Abstract
Ribonuclease III (RNase III) represents a highly conserved family of double-strand-specific endoribonucleases that are important for RNA processing and post-transcriptional gene regulation in both prokaryotes and eukaryotes. We constructed a single amino acid substitution (E38A) of RNase III that shows a unique and useful enzymatic activity. It produces a dsRNA product of a discrete size migrating as 23 base pairs (bp) when given a long dsRNA as a substrate in an easy-to-control reaction. We demonstrate that the RNase III(E38A) mutant produces the 23-bp dsRNA product by making a double-strand cleavage of the long dsRNA substrate with the product being protected from further digestion. Using the hairpin RNA R1.1 as a substrate, RNase III(E38A) cleaves at the primary site and remains bound to the RNA, thereby preventing cleavage at the secondary site. The 23-bp dsRNA product is demonstrated to be a pool of dsRNAs representative of the long dsRNA substrate and has RNA interference activity in mammalian tissue culture transfection experiments. The RNA interference activity suggests that the 23-bp dsRNA product has typical 2-nucleotide 3′ overhangs and behaves as siRNA thereby making it a useful tool in RNA interference experiments.
Keywords: RNA interference, Dicer, enzyme activity
INTRODUCTION
Ribonuclease III (RNase III) represents a highly conserved family of double-strand-specific endoribonucleases that are important for RNA processing and post-transcriptional gene regulation in both prokaryotes and eukaryotes (Court 1993; Lamontagne et al. 2001; Nicholson 2003). The family can be divided into three classes. Class 1 is the simplest in structure, having a single ribonuclease domain and a dsRNA-binding domain, and is the best characterized. Its members are found in eubacteria, archaebacteria, and yeast. Class 2 members have two ribonuclease domains and a single dsRNA-binding domain. These are found in eukaryotes with Drosha being a typical example (Lee et al. 2006). Class 3, also known as the Dicer family of enzymes, are the largest and typically contain two ribonuclease domains, a dsRNA-binding domain, a DEAD box helicase domain, and a PAZ domain (Lamontagne et al. 2001; Nicholson 2003; MacRae and Doudna 2007).
Each RNase III family member contains a characteristic ribonuclease domain, the RNase III domain. This catalytic domain cleaves dsRNA, producing a characteristic 5′ phosphate with a 2-nucleotide (nt) overhang at the 3′ end. The domain contains a stretch of conserved residues, ERLEFLGD (residues 38–45 of the Escherichia coli RNase III), known as the RNase III signature motif that makes up a significant portion of the active site. RNase III enzyme functions as a homodimer with dimerization occurring through the ribonuclease domain. The two ribonuclease domains combine to form a single processing center, with each domain contributing to the hydrolysis of one RNA strand of the duplex substrate. Structural studies of Aquifex aeolicus RNase III (Aa-RNase III) (Blaszczyk et al. 2001), and catalytic studies of E. coli RNase III (Ec-RNase III) reveal a symmetric RNA cleavage domain with catalytic valley that can accommodate two turns of the dsRNA helix (Nicholson 2003; Zhang et al. 2004; Pertzev and Nicholson 2006). A cluster of six acidic residues—E38, E41, D45, D114, and E117 (for Ec-RNase III) from one subunit and E65 from the partner subunit—is located at either end of the valley (Blaszczyk et al. 2001). E38 is involved in protein dimerization; E65 is involved in substrate recognition and scissile-bond selection; D45, D114, and D117 chelate the Mn2+ ion (Blaszczyk et al. 2001; Sun and Nicholson 2001); and E41, D45, D114, and E117 carry out the hydrolysis of the scissile bond (Gan et al. 2006).
In E. coli, cleavage of dsRNA by RNase III is an essential step in both maturation and degradation of many coding and noncoding RNAs (Nicholson 1999, 2003). Cellular substrates of E. coli RNase III typically possess a dsRNA element of about 20 base pairs (bp) that is cleaved in a site-specific manner and is required for the proper function of the mature species or for efficient degradation. In vitro, RNase III will cleave a dsRNA at random positions. It is this nonspecific cleavage of dsRNA that has made RNase III a useful tool for RNA interference experiments.
In the work described here, we focus our attention on one critical residue, E38, which comprises the active site and was originally thought to contact the dsRNA substrate (Blaszczyk et al. 2001). We created several substitutions at this position in hopes of creating an RNase III protein with altered enzymatic activity. The similarity of the cleavage domain of bacterial RNase III to the two RNase cleavage domains of Dicer suggested that one such activity would be the generation of small dsRNAs similar in size to that generated by Dicer. One substitution, E38A, has been described previously (Blaszczyk et al. 2001; Sun et al. 2004; Zhang et al. 2004; Gan et al. 2006). However, by using long dsRNAs as substrates, which were not used in any previous study, we were able to uncover a previously unknown activity for E38A. E38A produces a dsRNA product of a discrete size that comigrates with a 23-bp dsRNA when using a long dsRNA as a substrate. This product has not been observed before and allowed us to determine the mode of activity of this mutant. Significantly, the dsRNA product is similar in size to the product generated by Dicer, and we demonstrate that E38A can generate short dsRNA fragments suitable for RNA interference experiments.
