Abstract
Elongation factor G (EF-G) promotes the translocation step in bacterial protein synthesis and, together with ribosome recycling factor (RRF), the disassembly of the post-termination ribosome. Unlike translocation, ribosome disassembly strictly requires GTP hydrolysis by EF-G. Here we report that ribosome disassembly is strongly inhibited by vanadate, an analog of inorganic phosphate (Pi), indicating that Pi release is required for ribosome disassembly. In contrast, the function of EF-G in single-round translocation is not affected by vanadate, while the turnover reaction is strongly inhibited. We also show that the antibiotic fusidic acid blocks ribosome disassembly by EF-G/RRF at a 1000-fold lower concentration than required for the inhibition of EF-G turnover in vitro and close to the effective inhibitory concentration in vivo, suggesting that the antimicrobial activity of fusidic acid is primarily due to the direct inhibition of ribosome recycling. Our results indicate that conformational coupling between EF-G and the ribosome is principally different in translocation and ribosome disassembly. Pi release is not required for the mechanochemical function of EF-G in translocation, whereas the interactions between RRF and EF-G introduce tight coupling between the conformational change of EF-G induced by Pi release and ribosome disassembly.
Keywords: translation, rapid kinetics, phosphate release, translation factor, GTPase
INTRODUCTION
Among the translation factors assisting the ribosome in synthesizing proteins, elongation factor G (EF-G) is the only one that has two distinct functions in different phases of protein synthesis. During the elongation phase, EF-G promotes translocation—the movement of the tRNA-peptidyl-tRNA-mRNA complex from the pre- to the post-translocation position—after a peptide bond has been formed. Following termination, which entails the hydrolytic release of the completed nascent peptide from P site-bound peptidyl-tRNA and results in a ribosome with deacylated tRNA in the P site and an empty A site, EF-G, together with another factor, the ribosome recycling factor (RRF) (Hirashima and Kaji 1973), brings about the rapid dissociation of the post-termination ribosome into ribosomal subunits (Karimi et al. 1999; Ito et al. 2002; Fujiwara et al. 2004; Peske et al. 2005; Zavialov et al. 2005a). Subsequently, initiation factor 3 (IF3) binds to the 30S subunit, preventing reassociation of the subunits and preparing the 30S subunit for initiation by promoting the release of tRNA and mRNA (Karimi et al. 1999; Peske et al. 2005). RRF is an essential protein, indicating that rapid disassembly of the post-termination complex is essential for cell growth (Janosi et al. 1998).
During both translocation and ribosome disassembly, EF-G hydrolyzes GTP. When GTP hydrolysis is prevented by replacing GTP with nonhydrolyzable analogs, translocation still takes place, albeit slowly (Rodnina et al. 1997), whereas ribosome disassembly by EF-G/RRF is completely blocked (Karimi et al. 1999; Hirokawa et al. 2002; Peske et al. 2005; Zavialov et al. 2005a). In the absence of IF3, reassociation of subunits resumes when the GTP concentration decreases as a result of turnover GTP hydrolysis by EF-G (Peske et al. 2005); this “transient” behavior of the in vitro system was reanalyzed and confirmed recently (Hirokawa et al. 2008). The dependence of ribosome disassembly on GTP hydrolysis may reflect a requirement for GTP hydrolysis itself, as is the case with ribosome unlocking in translocation (Savelsbergh et al. 2003). Alternatively, it may reflect the necessity for phosphate (Pi) release in addition to GTP hydrolysis to drive the reaction. To clarify this question, Pi release and the effect of inhibiting Pi release were studied kinetically.
Fusidic acid (FA) is a well-known inhibitor of EF-G function. FA inhibits EF-G turnover, in either GTP hydrolysis or translocation (Bodley et al. 1969; Lee-Huang et al. 1974; Johanson et al. 1996). However, under single-round conditions, i.e., when EF-G is present in excess over ribosomes, FA does not block translocation or GTP hydrolysis (Bodley et al. 1969; Inoue-Yokosawa et al. 1974; Seo et al. 2004). It has been reported that FA inhibits the EF-G/RRF-dependent release of tRNA from post-termination complexes and the release of mRNA from post-termination polysomes, i.e., the formation of monosomes from polysomes as analyzed by sucrose gradient centrifugation (Hirashima and Kaji 1973; Hirokawa et al. 2002, 2005). While these results indicated that FA impaired ribosome disassembly, the effect was not studied further. In order to characterize the mechanism of inhibition, we have examined the effect of FA on ribosome disassembly in more detail.
RESULTS
Influence of RRF on GTP hydrolysis by EF-G and release of inorganic phosphate
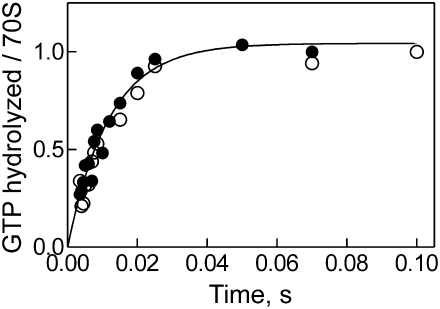
Upon binding to ribosomes (vacant, pre-translocation, post-translocation), EF-G undergoes a conformational change that leads to rapid GTP hydrolysis followed by slower release of inorganic phosphate (Pi) (Savelsbergh et al. 2003). The timing of GTP hydrolysis and Pi release during EF-G/RRF-dependent ribosome recycling is not known. We have used the quench-flow technique to measure the rate of GTP hydrolysis at near-saturating concentration of EF-G (3 μM) on post-termination complexes in the presence and absence of RRF (Fig. 1). At 37°C, the rate of GTP hydrolysis by EF-G on vacant or pre-translocation ribosomes is >250 sec−1 (Rodnina et al. 1997; Savelsbergh et al. 2003), which is close to the resolution limit of the quench-flow technique. Therefore, we carried out the experiments at 4°C to slow down the reaction such that accurate measurements could be performed. Under these conditions, the rate of GTP hydrolysis by EF-G on post-termination complex was 75 ± 25 sec−1 in the absence or presence of RRF (Fig. 1), indicating that the GTPase activity of EF-G is not influenced by the presence of RRF on the ribosome.
