) or NS3-specific (for NS3 RACE) primers. (Arrows) Primer binding sites used for PCR amplification. (B) RT-PCR amplification showing the (top) E2-specific and (middle) NS3-specific RACE products from FK/R2AN cells transfected with indicated siRNA- or tsiRNA-expression vectors. (Bottom) Amplification of β-actin was used as an internal loading control. Each product is marked with an arrow on the right of agarose gels stained with ethidium bromide. (C) Sequence analysis of (left) E2 and (right) NS3 RACE products. The HCV sequences of the 22-nt siE2 and 21-nt siN target sites are represented above. (Arrows) Predicted cleavage points within the viral RNA by (solid arrows) 21-nt and (dotted arrows) 22-nt siRNA. (Underlined) The extra sequence on 22-nt siE2, which is also identical with viral RNA sequence, for efficient transcription. (White boxes and both solid and dotted arrows on the sequences) The 5′-termini of correctly cleaved viral RNAs. (Gray arrow) The 3-nt-shifted cleavage position on the HCV E2 coding region by tsiE2N(s25s21).
The individual PCR product was inserted into a sequencing vector. The resulting sequencing analyses show that single siE2 (22 nt) cleaves the viral nucleotides paired to bases between positions 10 and 11 from the 5′-end of the antisense siRNA as predicted (Elbashir et al. 2001). siN (21 nt) also recognizes the expected cleavage site within the NS3-coding region (Fig. 6C). Importantly, these sequencing data provide a strong evidence that the 21 + 21 and 23 + 21 tsiRNAs, tsiE2N(s21s21) and tsiE2N(s231s21), are able to target and cut both the E2 and NS3 sites as precisely as single siRNAs. Interestingly, tsiE2N(s25s21) cleaves the predicted position on HCV NS3 RNA homogeneously, while its major cleavage sites within E2 viral RNA were located at two positions, between nucleotides 10 and 11 (∼50% of total sequencing samples, indicated with a solid arrow in Fig. 6C) and also between nucleotides 7 and 8 shifted by 3 bases to the 3′ direction on the viral RNA (∼50%, indicated with a gray arrow in Fig. 6C). Taken together, the results indicate that 21- to 23-mer is the optimal length for producing functional siRNA components from tsiRNAs, which can homogeneously cleave target RNAs at the predicted position by the cellular RNAi machinery.
Silencing of mutant sequences by tsiRNAs
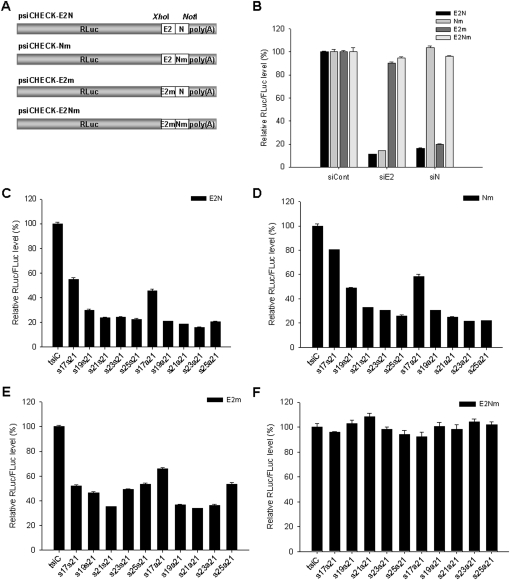
HCV is one of the highly escape-prone RNA viruses. There was an experimental evidence that viral genomic mutations could be elicited through repeated treatments with a single siRNA (Wilson and Richardson 2005). In this study, to evaluate the potency of dual functional tsiRNA for overcoming this limitation, target sequences, that is, wild-type E2 and mutant NS3 (Nm), mutant E2 and wild-type NS3 (E2m), and both E2 and NS3 mutations (E2Nm) as well as whole wild-type sequences (E2N), were prepared and cloned downstream from the RLuc-coding gene of psiCHECK-2 (Fig. 7A). First, we investigated the sequence-specific RNAi activity of single siRNA vectors using these targets (Fig. 7B). As expected, siE2 and siN inhibited only perfectly matched target gene expression but not the mismatched mutants. This means that the use of single siRNA-encoding plasmid could not overcome the emergence of HCV escape variants.
FIGURE 7.