RESULTS
E38A construction and activity
An alignment of amino acid sequences of RNase III enzymes from several representative bacterial species and Saccharomyces cerevisiae points to regions of interest that could be used to construct mutants of unique and biochemically useful activities. These amino acids are 100% conserved among the RNase III sequences, are contained within the active site, and have been proposed to contact dsRNA substrate (Blaszczyk et al. 2001) and therefore would be critical to the enzyme's activity. We focused our attention on one, designated E38 in E. coli. We made several substitutions at this position with three, E38A, E38T, and E38W, having a useful enzymatic activity that produces a dsRNA product that migrates as 23 bp when using a long dsRNA substrate. The report here focuses on E38A. Two substrates were used for most of the activity assays: a 900-bp dsRNA generated from the E. coli MalE gene and the 60-nt ssRNA R1.1 (Li and Nicholson 1996; Nicholson 2003).
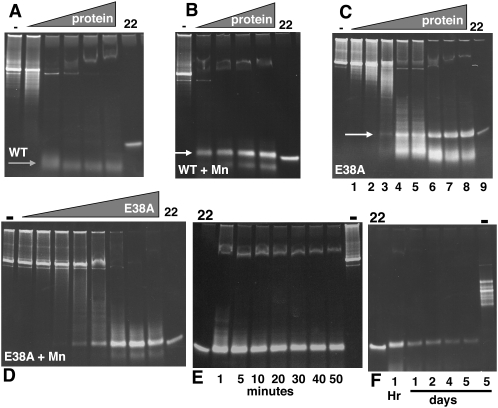
E38A was produced in E. coli as a His tag fusion protein and purified over nickel resin. As can be seen in Figure 1C, it has the ability to cleave a 900-bp dsRNA substrate to a dsRNA product that migrates approximately as 23 bp (hereafter referred to as the 23-bp dsRNA product). A smaller, diffuse band is also seen of 10–15 bp. This is in contrast to the wild-type (WT) activity (Fig. 1A) that cleaves the same substrate to 10-bp fragments; but similar to the WT enzyme with Mn2+ replacing Mg2+ as the divalent cation (Fig. 1B), which also yields a 23 bp dsRNA product (Morlighem et al. 2007). In the case of the WT enzyme with Mn2+, there is a different smaller species that appears to be slightly larger than that produced by E38A. We further explored the activity of E38A under different reaction conditions and with different substrates. Using an enzyme to substrate ratio as defined in lane 8 of the E38A panel in Figure 1C (in this particular instance, 4 μg of protein and 500 ng dsRNA in a 20 μL reaction volume), we followed the progress of digestion. The reaction appears to be complete by 10 min (Fig. 1E). However, the release of the cleaved dsRNA is extremely slow. While lesser amounts of enzyme result in longer products, these reactions reach an ending point within 10 min and do not appear to progress further (Fig. 1C). The products shown in the E38A panels of Figure 1 are relatively stable and are similar even after an overnight digestion (Fig. 1F). This is not true for the WT enzyme in Mn2+; whereas a short digestion results in a discrete product size, an overnight digestion results in the loss of the 23-bp dsRNA product (data not shown). The E38A-generated 23-bp product is fairly stable over time and is still detectable after 5 days of digestion with E38A (Fig. 1F).
FIGURE 1.
RNase activity for wild-type (WT) and E38A substitutions. A 900-bp dsRNA (500 ng) was cleaved with WT His-tagged RNase III or His-tagged E38A with a serial dilution of protein (illustrated by the gray triangle) using a starting amount of 4 μg. All reactions were performed as described in Materials and Methods. (A) WT RNase III. (B) WT RNase III with Mn2+ replacing Mg2+ in reaction buffer. (C) RNase III (E38A). (D) E38A with Mn2+ replacing Mg2+ in reaction buffer. (E) Reactions with E38A stopped at the times indicated in minutes. (F) Reactions with E38A stopped at the times indicated in days. “−” Indicates no protein in the reaction; “22” indicates a synthetic 22-nt dsRNA size marker. White arrow points to 23-bp dsRNA product; gray arrow points to smaller species product of WT RNase III.