FIGURE 1.
Single-round GTP hydrolysis by EF-G in the absence and presence of RRF. Post-termination complex (Materials and Methods; 1 μM final concentration after mixing) was rapidly mixed with IF3 (2 μM), [γ-32P]GTP (30 μM), and EF-G (3 μM) alone (●) or together with RRF (5 μM) (○). The extent of GTP hydrolysis is presented relative to the input of post-termination complex. Single-exponential fitting yielded a rate of GTP hydrolysis of 75 ± 25 sec−1 (4°C).
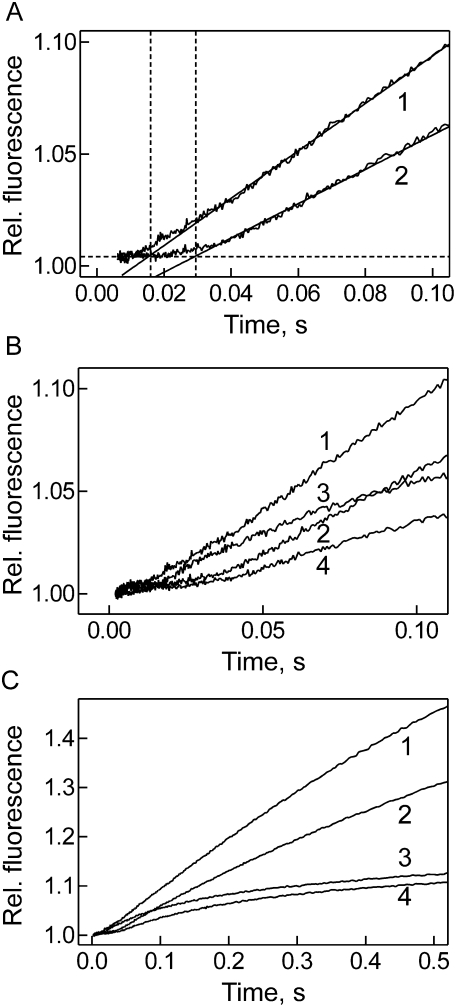
During translocation, the release of Pi from EF-G is rate-limited by ribosome unlocking, which in the presence of EF-G takes place with a rate constant of 35 sec−1 and is required for the rapid translocation movement of the tRNA-mRNA complex to take place (Savelsbergh et al. 2003, 2005). Unlocking represents a conformational change of the ribosome that follows the ratcheting movement of the subunits relative to one another as revealed by cryo-EM (Frank and Agrawal 2000; Agirrezabala et al. 2008; Julian et al. 2008). In order to examine whether RRF had an effect on Pi release from EF-G-bound to the post-termination complex, we monitored the kinetics of Pi release using fluorescence-labeled phosphate-binding protein (PBP) which shows a fluorescence increase upon Pi uptake (Fig. 2A; Brune et al. 1994; Savelsbergh et al. 2005). With EF-G alone (Fig. 2A, trace 1), Pi release on the post-termination complex showed the same characteristics as on vacant ribosomes or the pre-translocation complex (Savelsbergh et al. 2003, 2005), i.e., after a delay of ∼15 msec, Pi was released in a rapid burst which was followed by a quasi-linear turnover phase. When RRF was present, the initial lag phase of Pi release was longer, ∼30 msec (Fig. 2A, trace 2), indicating that Pi release is preceded by a step with a rate of ∼30 sec−1. As GTP hydrolysis is much faster than that (Fig. 1), the latter step is attributed to a conformational change that follows GTP hydrolysis and precedes, or enables, Pi release and may be related to unlocking. The turnover rate of Pi release, as deduced from the quasi-linear fluorescence increase following the delay (Fig. 2A, traces 1 and 2), was comparable in the absence and presence of RRF, 1.6 ± 0.1 sec−1 and 1.4 ± 0.1 sec−1, respectively. The smaller amplitude of turnover Pi release observed in the presence of RRF (Fig. 2A, trace 2) is attributed to the fact that the 50S subunits that formed progressively under these conditions did not stimulate the GTPase activity of EF-G (Voigt et al. 1974; data not shown).
FIGURE 2.
Effect of RRF on the release of inorganic phosphate from ribosome-bound EF-G. (A) Effect of RRF on the delay of Pi release. Post-termination complex (Materials and Methods; final concentration after mixing, 0.2 μM) with GTP (200 μM) was rapidly mixed in a stopped-flow apparatus with IF3 (2 μM), GTP (200 μM), and EF-G (2 μM) (1) or EF-G (2 μM) and RRF (5 μM) (2). The release of Pi from ribosome-bound EF-G was monitored by the fluorescence increase of MDCC-labeled PBP (2.5 μM) caused by the binding of Pi. The delays are indicated by vertical dotted lines. (B) Pi release in the presence of FA, short time window. Pi release was measured as in A (traces 1 and 2) or in the presence of FA (200 μM) without (trace 3) or with RRF (trace 4). (C) Pi release in the presence of FA, long time window.
In order to determine the rate of single-round Pi release from EF-G in the presence of RRF, the turnover reaction, which under the present experimental conditions masked the initial Pi release, had to be suppressed. This was accomplished by adding fusidic acid (FA), a well-known inhibitor of EF-G turnover. FA binds to EF-G on the ribosome and stabilizes the ribosome-EF-G complex, such that the dissociation of EF-G from the ribosome is inhibited. When FA was added, neither the initial delay nor the burst phase observed in the absence (Fig. 2B,C, trace 3) or presence (Fig. 2B,C, trace 4) of RRF were affected, while the turnover was strongly suppressed in both cases. The rate of the first round of Pi release, ∼6 sec−1, observed with the post-termination complex in the absence of RRF was somewhat lower than that observed previously with vacant ribosomes or pre-translocation complex at comparable conditions, which was 20 sec−1 (Savelsbergh et al. 2003, 2005). This difference may reflect different states of the ribosome in those complexes. In the presence of RRF, the rate of single-round Pi release was slowed down about twofold to ∼3 sec−1 (Fig. 2B).