Silencing of mutant HCV sequences by tsiRNAs. (A) Construction of target vectors. Wild-type or mutant E2 and NS3 siRNA-binding sequences were fused and cloned into XhoI and NotI sites downstream from the RLuc gene of the psiCHECK-2 vector. (E2N) Wild-type siE2 and siN-target sequence; (Nm) wild-type siE2 and mutant siN-target sequence; (E2m) mutant siE2 and wild-type siN-target sequence; (E2Nm) mutant siE2 and siN-target sequence. (B) Specific inhibition of HCV target sequences by pGD-siRNA vectors, pGD-siE2 and pGD-siN, relative to a control pGD-siC. (C–F) RLuc/FLuc activity in cells co-transfected with a series of HCV tsiRNA-expressing variants in the presence of psiCHECK-E2N, psiCHECK-Nm, psiCHECK-E2m, or psiCHECK-E2Nm 2 d post-transfection. Relative luciferase activity of cells treated with pGD-tsiC was set at 100%. The values represent the means (+SD) from three replicate transfections.
In a multitargeting approach, the length and orientation variant tsiRNA-expression vectors were co-transfected with psiCHECK-2-derived targets. The net silencing activity against wild-type E2N sequences was maximized when 21 + 21 and 23 + 21 tsiRNAs were treated, which was inpendent of sense + sense or sense + antisense orientation (Fig. 7C). Interestingly, E2 gene silencing increased in proportion to the length of the E2 siRNA unit from 17 nt to 25 nt (Fig. 7D), whereas NS3 silencing was maximized when siRNAs in tsiRNA were 21 + 21 nt long (Fig. 7E). In the double mutant targets used as a negative control, RNAi effect was not observed by any tsiRNA (Fig. 7F). These data clearly show that tsiRNAs of 21 + 21 or 23 + 21 nt are efficiently processed into separate siRNAs, consistent with Northern analysis (Fig. 4C), and eventually down-regulate their complementary sequences, possibly reducing the chance of viral escape.
Effect of tsiRNA on the intracellular miRNA processing and interferon response in mammalian cells
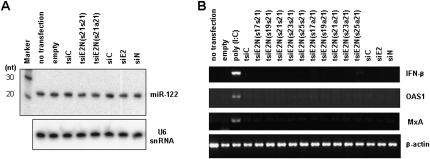
We asked whether overexpression of exogenous tsiRNA could affect cellular miRNA maturation machinery as tsiRNA should be transported from the nucleus to the cytoplasm by cellular Exportin5, which is a key protein to facilitate cytoplasmic migration of nuclear pre-miRNAs. To better understand the competition between these molecules for Exportin5, the level of miR-122 as a representative hepatic miRNA was measured in the absence and the presence of Pol III-expressed siRNA or tsiRNA in Huh7 cells. As shown in Figure 8A, both irrelevant and HCV-specific siRNA and tsiRNA (21 + 21) constructs had no effect on cellular miR-122 maturation and accumulation.
FIGURE 8.
Effect of tsiRNA expression on the hepatocyte metabolism. (A) Northern blot analysis for miR-122 in hepatocytes. Huh7 cells were treated with indicated siRNA or tsiRNA-expression vector. (Empty) pGD vector containing only the U6 promoter and termination sequences without insert for transcription (negative control). On day 2, total RNA samples were loaded onto 15% PAGE/7 M urea and hybridized with 32P-labeled complementary oligonucleotide for detection of miR-122. (Upper) The marker is a radio-labeled 10-nt ladder. (Lower) U6 snRNA was used as an internal loading control on 5% PAGE/7 M urea. (B) IFN response to cellular expressed tsiRNA. poly(I:C) as a strong IFN pathway stimulator was used as a positive control. Total RNA was isolated from cells on day 2 post-transfection and subjected to semiquantitative RT-PCR amplification of mRNAs from genes encoding IFN-β (35 cycles), OAS1 (20 cycles), and MxA (30 cycles). β-Actin mRNA expression was analyzed as an internal control.