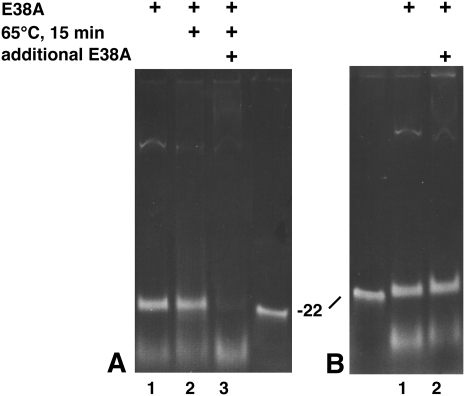
The above results suggest that the nuclease activity of E38A is not affected; but the reaction progress is considerably slower (on the order of days). To explain these results, we hypothesize that E38A once bound to the substrate makes one double-strand cleavage (i.e., cleavage of both strands of the dsRNA) and remains bound to the product for long periods of time, protecting the product from further cleavage. The 23-bp dsRNA product is only observed at a high enzyme to substrate ratio, suggesting that the length of helical dsRNA protected by the enzyme is equivalent to 23 bp. To test this hypothesis, we set up a digestion yielding a maximum amount of dsRNA product and removed one-third for gel electrophoresis (Fig. 2A, lane 1). The remaining two thirds were incubated for 15 min at 65°C to heat denature E38A. One-half of this (one-third of the original reaction) was removed for gel electrophoresis (Fig. 2A, lane 2), and additional E38A was added to the remaining one third of the original reaction, incubated for 30 min at 37°C, and electrophoresed (Fig. 2A, lane 3). Only after heat denaturation of E38A can the 23-bp dsRNA product be further digested (Fig. 2A, lane 3). Simply adding more E38A to the reaction does not result in a similar further digestion (Fig. 2B). We repeated the experiment using phenol denaturation instead of heat and obtained similar results (data not shown). A potential explanation for this is that the active E38A is protecting the dsRNA product from further digestion. But once removed from the dsRNA product (by denaturing with heat or phenol), the 23-bp dsRNA can be a substrate for further digestion. By extension, these results demonstrate that E38A, like WT RNase III, has the ability to bind and cleave a dsRNA substrate of 23 bp in size.
FIGURE 2.
E38A product release requires denaturation. (A) A 900-bp dsRNA was incubated with E38A for 30 min at 37°C in reaction buffer, and one third was removed for electrophoresis (lane 1). The remaining two thirds was incubated for 15 min at 65°C, and one-half was removed for electrophoresis (lane 2). Additional E38A was added to the remaining third and incubated for 30 min at 37°C (lane 3). (B) A 900-bp dsRNA was incubated with E38A for 30 min at 37°C in reaction buffer, and one-half was removed for electrophoresis (lane 1). Additional E38A was added to the remaining half and incubated for 30 min at 37°C (lane 2). “22” Designates a synthetic 22-nte dsRNA size marker.
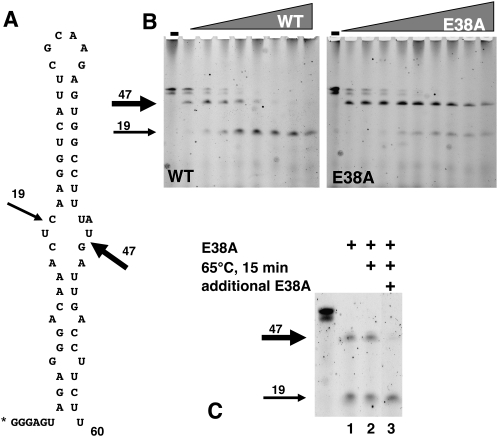
The natural substrates for RNase III are small hairpin RNA structures within E. coli, unlike the longer dsRNAs generated in vitro. To test the activity of E38A on such a substrate, we synthesized the phage T7 RNA processing signal, R1.1 (Li and Nicholson 1996; Nicholson 2003), with a fluorescein label at its 5′ end. This 60-nt RNA forms a hairpin that contains a primary and secondary RNase cleavage site (Fig. 3A). RNase III cleaves the hairpin substrate at the primary site preferentially, yielding a 47-nt product (Fig. 3B, WT panel.) Adding more enzyme to the reaction results eventually in all of the substrate being cleaved at the secondary site, yielding a 19-nt product (Fig. 3B, WT panel). In contrast, E38A cleaves the hairpin at the primary site but shows very little of the secondary site cleavage product, even with increasing amounts of protein. The lower band seen for R1.1 in the undigested lane of Figure 3, B and C, is either a smaller RNA molecule that is a result of the synthesis or represents a portion of the RNA that is not fully denatured. In either case, it does not affect the activity of either the WT RNase III or E38A. As with the long dsRNA substrate, the reactions are “complete” within 10 min, and the reaction products show no difference after an overnight digestion (data not shown). This suggests that the mode of action of E38A on R1.1 is similar as that on the dsRNA—i.e., E38A, once bound to R1.1, cleaves once and then remains bound to the cleaved product(s) for an extended period of time. To test this, we set up a similar disassociation experiment described in Figure 5 (below), this time with R1.1 as a substrate (Fig. 3C). R1.1 was digested with E38A, and one third of the reaction was removed for electrophoresis. The remaining two thirds was incubated for 15 min at 65°C, and one-half of that mixture was removed for gel electrophoresis. The final one third of the original reaction was incubated with additional E38A for 30 min at 37°C and then electrophoresed. As with the larger dsRNA as a substrate, heat denaturing the protein exposes the 47-nt abbreviated hairpin for further digestion. E38A behaves in a similar manner on R1.1 as it does on long dsRNA; i.e., once the digested R1.1 is released from E38A, it is available for further digestion.
FIGURE 3.