Ribosome disassembly depends on Pi release following GTP hydrolysis
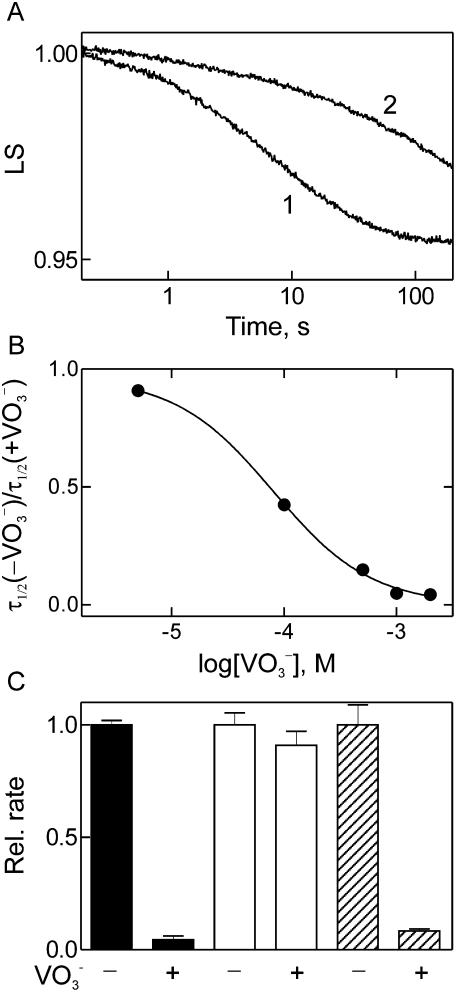
EF-G promotes translocation, albeit slowly, when GTP is replaced with a nonhydrolyzable analog, such as GDPNP (Inoue-Yokosawa et al. 1974; Rodnina et al. 1997; Katunin et al. 2002). Furthermore, translocation is independent of the release of Pi, which, however, limits the dissociation of EF-G from the ribosome (Savelsbergh et al. 2003, 2005). In contrast, as outlined above, the function of EF-G in ribosome disassembly strictly depends on GTP hydrolysis. Whether Pi release is also required to bring about the reaction is not known. Thus, we set out to examine the effect of inhibiting phosphate release on ribosome disassembly. In principle, Pi release may be suppressed by the addition of excess exogenous Pi, as has been shown for instance by inhibiting the function of kinesin (Schief et al. 2004). However, such experiments were not feasible in the buffer required for maintaining ribosome activity, due to the formation of insoluble complex salts containing phosphate. Therefore, we have used metavanadate (VO3 −), which can act as an analog of Pi by occupying the Pi binding site in the active site of enzymes. For instance, vanadate inhibits force generation in muscle actomyosin by binding to myosin·ADP and inhibiting the transition from the ADP-Pi state to the ADP state of myosin (Wilson et al. 1995). Metavanadate is readily soluble in translation buffer and forms a mixture of meta- and orthovanadate in aqueous solution around neutrality. Ribosome disassembly by EF-G/RRF, as monitored by light-scattering stopped-flow (Wishnia et al. 1975), was effectively inhibited by vanadate (Fig. 3A), indicating that Pi release following GTP hydrolysis is indeed required for EF-G function in disassembly. The full inhibitory effect (∼30-fold inhibition) was attained at ∼1 mM vanadate, and the IC50 (concentration for 50% inhibition) was ∼0.1 mM (Fig. 3B). In contrast, EF-G-catalyzed tRNA–mRNA translocation was not inhibited by vanadate (Fig. 3C), in keeping with the kinetic analysis which indicated that tRNA movement and Pi release are parallel, independent steps (Savelsbergh et al. 2003). In contrast, vanadate strongly inhibited the turnover of EF-G in multiple-round translocation (Fig. 3C), in keeping with the notion that Pi release is required for EF-G to dissociate from the ribosome (Savelsbergh et al. 2005). The inhibitory effects of vanadate are not due to an inhibition of GTP binding, as rapid kinetic experiments have not shown any significant effect of vanadate on GTP binding to EF-G (data not shown). Thus, although translocation and ribosome disassembly are both driven by EF-G, the requirements with respect to GTP hydrolysis and Pi release are different, ribosome disassembly requiring both.
FIGURE 3.
Effect of vanadate on EF-G function on the ribosome. (A) Inhibition by vanadate of post-termination complex disassembly. Post-termination complex was rapidly mixed with EF-G/RRF in the absence (1) or presence (2) of vanadate (2 mM), and complex disassembly was monitored by light scattering (LS) (Materials and Methods). (B) Concentration dependence of the inhibition by vanadate. Post-termination complex disassembly was measured as in A in the presence of increasing concentrations of vanadate. Plotted is the half-life (τ1/2) of the reaction measured in the absence relative to that measured in the presence of vanadate; τ1/2 was ∼5 sec in the absence and ∼150 sec in the presence of vanadate. (C) Comparison of post-termination complex disassembly and translocation. Rates of post-termination complex disassembly (black bars) were measured as in A. The rate of single-round translocation (∼15 sec−1, open bars) was measured by fluorescence stopped-flow, multiple-round translocation (hatched bars) by the puromycin assay (Materials and Methods); plotted is the relative initial rate of translocation measured in the absence (2 sec−1, set to 1.0) and presence (0.17 sec−1) of vanadate. The concentration of vanadate was 2 mM.
