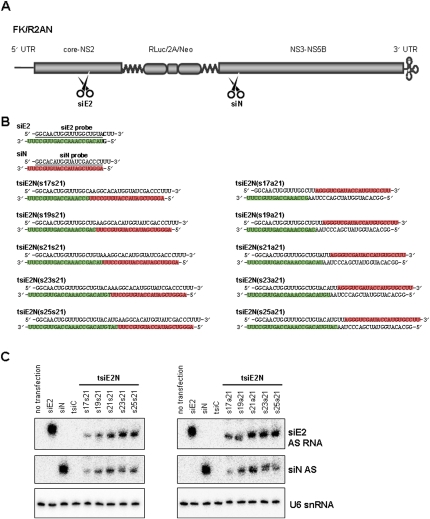
FIGURE 4.
Construction of tsiRNA-expression vectors targeting HCV genome. (A) Schematic representation of the HCV replicon FK-R2AN RNA and anti-HCV siRNA target sites against E2 (siE2) and NS3 (siN) coding genes. (5′ UTR) 5′-Untranslated region; (core-NS2) coding region for viral core, E1, E2, p7 to NS2; (RLuc) Renilla luciferase; (2A) foot-and-mouth disease virus (FMDV) 2A; (Neo) neomycin resistance gene; (NS3-NS5B) coding regions for viral proteins NS3, NS4A, NS4B, NS5A to NS5B; and (3′ UTR) 3′-untranslated region. (B) Sequence composition of HCV E2- (siE2) and NS3-specific (siN) siRNAs, as well as tsiE2N RNAs with sense + sense (s17s21, s19s21, s21s21, s23s21, and s25s21) and sense + antisense (s17a21, s19a21, s21a21, s23a21, and s25a21) alignments. In the pGD RNA-expression vector, the upper and lower strand RNAs are transcribed from the H1 and U6 promoters, respectively. (Green boxes) With E2 sequences and (red boxes) with NS3 sequences show the positions of antisense sequences in each duplex RNA. (Underlined) Probe sequences for Northern blot. (C) Northern blot analysis for detection of (AS) antisense strands of intracellularly processed siE2 and siN from tsiRNAs. Detection of cellular U6 snRNA was used as a loading control.