E38A digestion of hairpin RNA, R1.1. (A) R1.1 sequence and secondary structure are shown. Bold arrow indicates the primary cleavage site, and thin arrow indicates the secondary cleavage site. “*” indicates the fluorescein tag. (B) Fluorescein labeled R1.1 was incubated with increasing amounts of WT RNase III (left panel) or E38A (right panel) for 30 min at 37°C, electrophoresed on a denaturing polyacrylamide gel, and imaged with a PhosphorImager. Bold arrow indicates the product of cleavage at the primary site, and thin arrow indicates the product of cleavage at the secondary site. (C) Fluorescein labeled R1.1 was incubated with E38A for 30 min at 37°C in reaction buffer, and one-third was removed for electrophoresis (lane 1). The remaining two-thirds were incubated for 15 min at 65°C, and one-half was removed for electrophoresis (lane 2). Additional E38A was added to the remaining third and incubated for 30 min at 37°C (lane 3). “−” Indicates uncut R1.1.
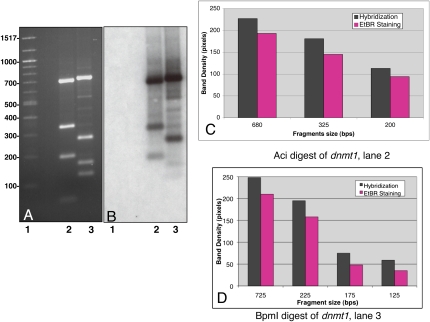
FIGURE 5.
dsRNA product generated with E38A represents a collection of 23-mers from its larger dsRNA substrate. (A) A 1112-bp fragment of dnmt1 was digested with AciI (lane 2) and BpmI (lane 3), electrophoresed on a 1.25% agarose gel, and imaged via EtBr staining. (Lane 1) Molecular weight size standards. (B) The gel was transferred to nitrocellulose and hybridized with 32P-labeled 23-bp dsRNA generated by E38A from a 1112-bp dsRNA transcribed from the same dnmt1 DNA fragment. Lanes 1–3 correspond to lanes 1–3 in A. (C) Bar graph of intensity (in pixels) of EtBr staining (purple) and hybridization intensity (gray) for each AciI-generated DNA fragment. (D) Bar graph of intensity (in pixels) of EtBr staining (purple) and hybridization intensity (gray) for each BpmI-generated DNA fragment.
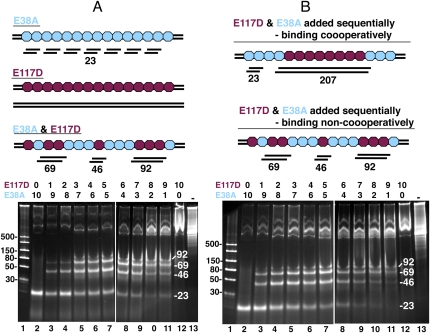
One would expect that if the binding of E38A along the length of the dsRNA substrate was at random positions, then the observed dsRNA products at low enzyme/substrate ratios would be larger, and shorten to 23 bp as the ratio increases. Alternatively, if binding of E38A to the dsRNA showed some cooperativity (i.e., each E38A dimer binding end-to-end with other E38A dimers on the dsRNA substrate), this would result in clusters of E38A dimers on the substrate dsRNA and preferentially yield 23-bp dsRNA products. In the various reactions performed with E38A, it seemed that the 23-bp dsRNA product was appearing at low protein/substrate ratios amid a background of larger digestion products, suggesting some level of cooperativity (Fig. 1C, for example). To test this idea, we used both E38A and E117D in a digestion reaction with a 900-bp dsRNA as substrate. The RNase III E117D mutant binds tightly to both R1.1 (Sun and Nicholson 2001) and to dsRNA (data not shown) but does not cleave. We conjecture for this experiment that E38A and E117D have the same cooperativity properties (i.e., they either bind to the dsRNA substrate cooperatively or they do not). We digested dsRNA with a mixture of both enzymes to see whether it affected the length of the dsRNA product. E117D would act to protect the dsRNA from cleavage by E38A and should result in a ladder pattern of 23-bp increments. Varying the ratio of E38A:E117D from 10:0 to 0:10 in the reactions should result in ladders of increasing size fragments and this is, in fact, what is observed (Fig. 4A). If E38A and E117D bind noncooperatively, the pattern should be the same whether the two proteins are added at the same time or in sequence (with E117D being added first). If the two RNase III mutants bind cooperatively, then the order of addition should affect the digestion pattern. E117D would cluster together resulting in segments of protected dsRNA before the addition of E38A. The more E117D added first, the larger the protected dsRNA segments. The pattern of digestion we observed from such an experiment is the same when the two proteins are added simultaneously (Fig. 4B), suggesting that E117D (and by extension, E38A and RNase III) does not bind dsRNA cooperatively.
FIGURE 4.
E38A and E117D combined digestion of dsRNA. E38A and E117D were used to digest a 900-bp dsRNA; total of 4 μg of the two proteins were incubated in the ratios indicated above the two gels (from 10:0 to 0:10). (Lane 1) dsRNA size markers. (Lanes 2–12) Digestions with E38A and E117D. (Lane 13) No enzyme. (A) Cartoon depicts mode of action of E38A alone (blue), E117D alone (maroon), and combined digestion when both proteins are added simultaneously to the reaction. Gel panel depicts results when both mutant enzymes are added to the substrate simultaneously at ratios indicated above the lanes. (B) Cartoon depicts mode of action for cooperative vs. noncooperative binding of RNase III to dsRNA when the two proteins are added sequentially, with E117D being added first. Top scenario shows the predicted results when binding is cooperative. Bottom scenario shows the predicted results when binding is noncooperative. Note that the predicted results for noncooperative binding are the same as if the two proteins are added simultaneously. The gel depicts the results when E117D is added first and E38A added 15 min later at ratios indicated above the lanes.