Fusidic acid effectively inhibits ribosome recycling
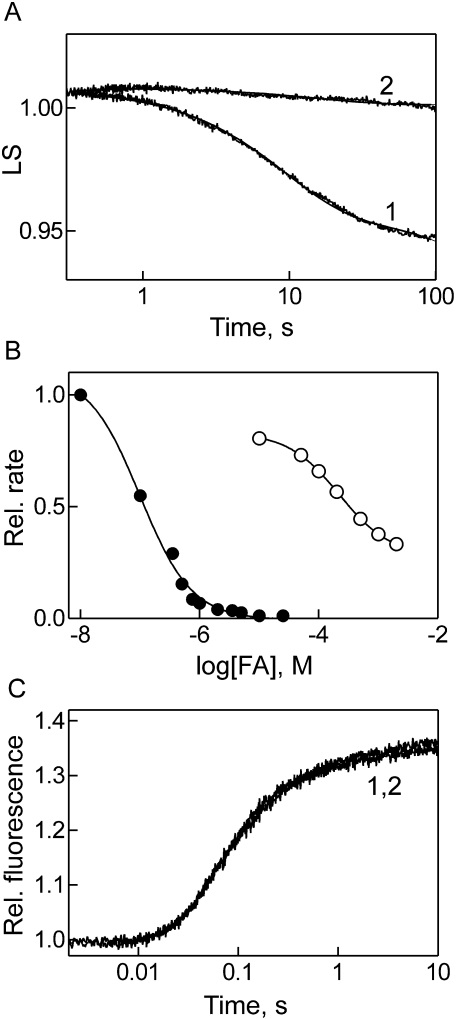
FA causes an inhibition of EF-G turnover, whereas single-round GTP hydrolysis and translocation are not blocked by the antibiotic (Bodley et al. 1969; Inoue-Yokosawa et al. 1974). We have examined the effect of FA on EF-G/RRF-dependent ribosome disassembly using the light-scattering stopped-flow assay introduced above, which—unlike sucrose gradient centrifugation (Hirashima and Kaji 1973; Hirokawa et al. 2002, 2005)—directly monitors the disassembly of the ribosome into subunits in real time. We observed that ribosome disassembly by EF-G·GTP/RRF was practically completely inhibited by FA even at single-round conditions (Fig. 4A). The inhibition was complete at unexpectedly low concentrations of FA (Fig. 4B). FA strongly inhibited subunit dissociation with an IC50 value ∼0.1 μM and caused a nearly 100-fold reduction of the reaction rate (to 0.003 sec−1) at a concentration of 1 μM FA, a concentration at which EF-G turnover, measured here by translocation, was not affected at all (Fig. 4B). The much higher IC50 of 15 μM FA reported previously (Hirokawa et al. 2005) probably has to be attributed to the assay (sucrose gradient centrifugation) used for monitoring the disassembly of a model post-termination complex derived from polysomes. The IC50 for the inhibition of EF-G turnover, assayed by translocation, was ∼200 μM, i.e., >1000-fold higher (Fig. 4B). The latter IC50 value is similar to previously published values determined by poly(Phe) synthesis and GTPase turnover assays, which were ∼100 μM (Okura et al. 1970) or 10 μM (Bodley et al. 1969), respectively. A high IC50 of FA in EF-G turnover is also indicated by recent data (Seo et al. 2004) which showed a further decrease of the GTPase turnover rate when the concentration of FA was increased from 50 μM to 500 μM. High IC50 values are consistent with FA acting as a “slow” inhibitor (Seo et al. 2006) which allows several rounds of EF-G turnover before the inhibition becomes effective (see Discussion). The rate of single-round translocation was not affected by FA (Fig. 4C). These results show that, unlike translocation, EF-G/RRF-dependent ribosome disassembly is directly inhibited by FA and—compared with the inhibition of EF-G turnover—rather effectively, i.e., at submicromolar concentration of the antibiotic.
FIGURE 4.
Effect of fusidic acid (FA) on EF-G function on the ribosome. (A) Inhibition of post-termination complex disassembly. Post-termination complex was rapidly mixed with EF-G/RRF as in Figure 3A in the absence (1) and presence (2) of FA (200 μM), and ribosome disassembly was monitored by light scattering (LS). (B) Dependence of the inhibition on FA concentration. Post-termination complex disassembly by EF-G/RRF was measured as in A at various concentrations of FA (●); 50% inhibition was observed at 0.1 μM FA. For comparison, the inhibition of EF-G turnover in translocation is shown (○); 50% inhibition was observed at ∼200 μM FA. (C) Time course of translocation in the absence and presence of FA. Translocation was monitored by the fluorescence of fluorescence-labeled peptidyl-tRNA (Materials and Methods).
DISCUSSION
Distinct functions of EF-G in translocation and ribosome disassembly
The functions of EF-G in translocation and ribosome disassembly are similar in several aspects, including effects of EF-G mutations or the inhibition by a number of antibiotics, but—as reported here—there are important differences in mechanistic detail. The main differences are the following: (1) Unlike translocation, which takes place reasonably fast with nonhydrolyzable GTP analogs, ribosome disassembly does not take place with GTP analogs (Karimi et al. 1999; Hirokawa et al. 2002; Peske et al. 2005; Zavialov et al. 2005a), indicating that it requires GTP hydrolysis. (2) In contrast to single-round translocation, ribosome disassembly is inhibited by vanadate, indicating that it requires Pi release. (3) In ribosome disassembly, Pi is released later and more slowly, compared with translocation. (4) Ribosome disassembly is blocked by FA, whereas translocation is not. Apparently, the differences in the behavior of EF-G in translocation and ribosome disassembly are to be attributed to the presence of RRF on the ribosome and its interactions with EF-G.