It has been reported that dsRNAs longer than 30 bp can induce IFN signaling in mammalian cells, resulting in nonspecific gene silencing (Manche et al. 1992). Although the mammalian cell line Huh7 is known to exhibit tolerance to dsRNA-induced IFN stimulation (Watanabe et al. 2006), it is important to determine whether any of the anti-HCV effect of tsiRNAs observed above was due to induction of IFN by long dsRNA. We performed a semi-quantitative RT-PCR assay to measure the mRNA expression levels of genes encoding IFN-β, OAS1, and MxA in FK/R2AN cells transfected with the siRNA- or tsiRNA-expression plasmids. No IFN pathway activation was observed following transfection with either the siRNA or longer tsiRNA constructs, whereas strong IFN stimulation was observed with poly(I:C) (Fig. 8B). This finding indicates that the in vitro gene silencing activity of tsiRNA against HCV was derived from sequence-specific RNAi activity rather than from an IFN response. In addition, these duplex RNA constructs exhibited a similar IFN-response profile in Huh7 cells (data not shown). However, they stimulated the IFN pathway as effectively as the positive control poly(I:C) in the human prostate cancer cell line PC3 (data not shown). This finding also indicates that there might be a variation among different cell lines with respect to IFN sensitivity dependent on dsRNA length. Taken together, our results suggest that expression of 21 + 21 or 23 + 21 HCV-specific tsiRNAs generates functional siRNA components and causes RNAi-mediated antiviral activity in a sequence-specific manner in HCV-replicating hepatocytes.
DISCUSSION
Previous reports have demonstrated that synthetic siRNA or plasmid-encoded shRNA can be used for sequence-specific viral inhibition particularly against RNA viruses such as HCV or HIV (Coburn and Cullen 2002; Lee et al. 2002; Novina et al. 2002; Kapadia et al. 2003; Randall et al. 2003; Yokota et al. 2003; Kim et al. 2006). This is because the viral RNA transcripts are produced or replicated in the cytoplasm, where cellular metabolic processes can also initiate the RNAi machinery. However, an important concern regarding the application of RNAi to treat viral infections is the generation of viral mutants that are resistant to the original siRNAs. Several lines of evidence have shown rapid emergence of HCV or HIV-1 escape variants following long-term and repeated treatment of homogeneous antiviral siRNAs, eventually leading to loss of gene silencing activity (Boden et al. 2003; Das et al. 2004; Wilson and Richardson 2005). Currently, diverse approaches have been used to overcome this problem by the use of synthetic siRNA mixture against different sites, Dicer-generated products from long dsRNAs, and expression of long hairpin RNAs (Wilson and Richardson 2005; Watanabe et al. 2006; Liu et al. 2007; Sano et al. 2008). In this study, we attempted to explore the basic principle of designing linear tandem siRNA (tsiRNA) that produces two effective siRNAs by targeting different reporter genes (EGFP and RLuc) or HCV genes (E2 and NS3). When compared with an approach for expression of two hairpin RNAs on the same vector, the linear tandem RNA expression using two convergent promoters has the advantage of improved genetic stability of plasmid DNA and reduced cost of oligomer synthesis.
In experiments to decide the optimal length required for inducing multitargeting RNAi triggers in HCV replicon cells, we observed that the most significant knockdown and efficient siRNA production was achieved using linear dsRNA (between 21 + 21 and 23 + 21 nt) with a 40–42-bp stem and 2-nt 3′-overhangs (Figs. 4, 5, 7). RACE data support their precise and concomitant cleavage of viral E2 and NS3 genes (Fig. 6). Interestingly, although the overall gene silencing by 25 + 21-nt (44-bp stem) tsiRNA and inhibition activity derived from 25-nt viral E2 sequence were significant, the 21-nt viral NS3-recognition binding site was marginally active (Fig. 7). In contrast, a previous report suggested that dual gene knockdown is maximized when 44-bp stem-sized duplex RNA-expressing plasmids were used for HIV-1 inhibition (Liu et al. 2007). This discrepancy between the optimal lengths of duplex lhRNA and tsiRNA is likely to result from differences in their siRNA processing mechanisms. In principle, Dicer could perform unidirectional and successive counting from the base of an lhRNA stem to the loop as suggested by Weinberg et al. (2007), whereas potentially both unidirectional and bidirectional processing could occur at the both exposed stem termini of tsiRNA. However, currently, it is not certain if the bidirectional cleavage within tsiRNA is processed primarily or if the biased or random one-side cleavage brings about two kinds of siRNA products. The important information from this study on silencing of viral mutant sequences is that treatment with length-optimized tsiRNA constructs (21 + 21 or 23 + 21) could reduce the chance of viral escape relative to single siRNA expression (Fig. 7). Our data indicate that the domains with unoptimized lengths, such as 17-nt and 25-nt fragments in 17 + 21 and 25 + 21, can negatively affect and reduce the processing correctness or RNAi activity of the remaining 21-nt partner (Fig. 7E). We also investigated the relationship between the polarity of Dicer processing and RNAi activity by inverting the orientation of one siRNA component within tsiRNAs (Figs. 2, 3, 5, 7). Results revealed that sense + sense and sense + antisense tsiRNAs have similar gene silencing effects. These findings reflect the fact that the contribution of thermodynamic stability of siRNA for selective RISC incorporation of antisense strand might be determined after Dicer cleavage (Khvorova et al. 2003; Schwarz et al. 2003). Moreover, the tsiRNA-expression vectors used in this study did not affect the native miRNA maturation machinery or the IFN response pathway (Fig. 8).