We next investigated the utility of the dsRNA product in RNA interference experiments. For the 23-bp dsRNA product to be useful in mammalian tissue culture RNAi experiments, it is desirable that it be representative of the dsRNA substrate. To test this we generated dsRNA from a 1112-bp fragment of the human DNA methylase, dnmt1. The E38A-generated 23-bp dsRNA product was radiolabeled with 32P at its 3′ termini with T4 RNA Ligase and 32P-pCp. These radiolabeled products were hybridized to two restriction enzyme digestions of dnmt1. The digests were electrophoresed and visualized by EtBr staining (Fig. 5). The gel was then transferred to nitrocellulose and hybridized with the labeled dsRNA (Fig. 5B). If the 23-bp dsRNA product is representative of the larger dsRNA, then the hybridization intensity should mimic the EtBr staining intensity. By simple inspection, the EtBr staining pattern is similar to, if not indistinguishable, from the hybridization pattern (Fig. 5A,B). No hybridization is seen to the molecular weight markers in lane 1 of Figure 5, A and B, indicating that the hybridization is specific, even after longer exposures. Both the EtBr-stained gel and hybridization autoradiogram were analyzed by densitometry (Fig. 5C,D). The intensity of both the EtBr staining (shown in purple) and the intensity of hybridization (shown in black) are shown for each DNA fragment observed. As the DNA fragments decrease in size for each digestion, so do the intensities of both EtBr and hybridization and in a very similar manner. This demonstrates that all regions of the 1112-bp dnmt1 fragment are represented in the pool of E38A-generated 23-bp dsRNA. We obtained the same results with a 737-bp fragment of GFP (data not shown).
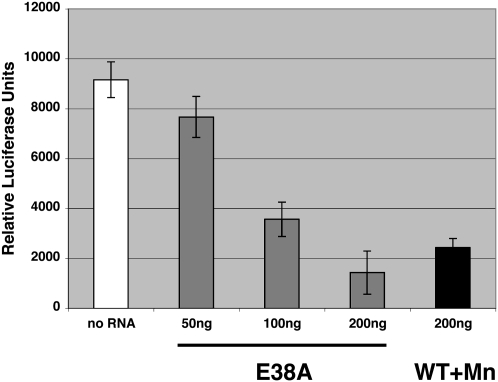
To test whether the 23-bp digestion product is suitable for RNAi, dsRNA was generated from a plasmid coding for the firefly luciferase. This dsRNA was then used as a substrate for E38A to generate a 23-bp product. NIH 3T3 cells were transfected with a plasmid expressing the firefly luciferase as well as a plasmid expressing the renilla luciferase. The cells were subsequently transfected with 23-bp dsRNA (siRNA) generated from the firefly luciferase by either E38A, or WT RNase III in the presence of Mn2+. Forty-eight hours later, the cells were assayed for both luciferase activities. As shown in Figure 6, increasing amounts of the E38A-generated firefly luciferase siRNA resulted in decreasing levels of firefly luciferase activity, with 200 ng showing a comparable decrease in activity as 200 ng of WT RNase III-generated siRNA. The activity was normalized for transfection efficiency using the renilla luciferase activity.
FIGURE 6.
RNA interference activity of E38A-generated luciferase siRNA. NIH 3T3 cells were transfected with a plasmid expressing firefly luciferase and subsequently transfected with either E38A-generated luciferase siRNA or WT RNase III/Mn++-generated luciferase siRNA. Cells were harvested 48 h after the addition of siRNA and assayed for luciferase activity. Activities were normalized for transfection efficiency as described in Materials and Methods. No siRNA added, white bar; E38A-generated siRNA, gray bars; WT RNase III-generated siRNA, black bar.
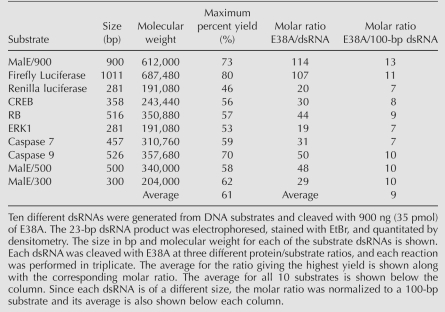
Because of the potential use of E38A in generating siRNAs for RNAi experiments, it is important to optimize the ratio of E38A to substrate dsRNA to obtain the maximum yield for different sequences of variable length. To practically assess both numbers, we calculated efficiencies from 10 different DNA fragments of different lengths (Table 1). dsRNA generated from each DNA fragment was used as a substrate in a titering reaction. The maximum percent yield for each individual dsRNA varied from 46% to 80% with an average of 61%. The molar ratio of E38A to dsRNA required to generate the maximum yield varied from 29 to 114. Normalizing this to 100 bp of dsRNA results in a ratio of E38A to 100-bp dsRNA of 7–13, with an average of nine.