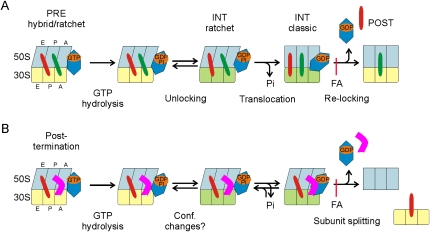
On the ribosome, GTP hydrolysis and Pi release induce different conformations of EF-G. GDP-GTP exchange is extremely rapid on unbound EF-G (Wilden et al. 2006), contrary to a recent report (Zavialov et al. 2005b). As there is much more GTP than GDP in the cell, the factor enters the ribosome predominantly in the GTP-bound form. The sequence of steps in translocation and ribosome disassembly is depicted in Figure 5. Upon binding to the ribosome, the nucleotide-binding pocket of EF-G closes (Wilden et al. 2006), presumably in conjunction with GTPase activation, and the ratcheted conformation of the ribosome with tRNAs in their hybrid states is induced or stabilized (Frank and Agrawal 2000; Spiegel et al. 2007; Agirrezabala et al. 2008; Julian et al. 2008). Upon GTP hydrolysis, EF-G changes conformation, thereby causing another conformational change of the ribosome (“unlocking”) (Savelsbergh et al. 2003), which in turn leads to tRNA-mRNA movement and, in parallel, reopening of the nucleotide-binding pocket and the rapid release of Pi (Fig. 5). The delay, i.e., the duration of the steps preceding Pi release, is ∼15 msec (Savelsbergh et al. 2003). In the presence of RRF, single-round Pi release is delayed somewhat longer, ∼30 msec, and slowed down to ∼3 sec−1. These observations indicate that the presence of RRF on the ribosome impairs opening of the nucleotide-binding pocket of EF-G and release of Pi. On the other hand, at 3 sec−1 single-round Pi release is still faster than subunit dissociation, which takes place at a rate of ∼0.2–0.3 sec−1. Thus, GTP hydrolysis and Pi release precede subunit dissociation. Pi release presumably induces a further conformational change of EF-G that is essential for ribosome disassembly to take place, in analogy to the initiation of the power stroke of motor ATPases by Pi release (Keller and Bustamante 2000). The conformational change appears to involve changes of the switch I and II regions in the G domain (Gao et al. 2007) that change the conformation upon loss of interactions with the γ-phosphate and relay the change to the neighboring domains. The low rate of ribosome disassembly, compared with unlocking in translocation (30 sec−1; Savelsbergh et al. 2003), presumably reflects the fact that the large-scale conformational changes of the ribosome that are required for disassembly take more time to accomplish than unlocking.
FIGURE 5.
Sequence of steps in translocation (A) and ribosome disassembly (B). The ribosomal subunits are depicted in light blue (50S) and yellow (30S) in the ratcheted (50S tilted) or nonratcheted (50S straight) conformation with the tRNAs (red, P site; green, A site) in hybrid (tilted) or classic (upright) positions. EF-G is colored in blue, RRF in magenta. Different conformations of EF-G are not indicated. In B, it is assumed that ribosome disassembly proceeds through the ratcheted conformation of the ribosome. The inhibition by fusidic acid (FA) of ribosome relocking following tRNA-mRNA movement (A) or of subunit splitting (B) is indicated. The mRNA bound to the 30S subunit is omitted for clarity.
The turnover rate of Pi release, which was ∼1.5 sec−1 at saturation with both EF-G and RRF, was higher than the rate of ribosome disassembly, 0.2–0.3 sec−1. This difference may indicate that, on the average, more than one molecule of GTP is hydrolyzed until ribosome disassembly takes place, i.e., that the two processes are not tightly coupled. The same is probably true for translocation, as a twofold difference between the turnover numbers of GTP hydrolysis and translocation was observed (Savelsbergh et al. 2005), and some extent of uncoupled GTP hydrolysis was consistently observed during translation in purified systems (e.g., Chinali and Parmeggiani 1980). Loose coupling of the GTPase cycle of EF-G and ribosome disassembly may lower the risk of premature ribosome disassembly in cases where RRF has entered a post-translocation ribosome, despite the inhibition of RRF binding by peptidyl-tRNA in the P site (P/P state) that is suggested by chemical probing (Lancaster et al. 2002), structural data (Wilson et al. 2005; Weixlbaumer et al. 2007), and biochemical recycling assays (Peske et al. 2005).
Ribosome disassembly as an in vivo target of fusidic acid
A striking, unexpected result of this study is the inhibition of EF-G/RRF-dependent ribosome disassembly by FA at very low concentrations. The inhibitory concentration is close to MIC50 values observed in vivo, suggesting that the inhibition of EF-G/RRF-dependent disassembly of the post-termination complex may be the mechanism by which FA inhibits cell growth, rather than the inhibition of EF-G turnover in translation, as is generally assumed. The IC50 value observed for EF-G/RRF-dependent ribosome disassembly, ∼0.1 μM, is very close to the affinity of FA binding to EF-G on the ribosome, ∼0.2–0.4 μM, as determined by direct binding assays (Okura et al. 1970; Willie et al. 1975). Binding of FA to EF-G on the ribosome seems to block a conformational change of EF-G that follows GTP hydrolysis and Pi release—both unaffected by FA—and is required for EF-G to dissociate from the ribosome. Assuming that the same or a similar conformational change of EF-G, enhanced by RRF, drives subunit dissociation, the inhibition by FA of ribosome disassembly may be explained by an inhibition of that transition.
An alternative possibility to explain the extraordinary sensitivity against FA of the recycling reaction could be that EF-G undergoes turnover before ribosome disassembly takes place and that it is the turnover reaction, i.e., the dissociation of EF-G from the ribosome, that is inhibited, rather than the function in ribosome disassembly. We consider such a scenario unlikely, because (1) a twofold turnover can hardly produce the observed 100- to 1000-fold difference in the effective inhibitory FA concentrations, and (2) the turnover—at least in the absence of RRF—is not affected at all at a concentration of FA (1 μM) that completely blocks ribosome disassembly.
One possibility to explain the grossly different effective inhibitory FA concentrations observed in ribosome disassembly and translocation is kinetic partitioning. Assuming that strong binding of FA to ribosome-bound EF-G takes place only after Pi release, the extent of FA binding and, with that, of inhibition may be determined by a kinetic competition between the binding of FA and a rearrangement of the ribosome-EF-G complex to a conformation that is refractory to FA binding (Fig. 5). During translocation, this rearrangement, which is followed by the dissociation of EF-G from the ribosome, is relatively rapid, ∼5 sec−1, whereas in ribosome disassembly the rearrangement is much slower, 0.2–0.3 sec−1. Thus, in the latter case the time window for FA binding is longer and, as a consequence, a lower concentration of FA may be required to achieve full binding, i.e., full inhibition. Alternatively, or in addition, FA may inhibit a conformational transition of EF-G that is essential for ribosome disassembly, but not for unlocking in translocation, and may be induced by the presence of RRF.