To our knowledge, this is the first report suggesting actual multidicing activity of tandem siRNAs expressed from convergent Pol III promoters in cells. As a further study using the liver-targeted gene delivery technology, multisilencing activity of unmodified or chemically modified, synthetic tsiRNAs could also be evaluated (Kim et al. 2007). In summary, our results propose that different species of siRNA processed from a vector-based, constitutively expressed linear tsiRNA are able to induce sequence-specific cleavage and silencing of cognate mRNAs. This study provides the basic principle for the design of tsiRNAs for multiple cleavage of viral RNAs as well as for simultaneous knockdown of both pathogenic and host mRNAs to overcome problems inherent in the use of a single siRNA.
MATERIALS AND METHODS
Construction of DNA plasmids
A linear dsRNA expression cassette was prepared by PCR amplification of the human RNA polymerase III U6 and H1 promoters from pRNAiDu using their respective forward (f) primers: U6f, 5′-CGGAATTCCCCAGTGGAAAGAC-3′ and H1f, 5′-CGGAATTCATATTTGCATGTCGC-3′ (Zheng et al. 2004; Shin et al. 2006). The PCR product (∼470 bp) was purified from a 3% agarose gel and cloned with the pGEM-T Easy Vector System (Promega), generating the siRNA control plasmid pGD-siC (Fig. 1A). For cloning of siRNA- or tsiRNA-expression plasmids, complementary oligonucleotides were annealed and ligated into the BamHI and HindIII sites of pGD-siC.
The psiCHECK target vectors were prepared by cloning of wild-type or mutant E2-NS3 siRNA-binding oligonucleotides into XhoI and NotI enzyme sites downstream from the RLuc gene of psiCHECK-2 (Promega). Complementary oligonucleotides for cloning of psiCHECK vectors were as follows:
psiCHECK-E2N, E2N-fwd, 5′-TCGAG AAGGCAACTGGTTTGGCTGTACATGGTCTAAGGCACATGGTATCGACCCT GC-3′ and
E2N-rev, 5′-GGCCGCAGGGTCGATACCATGTGCCTTAGACCATGTACAGCCAAACCAGTTGCCTTC-3′;
psiCHECK-Nm, Nm-fwd, 5′-TCGAG AAGGCAACTGGTTTGGCTGTACATGGTCTAAGGCACATATGATCGAGAGT GC-3′ and
Nm-rev, 5′-GGCCGCACTCTCGATACATTGTGCCTTAGACCATGTACAGCCAAACCAGTTGCCTTC-3′;
psiCHECK-E2m, E2m-fwd, 5′-TCGAG AAGGCAACTTAGTTGGCAAGACATGGTCTAAGGCACATGGTATCGACCCT GC-3′ and
E2m-rev 5′-GGCCGCAGGGTCGATACCATGTGCCTTAGACCATGTCTTGCCAACTAAGTTGCCTTC-3′; and
psiCHECK-E2Nm, E2Nm-fwd, 5′-TCGAG AAGGCAACTTAGTTGGCAAGACATGGTCTAAGGCACATATGATCGAGAGT GC-3′ and
E2Nm-rev 5′-GGCCGCACTCTCGATACATTGTGCCTTAGACCATGTCTTGCCAACTAAGTTGCCTTC-3′.
Enzyme sites are in italics, mutant sequences are bold, and the binding sites of 21-nt antisense strands of siRNAs are underlined. All sequences were verified using the BigDye Terminator Cycle Sequencing kit (ABI).