TABLE 1.
Percent yield of E38A-generated 23-bp dsRNA
DISCUSSION
We describe an E. coli RNase III mutant, E38A that demonstrates a unique and useful ability to cleave long dsRNA molecules into 23-bp dsRNA product. We also demonstrate that the 23-bp dsRNA products generated by E38A are representative of a long dsRNA substrate used in the digestion and that these products are effective in RNA interference experiments. When long dsRNA is used as a substrate, E38A cleaves both strands of a dsRNA; when the natural hairpin substrate, R1.1, is used as a substrate, E38A cleaves the primary site. Both dsRNA and ssRNA products, respectively, are protected from further digestion. The products are capable of being further digested if E38A is denatured by either heat or phenol extraction post reaction. This suggests that E38A remains bound to its product, thereby protecting it. Gan et al. (2006) proposed that for the Aa-RNase III, E37 (E38 in Ec-RNase III) is involved in protein dimerization and not with phosphodiester bond cleavage. This is consistent with the activity we observe for E38A, which retains its ability to cleave dsRNA but may disassociate slowly from its product subsequent to the cleavage reaction. One explanation for the observance of a 23-bp dsRNA product when using long dsRNA as a substrate and an excess of enzyme to substrate could be the length of the dsRNA protected by the bound E38A homodimer (Gan et al. 2006). In fact, we observe that the maximum yield of 23-bp product is produced with an average molar ratio of ∼5:1 of E38A dimer to 100-bp dsRNA. This is close to what would be predicted if each E38A dimer covers 23 bp of dsRNA. The E38A substitution of Ec-RNase III has been reported previously (Sun et al. 2004; Zhang et al. 2004; Gan et al. 2006). In all of those studies, E38A had been reported to have essentially WT activity both in vivo and in vitro with the exception of a slightly lower binding affinity for both the R1.1 substrate (Zhang et al. 2004) and Mg2+ ion (Sun et al. 2004). However those studies did not examine long dsRNA as a substrate and therefore did not observe this activity of E38A. The use of a long dsRNA as a substrate resulting in a 23-bp dsRNA product revealed this property of the E38A substitution. It was our initial observation that the discreet 23-bp dsRNA product started accumulating in low enzyme to substrate ratios, suggesting that E38A (and by extension WT RNase III) was binding cooperatively to the dsRNA substrate. To test this we designed an experiment that utilized the activity of another mutant, E117D, which binds dsRNA (data not shown) or R1.1 (Sun and Nicholson 2001) tightly but does not cleave, thereby acting to protect the bound RNA. Both of the E117D and E38A substitutions are part of the internal portion of the enzyme and therefore would not be expected to contribute to any protein/protein interactions or any cooperativity. Combining E38A and E117D in different ratios in a reaction with long dsRNA as a substrate generates dsRNA products that resemble a ladder of 23-bp increments. We saw no difference in the ladder pattern regardless of whether the two proteins were added simultaneously or sequentially, and we concluded from this that there is no cooperative binding of the RNase III to its substrate (Fig. 4).
RNA interference has become a powerful technique for the study of gene function both in tissue culture and in whole organisms in an increasing number of metazoan species. In addition, it has great potential as an antiviral and anti-cancer therapeutic (Stevenson 2004; Ketzinel-Gilad et al. 2006; Putral et al. 2006; Kim and Rossi 2007). However the specific protocols for RNA interference required for different species and even for tissue culture are still quite varied and there is still much room for optimization. Tools that allow researchers to efficiently generate siRNAs are increasingly important. The bacterial RNase III enzyme has the ability to digest long, in vitro–generated dsRNA molecules to a size useful for RNAi. Unfortunately it is very difficult to control the reaction conditions to obtain enough dsRNA material of the functional size. Replacing Mg2+ with Mn2+ as a cofactor greatly increases the efficiency of generating the appropriate size RNA for RNAi experiments (Morlighem et al. 2007) and is the basis for a commercial product, ShortCut RNase III (New England Biolabs). RNase III(E38A) is an alternative for generating dsRNA of a size suitable for RNAi that is a representative mixture of the longer substrate dsRNA and should therefore be a useful tool for RNAi experiments.
MATERIALS AND METHODS
Construction of RNase III WT and mutant expression clones, and protein purification
WT RNase III
E. coli RNase III was amplified from an MBP/RNase III fusion construct (gift of C. Guan, New England Biolabs) with the following primers:
284-153, ACAGGATCCCATGAACCCCATCGTAATTAAT; and
284-154, ACAGGATCCTCTAGAGTCATTCCAGCTCCAGTTTTT.
The PCR product was cleaved with BamHI and inserted into the BamHI site of pET16b. Transformants were screened for RNase III inserts in the correct orientation. Positive clones were confirmed by DNA sequencing, resulting in a plasmid that produced a His-tag WT RNase III.
RNase III single amino acid substitutions
The DNA sequence of one His-tag RNase III clone revealed a PCR-introduced single nucleotide missense mutation that resulted in an E to D substitution at position 117 (E117D).