The interaction of RRF with the ribosome has been studied by directed hydroxyl radical footprinting on ribosomal RNA (Lancaster et al. 2002), by cryo-EM (Agrawal et al. 2004; Gao et al. 2005; Barat et al. 2007; Gao et al. 2007), and by X-ray crystallography (Wilson et al. 2005; Borovinskaya et al. 2007; Weixlbaumer et al. 2007; Pai et al. 2008). RRF is bound at the interface between the subunits where it interacts mainly with the 50S subunit. RRF contacts helix 69 (H69) of 23S rRNA on the 50S subunit, which connects to helix 44 (h44) of 16S rRNA on the 30S subunit to form bridge 2a. Bridge 2a is most important for holding the subunits together, as the deletion of H69 destabilizes subunit interaction considerably (Ali et al. 2006). A displacement of H69 by RRF bound to 70S ribosomes from Escherichia coli has been observed (Gao et al. 2005; Wilson et al. 2005; Borovinskaya et al. 2007; Gao et al. 2007), whereas on ribosomes from Thermus thermophilus the binding of RRF did not have this effect (Weixlbaumer et al. 2007). These observations suggest that EF-G, driven by GTP hydrolysis and, as shown here, by Pi release induces a movement of RRF that, in turn, displaces H69 and possibly other bridges, thereby destabilizing the interactions between the subunits such that disassembly is promoted. A movement of domain II of RRF, which is flexibly attached to domain I, seems to play an important role (Gao et al. 2005, 2007).
The structure of the post-termination complex containing both RRF and EF-G is not known. There is evidence, however, indicating that the two factors interact on the ribosome. Interactions have been demonstrated by mutational analysis of EF-G and RRF (Ito et al. 2002), by monitoring factor binding to the ribosome (Seo et al. 2004), or by cryo-EM reconstructions of complexes of 50S ribosomal subunits with the two factors (Gao et al. 2005, 2007). According to these data, the interaction between the two factors involves residues in domain III of EF-G and the hinge region in RRF as well as in domain IV of EF-G and domain II of RRF. The conformation of EF-G bound to FA on the ribosome has been characterized by cryo-EM of ribosome–EF-G complexes stalled by FA (Agrawal et al. 1998; Stark et al. 2000). Compared to the conformation of unbound EF-G, determined crystallographically (Ævarsson et al. 1994; Czworkowski et al. 1994; Al-Karadaghi et al. 1996; Hansson et al. 2005), in the ribosome-bound conformation domain IV of EF-G is tilted relative to the body of the molecule formed of domains I and II. Based on cryo-EM, it has been suggested that the rearrangement of EF-G that is induced by GTP hydrolysis consists of a movement of domain IV. The movement may be transmitted to RRF and drive the ribosome rearrangement that leads to dissociation of the subunits. With FA bound to EF-G, the movement would be precluded, explaining the strong inhibition of ribosome disassembly by FA. The latter scenario is consistent with a function in ribosome disassembly of EF-G as a molecular motor that brings about subunit dissociation at the expense of GTP hydrolysis and Pi release, rather than its function in translocation, where EF-G—after unlocking the ribosome at the expense of GTP hydrolysis—appears to act as a Brownian ratchet that biases tRNA-mRNA movement toward the post-translocation state.
MATERIALS AND METHODS
Buffers and reagents
All measurements were done in buffer A: 50 mM Tris-HCl at pH 7.5, 70 mM NH4Cl, 30 mM KCl, and 7 mM MgCl2. GTP, phosphoenolpyruvate, and pyruvate kinase were purchased from Roche Diagnostics. 7-methylguanosine and purine nucleoside phosphorylase were from Sigma. Radioactive compounds were from Hartmann Analytics. All other chemicals were from Merck.
Ribosomes, mRNAs, tRNAs, and translation factors
Ribosomes from E. coli MRE 600 were prepared as described (Rodnina et al. 1995). f[3H]Met-tRNAfMet, [14C]Phe-tRNAPhe, EF-Tu, EF-G, MFTI-mRNA, phosphate-binding protein, and initiation factors were prepared as described (Rodnina et al. 1994, 1997, 1999; Savelsbergh et al. 2003). RRF was prepared as described (Peske et al. 2005).
Ribosome complexes
Pre-translocation complex: Ribosomes (1 μM) were incubated with a threefold excess of MFTI-mRNA (Rodnina et al. 1999) and a 1.5-fold excess each of IF1, IF2, IF3, and f[3H]Met-tRNAfMet in buffer A containing 1 mM GTP for 30 min at 37°C. Ternary complex EF-Tu·[14C]Phe-tRNAPhe·GTP was prepared by incubating EF-Tu (2 μM) with GTP (1 mM), phosphoenolpyruvate (3 mM), and pyruvate kinase (0.5 μg/mL) for 30 min at 37°C and then with [14C]Phe-tRNAPhe (1 μM) for an additional 5 min. Equal volumes of initiation complex and ternary complex were mixed and incubated for 1 min at 37°C to form pre-translocation complex carrying tRNAfMet in the P site and f[3H]Met[14C]Phe-tRNAPhe in the A site. Pre-translocation complexes used for stopped-flow experiments on translocation contained fluorescent f[3H]Met[14C]Phe-tRNAPhe(Prf16/17) in the A site. Ribosome complexes were purified from excess factors, mRNA, and tRNAs by centrifugation through a 1.1 M sucrose cushion in buffer A with 20 mM MgCl2. Pellets were dissolved in buffer A with 7 mM MgCl2 and tRNA binding was verified by nitrocellulose filtration.
Post-termination complex: Ribosomes (1 μM) were incubated with MFTI-mRNA (2 μM) and tRNAPhe (4 μM) for 30 min at 37°C in buffer A with 20 mM MgCl2. The MgCl2 concentration was reduced to 7 mM before using the complexes for subunit disassembly.
Translocation
Multiple-turnover translocation was induced by adding EF-G (final concentration 0.02 μM) and GTP (1 mM) to the pre-translocation complex (0.2 μM) and incubating for 1 min at 37°C. The extent of translocation was determined by the reactivity of P site peptidyl–tRNA with puromycin (Rodnina et al. 1997). Single-round translocation was measured by fluorescence stopped-flow using a pre-translocation complex with f[3H]Met[14C]Phe-tRNAPhe(Prf16/17) in the A site, EF-G (3 μM), and GTP (1 mM) (Savelsbergh et al. 2003).
