Abstract
Cordycepin (3′ deoxyadenosine) is a biologically active compound that, when incorporated during RNA synthesis in vitro, provokes chain termination due to the absence of a 3′ hydroxyl moiety. We were interested in the effects mediated by this drug in vivo and analyzed its impact on RNA metabolism of yeast. Our results support the view that cordycepin-triphosphate (CoTP) is the toxic component that is limiting cell growth through inhibition of RNA synthesis. Unexpectedly, cordycepin treatment modulated 3′ end heterogeneity of ACT1 and ASC1 mRNAs and rapidly induced extended transcripts derived from CYH2 and NEL025c loci. Moreover, cordycepin ameliorated the growth defects of poly(A) polymerase mutants and the pap1-1 mutation neutralized the effects of the drug on gene expression. Our observations are consistent with an epistatic relationship between poly(A) polymerase function and cordycepin action and suggest that a major mode of cordycepin activity reduces 3′ end formation efficiency independently of its potential to terminate RNA chain elongation. Finally, chemical-genetic profiling revealed genome-wide pathways linked to cordycepin activity and identified novel genes involved in poly(A) homeostasis.
Keywords: cordycepin, ATP, poly(A), 3′ end formation, transcription, RNA, Saccharomyces cerevisiae
INTRODUCTION
The adenosine analog cordycepin (3′ deoxyadenosine) was first isolated from the ascomycete fungus Cordyceps militaris (Cunningham et al. 1950). Due to the absence of a 3′-hydroxyl moiety, the incorporation of cordycepin during RNA synthesis will result in termination of chain elongation. This activity has been well described in vitro with purified RNA polymerases and poly(A) polymerases from a number of organisms including yeast (Horowitz et al. 1976) and mammals (Muller et al. 1977). Furthermore, cordycepin exhibits well-established anti-bacterial, anti-fungal, and anti-parasitic properties (Sugar and McCaffrey 1998; Ahn et al. 2000; Rottenberg et al. 2005). In humans, anti-oxidant, anti-inflammatory, and anti-proliferative activities have been attributed to the drug (Zhu et al. 1998a,b). Indeed, for centuries, the fungus Cordyceps sinensis has been a widely administered traditional Chinese medicine, and cordycepin is thought to be one of the bioactive components mediating a host of beneficial effects (Paterson 2008). While the therapeutic potential of cordycepin underscores its unique properties, the effects of the drug on RNA metabolism are not completely understood. However, cordycepin was previously used to screen for novel genes involved in pre-mRNA polyadenylation (Bloch et al. 1978). This identified RNA14 and RNA15 genes, which encode core components of the 3′ end formation machinery in yeast (Minvielle-Sebastia et al. 1994). Thus, cordycepin responsive growth phenotypes may be useful indicators to detect deficiencies in pre-mRNA processing.
In this work, we present the first comprehensive analysis of the molecular effects of cordycepin in the budding yeast Saccharomyces cerevisiae. We provide evidence that the drug interfered with RNA synthesis and that this effect is mediated by CoTP, the toxic derivative of cordycepin. Unexpectedly, we observed defects during 3′ end formation of transcripts that occurred independently of a possible role of CoTP in terminating RNA chain elongation. Genome-wide chemical genetic profiling identified mutations that mediated cordycepin-responsive growth and linked novel genes to poly(A) metabolism. Our results extend the understanding of the molecular effects of the drug and establish cordycepin responsive growth as a functional test for the characterization of unknown genes.