The starting construct for the remaining RNase III mutations was the WT His-tag RNase III clone. All mutations [except for Ec-RNase III(E117D)] were constructed by PCR-sewing. Using E38A as an example, during the first round of PCR, the WT RNase III clone was amplified with the following primer sets in two separate amplification reactions.
New England Biolabs primer catalog no. S1248 and 304-269: AATTCTAAACGCGCGTTATGTTTACT
284-154 and 304-268: CAGTAAACATAACGCGCGTTTAGAAT
The two PCR products were then used as substrates in a subsequent PCR with the following primers: New England Biolabs no. S1248 and 284-154. The resulting PCR product was cleaved with XbaI and cloned into the XbaI site of a modified pET16b vector created by cutting the WT RNase III clone with XbaI and retaining the vector fragment. E. coli transformants were screened for those containing the RNase III fragment in the correct orientation. Positive clones were confirmed by DNA sequencing.
Purification of His-tag WT and mutant RNase III proteins
Thirty milliliter cultures of each RNase III construct were grown in E. coli T7 Express (New England Biolabs) to mid-log phase, induced by the addition of IPTG to a final concentration of 100 μM and shaken overnight at 15°C. Induced cultures were lysed by sonication and His-tagged fusion proteins purified from the cleared lysates by affinity to nickel resin (Qiagen) according to the manufacturer's instructions. Purified His-tag RNase mutant proteins were then dialyzed vs. 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, 0.5 mM DTT, 50% glycerol and stored at −20°C.
Enzyme assays and electrophoresis
Unless otherwise noted, the dsRNA substrate used in all cleavage reactions was generated from the pMalC positive control plasmid of the HiScribe RNAi Transcription Kit (New England Biolabs) as per the manufacturer's instructions. Hairpin RNA substrate R1.1, GGGAGUAGAGGGACAAACUCAAGGUCAUUCGCAAGAGUGGCCUUUAUGAUUGACCUUCUU (Li and Nicholson 1996; Nicholson 2003), and fluorescein tag (Glen Research Corp.) were synthesized on an Applied Biosystems Model 394 using the protocols supplied by the manufacturer.
Unless otherwise noted, all RNase III cleavage reactions were done using 4 μg of purified His-tagged protein and either 500 ng of MalE dsRNA or 400 ng R1.1 as the substrate in New England Biolabs Buffer 2 (10 mM Tris-HCl, 10 mM MgCL2, 50 mM NaCl, 1 mM dithiothreitol at pH 7.9) and incubated for 30 min at 37°C. For protein titration reactions, 4 μg of purified His-tagged protein was used for the highest amount and serial diluted. Reactions were stopped by the addition of DNA loading dye (20 mM EDTA at pH 8.0, 1.25% SDS, 10% glycerol) and incubated for 10 min at 65°C before loading onto a 20% polyacrylamide/TBE gel (Invitrogen). For reactions using the 900-bp MalE dsRNA as a substrate, following electrophoresis, gels were stained with EtBr and imaged. Reactions using the hairpin RNA substrate were electrophoresed on a 15% TBE-Urea gel (Invitrogen) and imaged using an FX Pro Plus Molecular Imager (Bio-Rad).
RNA interference experiments
An E38A-generated 23 bp dsRNA product was produced from the firefly luciferase (1000 bp) as described above using pGL3/Luc as a substrate. The luciferase dsRNA product was ethanol precipitated, redisolved in water, and further purified using siRNA Purification Unit (Ambion) according to the manufacturer's instructions. A luciferase 23-bp dsRNA product was also produced from pGL3/Luc using ShortCut RNAi Kit (New England Biolabs) according to the manufacturer's instructions. NIH 3T3 cells were transiently transfected with 300 ng/well of pGL3/Luc and 300 ng/well of pBind vector (which contains the renilla luciferase under control of an SV40 Early promoter; Promega.) in 24-well plates with TransPass D2 (New England Biolabs) according to the manufacturer's instructions. After a 3-h incubation at 37°C, E38A-generated luciferase or ShortCut-generated luciferase siRNA was transfected with TransPass R2 (New England Biolabs) according to the manufacturer's instructions at 50, 100, or 200 ng/well. Two days post transfection, the cells were harvested and both firefly and renilla luciferase activity assayed with Dual-Glo Luciferase Assay System (Promega) according to the manufacturer's instructions. All transfections were done in quadruplicate, and the firefly luciferase activity was normalized for transfection efficiency using the renilla luciferase activity.
Southern blotting and RNA/DNA hybridization
As described above, 23-bp dsRNA was produced from a 1112-bp clone of the human DNA methylase, dnmt1 (Jeltsch 2006). The 23-bp dsRNA product was electrophoresed, gel purified, and radiolabeled by the addition of 32P pCp (PerkinElmer) to the 3′ end with T4 RNA Ligase I (New England Biolabs) according to the manufacturer's instructions. The dnmt1 insert DNA was amplified and digested with AciI and BpmI, electrophoresed on a 1.25% agarose gel, and transferred to nitrocellulose (Sambrook and Russel 2007). The blot was then probed at high stringency with the radiolabeled dsRNA (Sambrook and Russel 2007). The washed blot was autoradiographed and hybridizated, along with the EtBr-stained gel, and quantitated by densitometry using Quantity 1 Software (Bio-Rad).