Ribosome recycling
Subunit disassembly was measured in a stopped-flow system (Applied Photophysics) by rapidly mixing post-termination complex (0.05 μM) with EF-G (3 μM), RRF (5 μM), IF3 (2 μM), and GTP (1 mM) in buffer A and monitoring light-scattering at 435 nm (all concentrations after mixing).
GTP hydrolysis
Single-round GTP hydrolysis was measured by quench-flow, using [γ-32P]GTP, and liberated 32Pi was determined as previously described (Savelsbergh et al. 2005).
Release of inorganic phosphate
Pi release from EF-G after GTP hydrolysis was measured by the fluorescence change of MDCC-labeled phosphate-binding protein (PBP-MDCC) (Brune et al. 1994) in buffer A at 37°C (Mohr et al. 2000; Savelsbergh et al. 2003, 2005). Post-termination complex (final concentration, 0.2 μM) was mixed in the stopped-flow apparatus with EF-G (2 μM) and GTP (30 μM). Where indicated, RRF (5 μM) and/or FA (200 μM) were present. PBP-MDCC (2.5 μM), 7-methylguanosine (100 μM), and purine nucleoside phosphorylase (0.1 U/mL) were present in all reactions.
ACKNOWLEDGMENTS
We thank Frank Peske for performing initial experiments and critically reading the manuscript, and Carmen Schillings, Astrid Böhm, Simone Möbitz, and Petra Striebeck for expert technical assistance. The work was supported by grants of the Deutsche Forschungsgemeinschaft to M.V.R and W.W.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1592509.
REFERENCES
- Ævarsson A., Brazhnikov E., Garber M., Zheltonosova J., Chirgadze Y., al-Karadaghi S., Svensson L.A., Liljas A. Three-dimensional structure of the ribosomal translocase: Elongation factor G from Thermus thermophilus. EMBO J. 1994;13:3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X., Lei J., Brunelle J.L., Ortiz-Meoz R.F., Green R., Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol. Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R.K., Penczek P., Grassucci R.A., Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: The mechanism of translocation. Proc. Natl. Acad. Sci. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R.K., Sharma M.R., Kiel M.C., Hirokawa G., Booth T.M., Spahn C.M., Grassucci R.A., Kaji A., Frank J. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc. Natl. Acad. Sci. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali I.K., Lancaster L., Feinberg J., Joseph S., Noller H.F. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell. 2006;23:865–874. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Al-Karadaghi S., Ævarsson A., Garber M., Zheltonosova J., Liljas A. The structure of elongation factor G in complex with GDP: Conformational flexibility and nucleotide exchange. Structure. 1996;4:555–565. doi: 10.1016/s0969-2126(96)00061-5. [DOI] [PubMed] [Google Scholar]
- Barat C., Datta P.P., Raj V.S., Sharma M.R., Kaji H., Kaji A., Agrawal R.K. Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits. Mol. Cell. 2007;27:250–261. doi: 10.1016/j.molcel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Bodley J.W., Zieve F.J., Lin L., Zieve S.T. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem. Biophys. Res. Commun. 1969;37:437–443. doi: 10.1016/0006-291x(69)90934-6. [DOI] [PubMed] [Google Scholar]
- Borovinskaya M.A., Pai R.D., Zhang W., Schuwirth B.S., Holton J.M., Hirokawa G., Kaji H., Kaji A., Cate J.H. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- Brune M., Hunter J.L., Corrie J.E., Webb M.R. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- Chinali G., Parmeggiani A. The coupling with polypeptide synthesis of the GTPase activity dependent on elongation factor G. J. Biol. Chem. 1980;255:7455–7459. [PubMed] [Google Scholar]
- Czworkowski J., Wang J., Steitz T.A., Moore P.B. The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Agrawal R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Ito K., Yamami T., Nakamura Y. Ribosome recycling factor disassembles the post-termination ribosomal complex independent of the ribosomal translocase activity of elongation factor G. Mol. Microbiol. 2004;53:517–528. doi: 10.1111/j.1365-2958.2004.04156.x. [DOI] [PubMed] [Google Scholar]
- Gao N., Zavialov A.V., Li W., Sengupta J., Valle M., Gursky R.P., Ehrenberg M., Frank J. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol. Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Gao N., Zavialov A.V., Ehrenberg M., Frank J. Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits. J. Mol. Biol. 2007;374:1345–1358. doi: 10.1016/j.jmb.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson S., Singh R., Gudkov A.T., Liljas A., Logan D.T. Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Lett. 2005;579:4492–4497. doi: 10.1016/j.febslet.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Hirashima A., Kaji A. Role of elongation factor G and a protein factor on the release of ribosomes from messenger ribonucleic acid. J. Biol. Chem. 1973;248:7580–7587. [PubMed] [Google Scholar]
- Hirokawa G., Kiel M.C., Muto A., Selmer M., Raj V.S., Liljas A., Igarashi K., Kaji H., Kaji A. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 2002;21:2272–2281. doi: 10.1093/emboj/21.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa G., Nijman R.M., Raj V.S., Kaji H., Igarashi K., Kaji A. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA. 2005;11:1317–1328. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa G., Iwakura N., Kaji A., Kaji H. The role of GTP in transient splitting of 70S ribosomes by RRF (ribosome recycling factor) and EF-G (elongation factor G) Nucleic Acids Res. 2008;36:6676–6687. doi: 10.1093/nar/gkn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue-Yokosawa N., Ishikawa C., Kaziro Y. The role of guanosine triphosphate in translocation reaction catalyzed by elongation factor G. J. Biol. Chem. 1974;249:4321–4323. [PubMed] [Google Scholar]
- Ito K., Fujiwara T., Toyoda T., Nakamura Y. Elongation factor G participates in ribosome disassembly by interacting with ribosome recycling factor at their tRNA-mimicry domains. Mol. Cell. 2002;9:1263–1272. doi: 10.1016/s1097-2765(02)00547-6. [DOI] [PubMed] [Google Scholar]