Cell culture and transfection
The human hepatoma cell line Huh7 was maintained in Dulbecco's modified Eagle's medium (DMEM; HyClone) supplemented with 10% fetal bovine serum (FBS; HyClone). The day before transfection, Huh7 cells were seeded into 12-well plates at a density of 1.5 × 105 per well in all experiments. To determine the gene silencing effect derived from siRNA- or tsiRNA-expressing plasmids, Lipofectamine 2000 (Invitrogen) was used to co-transfect Huh7 cells with 10 ng of pEGFPLuc (BD Biosciences Clontech) and 1 ng of phRL-CMV (Promega) as targets in the presence of 1 μg of a series of pGD constructs, according to the manufacturers’ instructions. For examination of multiple gene silencing, 0.9 μg of psiCHECK and 0.1 μg of pGD-siRNA or -tsiRNA vectors were co-transfected into Huh7 cells. RLuc expression was normalized to internal FLuc activity 2 d post-transfection. For monitoring miRNA levels in hepatocytes, pGD vectors (1 μg) were transfected.
The HCV genotype 1b replicon-containing cell line FK/R2AN, which was constructed by insertion of the fused Renilla luciferase and FMDV 2A genes upstream of the neomycin resistance cassette within the original replicon (accession number AJ238799), was obtained from R. Bartenschlager (University of Heidelberg, Germany) and modified by Sung K. Jang (Pohang University of Science and Technology, South Korea) (Lohmann et al. 1999; Kim et al. 2006). Cells were maintained in DMEM (HyClone) containing 10% FBS (HyClone) and 0.6 mg/mL G418 (Calbiochem). Prior to transfection, viral replication in replicon-containing cells was validated by luciferase assay and Western blot analysis as described previously (Kim et al. 2006). To test the RNAi activity of expressed siRNA or tsiRNA, 1.5 × 105 cells were seeded into 12-well plates, transfected with 1 μg of pGD-derived plasmids using Lipofectamine 2000 (Invitrogen) and then incubated for 2 d at 37°C. For IFN response analysis, cells were treated with 1 μg of poly(I:C), or siRNA- or tsiRNA-encoding plasmid DNA, using the same reagent.
Luciferase assays
On day 2, Huh7 cells transfected with plasmids encoding luciferases were lysed with 1× Passive Lysis Buffer (Promega) by shaking at room temperature for 30 min. Both firefly and Renilla luciferase expression levels were determined using the Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer's instructions. In FK/R2AN cells, HCV replication efficiency was determined by measuring reporter protein levels with the Renilla Luciferase Assay System (Promega) at day 2 post-transfection.
Northern blot analysis
For Northern blot analysis of expressed or processed siRNAs or cellular miR-122, Huh7 cells or FK/R2AN cells were transfected with pGD vectors and incubated for 2 d, and then total RNA was extracted using Trizol reagent (Invitrogen). RNA (5 μg per lane) was separated on a 7 M urea/15% (for siRNA or miRNA detection) or 5% (for U6 snRNA detection) polyacrylamide gel and then transferred to a Hybond N+ nylon membrane (Amersham Bioscience). The membrane was hybridized with [γ-32P]-labeled DNA probes complementary to either the siRNA antisense strand (or miRNA) or U6 snRNA, and then hybridization was performed in ExpressHyb Hybridization Solution (Clontech Laboratories Inc.), according to the manufacturer's protocol. The following probe sequences were used:
EGFP siRNA, 5′-GCAGCACGACTTCTTCAAG-3′;
RLuc siRNA, 5′-GGGCGAGGTTAGACGGCCT-3′;
E2 siRNA, 5′-GGCAACTGGTTTGGCTGTA-3′;
NS3 siRNA, 5′-GGCACATGGTATCGACCCT-3′; and
U6 snRNA, 5′-TAGTATATGTGCTGCCGAAGCGAGCA-3′.
The cellular miR-122 level in Huh7 cells was determined using its specific probe, 5′-ACAAACACCATTGTCACACTCCA-3′. The radiolabeled Decade Marker System (Ambion) was used as an RNA marker.
Western blot analysis
To measure HCV protein expression, FK/R2AN cells alone or transfected with pGD vectors were lysed using the M-PER Mammalian Protein Extraction Reagent (Pierce). Total cell lysates (30 μg) were resolved in a 12% SDS polyacrylamide gel and transferred to an Immobilon-P PVDF membrane (Millipore). HCV core protein was identified using its specific primary antibody purchased from Affinity Bioreagents. Cellular β-actin protein was detected using anti-human β-actin antibody (Sigma). Band intensities were quantified with the public domain software ImageJ from the National Institutes of Health (http://rsb.info.nih.gov/ij/).