Quantitation of E38A-generated dsRNA
Percentage yields were quantitated as follows: 900 ng (35 pmol) of E38A and three twofold dilutions of each dsRNA substrate prepared using HiScribe RNAi Transcription Kit (New England Biolabs) as per manufacturer's instructions, in ranges between 130 ng and 480 ng, were used in each reaction. The ten dsRNA substrates were prepared as described above. Reaction products were electrophoresed and imaged as described above. The amount of 23-bp dsRNA product from each reaction was then determined by densitometry using Quantity 1 Software (Bio-Rad) and a standard curve using known amounts of a synthetic 22-nt dsRNA. All reactions and quantitations were done in triplicate.
ACKNOWLEDGMENTS
We thank Bill Jack, Larry McReynolds, Chudi Guan, and Tom Evans for helpful discussions and critique of the manuscript; Cathy Shea and Halli Benson for technical assistance; and we greatly appreciate the support of Don Comb.
Footnotes
Abbreviations: kB, kilobase pair; kD, kilodalton; PCR, polymerase chain reaction; RT-PCR, reverse transcriptase polymerase chain reaction; bp, base pair(s); EtBr, ethidium bromide; RNAi, RNA interference.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1196509.
REFERENCES
- Blaszczyk J., Tropeae J.E., Bubunenko M., Routzahn K.M., Waugh D.S., Court D.L., Ji X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001;9:1225–1236. doi: 10.1016/s0969-2126(01)00685-2. [DOI] [PubMed] [Google Scholar]
- Court D.L. RNA processing and degradation by RNase III. In: Belasco J.G., Brawerman G., editors. Control of messenger RNA stability. Academic; New York: 1993. pp. 71–116. [Google Scholar]
- Gan J., Tropea J.E., Austin B.P., Court D.L., Waugh D.S., Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Jeltsch A. On the enzymatic properties of Dnmt1: Specificity, processivity, mechanism of linear diffusion, and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–66. doi: 10.4161/epi.1.2.2767. [DOI] [PubMed] [Google Scholar]
- Ketzinel-Gilad M., Shaul Y., Galun E. RNA interference for antiviral therapy. J. Gene Med. 2006;8:933–950. doi: 10.1002/jgm.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Rossi J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- Lamontagne B., Larose S., Boulanger J., Elela S.A. The RNase III family: A conserved structure and expanding functions in eukaryotic dsRNA metabolism. Curr. Issues Mol. Biol. 2001;3:71–78. [PubMed] [Google Scholar]
- Lee Y., Han J., Yeom K.H., Jin H., Kim V.N. Drosha in primary microRNA processing. Cold Spring Harb. Symp. Quant. Biol. 2006;71:51–57. doi: 10.1101/sqb.2006.71.041. [DOI] [PubMed] [Google Scholar]
- Li H., Nicholson A.W. Defining the enzyme binding domain of a ribonuclease III processing signal. Ethylation interference and hydroxyl radical footprinting using catalytically inactive RNase III mutants. EMBO J. 1996;15:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- MacRae I.J., Doudna J.A. Ribonuclease revisited: Structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Morlighem J.E., Petit C., Tzertzinis G. Determination of silencing potency of synthetic and RNase III-generated siRNA using a secreted luciferase assay. Biotechniques. 2007;42:599–600. doi: 10.2144/000112444. 602, 604–606. [DOI] [PubMed] [Google Scholar]
- Nicholson A.W. Function, mechanism, and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Nicholson A.W. The ribonuclease III subfamily: Forms and functions in RNA maturation, decay, and gene silencing. In: Hannon G.J., editor. RNAi: A guide to gene silencing. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. pp. 149–174. [Google Scholar]
- Pertzev A.V., Nicholson A.W. Characterization of RNA sequence determinants and antideterminants of processing reactivity for a minimal substrate of Escherichia coli ribonuclease III. Nucleic Acids Res. 2006;34:3708–3721. doi: 10.1093/nar/gkl459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putral L.N., Gu W., McMillan N.A. RNA interference for the treatment of cancer. Drug News Perspect. 2006;19:317–324. doi: 10.1358/dnp.2006.19.6.985937. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russel D.W. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. [Google Scholar]
- Stevenson M. Therapeutic potential of RNA interference. N. Engl. J. Med. 2004;351:1772–1777. doi: 10.1056/NEJMra045004. [DOI] [PubMed] [Google Scholar]
- Sun W., Nicholson A.W. Mechanism of action of Escherichia coli ribonuclease III. Stringent chemical requirement for the glutamic acid 117 side chain and Mn2+ rescue of the Glu117Asp mutant. Biochemistry. 2001;40:5102–5110. doi: 10.1021/bi010022d. [DOI] [PubMed] [Google Scholar]
- Sun W., Li G., Nicholson A.W. Mutational analysis of the nuclease domain of Escherichia coli ribonuclease III. Identification of conserved acidic residues that are important for catalytic function in vitro. Biochemistry. 2004;43:13054–13062. doi: 10.1021/bi049258i. [DOI] [PubMed] [Google Scholar]
- Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]