- Janosi L., Mottagui-Tabar S., Isaksson L.A., Sekine Y., Ohtsubo E., Zhang S., Goon S., Nelken S., Shuda M., Kaji A. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 1998;17:1141–1151. doi: 10.1093/emboj/17.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., Ævarsson A., Liljas A., Hughes D. The dynamic structure of EF-G studied by fusidic acid resistance and internal revertants. J. Mol. Biol. 1996;258:420–432. doi: 10.1006/jmbi.1996.0259. [DOI] [PubMed] [Google Scholar]
- Julian P., Konevega A.L., Scheres S.H., Lazaro M., Gil D., Wintermeyer W., Rodnina M.V., Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc. Natl. Acad. Sci. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R., Pavlov M.Y., Buckingham R.H., Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- Katunin V.I., Savelsbergh A., Rodnina M.V., Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry. 2002;41:12806–12812. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- Keller D., Bustamante C. The mechanochemistry of molecular motors. Biophys. J. 2000;78:541–556. doi: 10.1016/S0006-3495(00)76615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster L., Kiel M.C., Kaji A., Noller H.F. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell. 2002;111:129–140. doi: 10.1016/s0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- Lee-Huang S., Lee H., Ochoa S. Inhibition of polypeptide chain initiation in Escherichia coli by elongation factor G. Proc. Natl. Acad. Sci. 1974;71:2928–2931. doi: 10.1073/pnas.71.8.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D., Wintermeyer W., Rodnina M.V. Arginines 29 and 59 of elongation factor G are important for GTP hydrolysis or translocation on the ribosome. EMBO J. 2000;19:3458–3464. doi: 10.1093/emboj/19.13.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura A., Kinoshita T., Tanaka N. Complex formation of fusidic acid with G factor, ribosome and guanosine nucleotide. Biochem. Biophys. Res. Commun. 1970;41:1545–1550. doi: 10.1016/0006-291x(70)90563-2. [DOI] [PubMed] [Google Scholar]
- Pai R.D., Zhang W., Schuwirth B.S., Hirokawa G., Kaji H., Kaji A., Cate J.H. Structural Insights into ribosome recycling factor interactions with the 70S ribosome. J. Mol. Biol. 2008;376:1334–1347. doi: 10.1016/j.jmb.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F., Rodnina M.V., Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol. Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Fricke R., Wintermeyer W. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry. 1994;33:12267–12275. doi: 10.1021/bi00206a033. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Fricke R., Kuhn L., Wintermeyer W. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J. 1995;14:2613–2619. doi: 10.1002/j.1460-2075.1995.tb07259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh A., Katunin V.I., Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh A., Matassova N.B., Katunin V.I., Semenkov Y.P., Wintermeyer W. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl. Acad. Sci. 1999;96:9586–9590. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A., Katunin V.I., Mohr D., Peske F., Rodnina M.V., Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol. Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A., Mohr D., Kothe U., Wintermeyer W., Rodnina M.V. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schief W.R., Clark R.H., Crevenna A.H., Howard J. Inhibition of kinesin motility by ADP and phosphate supports a hand-over-hand mechanism. Proc. Natl. Acad. Sci. 2004;101:1183–1188. doi: 10.1073/pnas.0304369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Kiel M., Pan D., Raj V.S., Kaji A., Cooperman B.S. Kinetics and thermodynamics of RRF, EF-G, and thiostrepton interaction on the Escherichia coli ribosome. Biochemistry. 2004;43:12728–12740. doi: 10.1021/bi048927p. [DOI] [PubMed] [Google Scholar]
- Seo H.S., Abedin S., Kamp D., Wilson D.N., Nierhaus K.H., Cooperman B.S. EF-G-dependent GTPase on the ribosome. Conformational change and fusidic acid inhibition. Biochemistry. 2006;45:2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- Spiegel P.C., Ermolenko D.N., Noller H.F. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H., Rodnina M.V., Wieden H.-J., van Heel M., Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- Voigt J., Sander G., Nagel K., Parmeggiani A. Effect of NH+4 and K+ on the activity of the ribosomal subunits in the EF-G- and EF-T-dependent GTP hydrolysis. Biochem. Biophys. Res. Commun. 1974;57:1279–1286. doi: 10.1016/0006-291x(74)90834-1. [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A., Petry S., Dunham C.M., Selmer M., Kelley A.C., Ramakrishnan V. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat. Struct. Mol. Biol. 2007;14:733–737. doi: 10.1038/nsmb1282. [DOI] [PubMed] [Google Scholar]
- Wilden B., Savelsbergh A., Rodnina M.V., Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc. Natl. Acad. Sci. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willie G.R., Richman N., Godtfredsen W.P., Bodley J.W. Some characteristics of and structural requirements for the interaction of 24,25-dihydrofusidic acid with ribosome—Elongation factor G complexes. Biochemistry. 1975;14:1713–1718. doi: 10.1021/bi00679a025. [DOI] [PubMed] [Google Scholar]
- Wilson G.J., Shull S.E., Cooke R. Inhibition of muscle force by vanadate. Biophys. J. 1995;68:216–226. doi: 10.1016/S0006-3495(95)80177-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.N., Schluenzen F., Harms J.M., Yoshida T., Ohkubo T., Albrecht R., Buerger J., Kobayashi Y., Fucini P. X-ray crystallography study on ribosome recycling: The mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishnia A., Boussert A., Graffe M., Dessen P.H., Grunberg-Manago M. Kinetics of the reversible association of ribosomal subunits: Stopped-flow studies of the rate law and of the effect of Mg2+ J. Mol. Biol. 1975;93:499–515. doi: 10.1016/0022-2836(75)90242-9. [DOI] [PubMed] [Google Scholar]
- Zavialov A.V., Hauryliuk V.V., Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol. Cell. 2005a;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Zavialov A.V., Hauryliuk V.V., Ehrenberg M. Guanine-nucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs. J. Biol. 2005b;4:9.1–9.19. doi: 10.1186/jbiol24. [DOI] [PMC free article] [PubMed] [Google Scholar]