RESULTS
Cordycepin toxicity in yeast is due to the inhibition of RNA synthesis
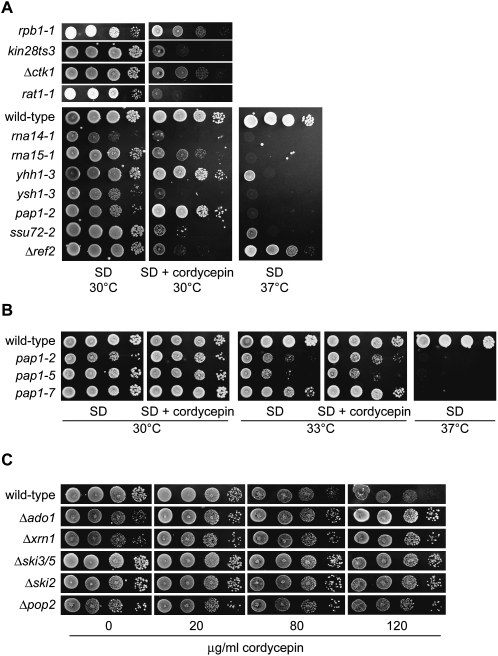
To analyze the molecular effects of cordycepin, we initially tested mutants in RNA synthesis pathways for hypersensitive growth in the presence of the drug (Fig. 1A). In these experiments, we employed a concentration of 20 μg/mL cordycepin, which only mildly affected growth of a wild-type strain (10% increased doubling time in liquid culture) (data not shown). We observed hypersensitive growth associated with mutants in the largest subunit of RNAP II (rpb1-1), kinases that phosphorylate the serine 5 (kin28-ts3) and serine 2 residues (Δctk1) of the RNAP II carboxy-terminal domain (CTD), a serine 5 CTD phosphatase (ssu72-2), and the nuclear 5′–3′ exonuclease Rat1 (rat1-1). Furthermore, mutants in certain 3′ end formation components responded strongly to the drug. This included the rna14-1 and rna15-1 strains, which were initially identified based on cordycepin sensitivity (Bloch et al. 1978), a mutation in the putative 3′ end nuclease Ysh1/Brr5 (ysh1-3), and the 3′ end factor Ref2 (Δref2). In contrast, mutation of Yhh1/Cft1 (yhh1-3) was without effect, and a mutant in poly(A) polymerase (pap1-2) showed increased resistance to cordycepin. To corroborate the latter observation, we tested additional mutant alleles in poly(A) polymerase and found that the pap1-2 and pap1-7 mutants reproducibly displayed a distinct growth advantage at 33°C on cordycepin medium (Fig. 1B). With the exception of the paradoxical behavior of pap1 mutants, cordycepin enhanced the growth defects of mutations in transcription and pre-mRNA 3′ end formation, consistent with a negative impact on RNA synthesis.
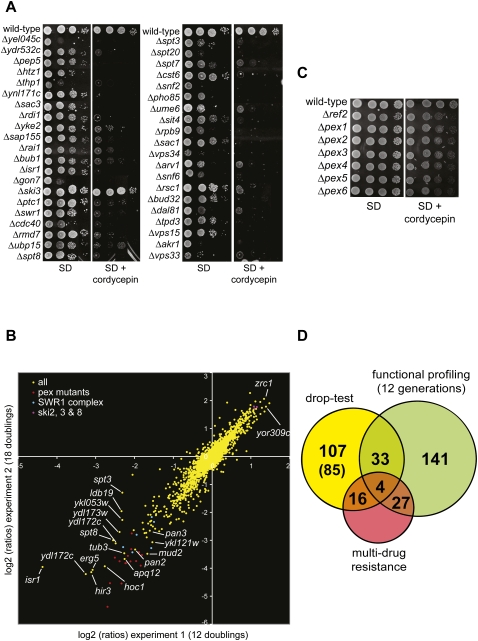
FIGURE 1.
Cordycepin-responsive mutant yeast strains involved in RNA synthesis and turn-over. (A–C) The indicated yeast strains were grown overnight in synthetic complete (SD) medium at the appropriate temperature. Tenfold serial dilutions were produced and spotted on media lacking or containing cordycepin and grown for 3–5 d. Unless indicated otherwise, the cordycepin concentration used was 20 μg/mL.