5′ Rapid amplification of cDNA ends (5′-RACE)
To evaluate viral RNA cleavage by processed siRNAs and to determine their cleavage sites, 2 μg of total RNA from transfected FK/R2AN cells was ligated with a 5′-RACE RNA Adapter and then subjected to reverse-transcription using the FirstChoice RLM-RACE Kit (Ambion), according to the modified manufacturer's instructions. Two sets of outer primers were used for primary amplification of the E2 and NS3, the 5′-RACE Outer Primer from the Ambion RACE kit, and an E2 outer primer, 5′-CCGTACAGGTATTGTACGTCC-3′ or NS3 outer primer, 5′-TTGGTCCAGGACTGTGCCGAT-3′, respectively. The E2 and NS3 DNA pools were reamplified with two additional sets of inner primers, the 5′-RACE Inner Primer from the Ambion RACE kit and an E2 inner primer, 5′-CTAAGCTCTGATCTGTCCCTG-3′ or NS3 inner primer, 5′-GTCAGTTGAGTGGCACTCAT-3′, respectively. As an internal control, cellular β-actin was amplified with gene-specific primers (Shin et al. 2007). The PCR products were resolved on a 2% agarose gel, and the E2 (340 bp) and NS3 (177 bp) fragments were eluted separately. Following cloning of amplified DNA into pCR2.1-TOPO (Invitrogen), cleavage sites were determined by sequencing analysis with the universal T7 primer.
Interferon assay
Total RNA isolated from FK/R2AN cells was reverse-transcribed into cDNA using oligo(dT)12–18 primer (Invitrogen) 2 d after transfection with 1 μg of the different pGD vectors. Poly(I:C) dsRNA was used as a positive control to induce the IFN response. And human IFN-β, OAS, and MxA genes were amplified with their respective forward and reverse primers: IFN-β, 5′-ATGACCAACAAGTGTCTCCT-3′ and 5′-TCAGTTTCGGAGGTAACCTG-3′ (35 cycles); OAS, 5′-TCAGAAGAGAAGCCAACGTGA-3′ and 5′-CGGAGACAGCGAGGGTAAAT-3′ (20 cycles); and MxA, 5′-AGTATGGTGTCGACATACCGGA-3′ and 5′-GAGTCTGGTAAACAGCCGAATG-3′ (20 cycles) (Shin et al. 2007; Weinberg et al. 2007). Cellular β-actin mRNA was also amplified with specific primers (35 cycles) and then used for normalization (Shin et al. 2007). The PCR products were analyzed on a 2% agarose gel.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Young Joo Kim (Korea Research Institute of Bioscience and Biotechnology) for calculation of the internal stability profile of siRNA duplexes. We are grateful to Sihyun Kim for critical readings of the manuscript and constructive comments. This work was supported by funds from the Green Cross Corporation, South Korea.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1268209.
REFERENCES
- Alter M.J. Epidemiology of hepatitis C. Hepatology. 1997;26:62S–65S. doi: 10.1002/hep.510260711. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R., Lohmann V. Replication of hepatitis C virus. J. Gen. Virol. 2000;81:1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- Boden D., Pusch O., Lee F., Tucker L., Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosher J.M., Labouesse M. RNA interference: Genetic wand and genetic watchdog. Nat. Cell Biol. 2000;2:E31–E36. doi: 10.1038/35000102. [DOI] [PubMed] [Google Scholar]
- Capodici J., Kariko K., Weissman D. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 2002;169:5196–5201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- Chander G., Sulkowski M.S., Jenckes M.W., Torbenson M.S., Herlong H.F., Bass E.B., Gebo K.A. Treatment of chronic hepatitis C: A systematic review. Hepatology. 2002;36:S135–S144. doi: 10.1053/jhep.2002.37146. [DOI] [PubMed] [Google Scholar]
- Coburn G.A., Cullen B.R. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 2002;76:9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A.T., Brummelkamp T.R., Westerhout E.M., Vink M., Madiredjo M., Bernards R., Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y., Tuschl T. siRNAs: Applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn D.M., Novina C.D., Sharp P.A. Killing the messenger: Short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Wychowski C., Lin C., Feinstone S.M., Rice C.M. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia S.B., Brideau-Andersen A., Chisari F.V. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. 2003;100:2014–2018. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaykas A., Moon R.T. A plasmid-based system for expressing small interfering RNA libraries in mammalian cells. BMC Cell Biol. 2004;5:16. doi: 10.1186/1471-2121-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim M., Shin D., Kim S.I., Park M. Inhibition of hepatitis C virus gene expression by small interfering RNAs using a tri-cistronic full-length viral replicon and a transient mouse model. Virus Res. 2006;122:1–10. doi: 10.1016/j.virusres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Kim S.I., Shin D., Choi T.H., Lee J.C., Cheon G.J., Kim K.Y., Park M., Kim M. Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I. Mol. Ther. 2007;15:1145–1152. doi: 10.1038/sj.mt.6300168. [DOI] [PubMed] [Google Scholar]
- Lee N.S., Dohjima T., Bauer G., Li H., Li M.J., Ehsani A., Salvaterra P., Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Lee N.S., Kim D.H., Alluin J., Robbins M., Gu S., Li H., Kim J., Salvaterra P.M., Rossi J.J. Functional and intracellular localization properties of U6 promoter-expressed siRNAs, shRNAs, and chimeric VA1 shRNAs in mammalian cells. RNA. 2008;14:1823–1833. doi: 10.1261/rna.1014008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Feng X.-H. Design and application of a versatile expression vector for RNAi in mammalian cells. J RNAi Gene Silencing. 2005;1:38–43. [PMC free article] [PubMed] [Google Scholar]
- Liu Y.P., Haasnoot J., Berkhout B. Design of extended short hairpin RNAs for HIV-1 inhibition. Nucleic Acids Res. 2007;35:5683–5693. doi: 10.1093/nar/gkm596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V., Korner F., Koch J., Herian U., Theilmann L., Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Manche L., Green S.R., Schmedt C., Mathews M.B. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novina C.D., Murray M.F., Dykxhoorn D.M., Beresford P.J., Riess J., Lee S.K., Collman R.G., Lieberman J., Shankar P., Sharp P.A. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- Randall G., Grakoui A., Rice C.M. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M., Li H., Nakanishi M., Rossi J.J. Expression of long anti-HIV-1 hairpin RNAs for the generation of multiple siRNAs: Advantages and limitations. Mol. Ther. 2008;16:170–177. doi: 10.1038/sj.mt.6300298. [DOI] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Shin D., Kim S.I., Kim M., Park M. Efficient inhibition of hepatitis B virus replication by small interfering RNAs targeted to the viral X gene in mice. Virus Res. 2006;119:146–153. doi: 10.1016/j.virusres.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Shin D., Kim S.I., Park M., Kim M. Immunostimulatory properties and antiviral activity of modified HBV-specific siRNAs. Biochem. Biophys. Res. Commun. 2007;364:436–442. doi: 10.1016/j.bbrc.2007.10.012. [DOI] [PubMed] [Google Scholar]
- ter Brake O., ’t Hooft K., Liu Y.P., Centlivre M., von Eije K.J., Berkhout B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol. Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Sudoh M., Miyagishi M., Akashi H., Arai M., Inoue K., Taira K., Yoshiba M., Kohara M. Intracellular-diced dsRNA has enhanced efficacy for silencing HCV RNA and overcomes variation in the viral genotype. Gene Ther. 2006;13:883–892. doi: 10.1038/sj.gt.3302734. [DOI] [PubMed] [Google Scholar]
- Weinberg M.S., Ely A., Barichievy S., Crowther C., Mufamadi S., Carmona S., Arbuthnot P. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol. Ther. 2007;15:534–541. doi: 10.1038/sj.mt.6300077. [DOI] [PubMed] [Google Scholar]
- Wilson J.A., Richardson C.D. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J. Virol. 2005;79:7050–7058. doi: 10.1128/JVI.79.11.7050-7058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T., Sakamoto N., Enomoto N., Tanabe Y., Miyagishi M., Maekawa S., Yi L., Kurosaki M., Taira K., Watanabe M., et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 2003;4:602–608. doi: 10.1038/sj.embor.embor840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Liu J., Batalov S., Zhou D., Orth A., Ding S., Schultz P.G. An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc. Natl. Acad. Sci. 2004;101:135–140. doi: 10.1073/pnas.2136685100. [DOI] [PMC free article] [PubMed] [Google Scholar]