While the enhancement of growth defects linked cordycepin activity to RNA metabolism, it is less clear whether inhibition of RNA synthesis is the reason for reduced cell growth. To get a better understanding of cordycepin toxicity, we identified mutations that ameliorated the inhibitory effects of the drug (see below). Consistent with previous results (Lecoq et al. 2001), we found that the absence of the adenosine kinase Ado1 suppressed cordycepin toxicity at high cordycepin concentrations that reduced growth of a wild-type strain (Fig. 1C, 120 μg/mL). Ado1 converts cordycepin to cordycepin-monophosphate (CoMP), which is transformed into cordycepin-triphosphate (CoTP) by adenylate kinase Adk1 (Konrad 1988). Since Δadk1 cells were also resistant to cordycepin (see below), we conclude that CoTP, rather than cordycepin, is the toxic component. Strikingly, we found that the absence of various components involved in cytoplasmic mRNA turn-over also suppressed cordycepin toxicity. This included the 5′–3′ exonuclease Xrn1 (Muhlrad et al. 1994), the Pop2 deadenylase (Daugeron et al. 2001), and Ski2 and Ski3/5 proteins that aid exosome mediated (3′–5′ directed) RNA turn over (Brown et al. 2000). The observation that RNA stabilization suppressed cordycepin toxicity is consistent with the idea that RNA synthesis is limiting cell growth in the presence of the drug.
Effects of cordycepin on mRNA synthesis in wild-type strains
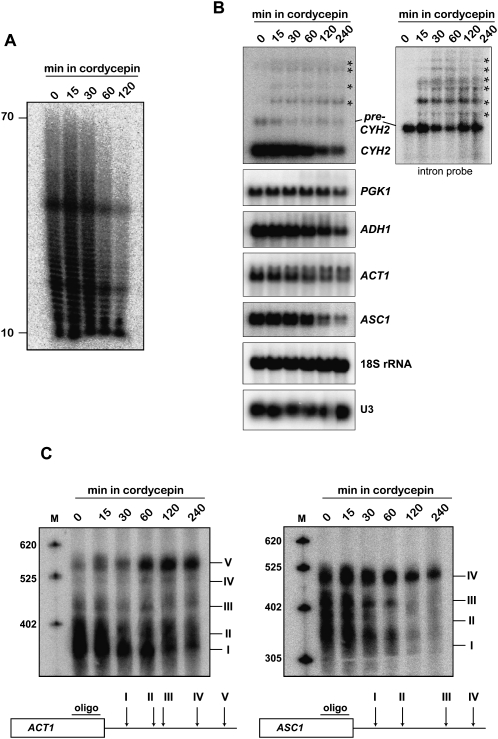
Next, we exposed wild-type strains to 20 μg/mL cordycepin and performed RNA analyses. Cellular poly(A) content was strongly reduced after 2 h, consistent with a strong inhibition of RNA synthesis (Fig. 2A). Probing of individual housekeeping mRNAs by Northern blot revealed reduced steady-state levels for CYH2, ASC1, and ADH1 and a less severe effect on ACT1 and the stable PGK1 mRNA (Fig. 2B); levels of 18S rRNA, U3 snoRNA, and tRNA3 LEU remained unchanged (Fig. 2B; data not shown). Most interestingly, we observed additional, longer RNAs for CYH2, which appeared as early as 15 min after cordycepin addition. These RNAs extended more than 3 kb beyond the normal 3′ end of the mRNA. Furthermore, a probe directed against the CYH2 intron readily detected the pre-mRNA and extended transcripts (Fig. 2B).
FIGURE 2.
Molecular effects of cordycepin on RNA metabolism. (A) Cellular poly(A) length distribution following cordycepin treatment. Total RNA was extracted from wild-type strains following growth in the presence of 20 μg/mL cordycepin for the indicated times. RNAs were labeled with [α-32P]cordycepin and poly(A) polymerase, and digested with RNases A and T1. RNase-resistant poly(A) tails were resolved on denaturing 15% polyacrylamide/8.3 M urea gels and visualized by PhosphorImaging (Fuji FLA7000). (B) Northern analysis of RNA obtained from a wild-type strain treated with cordycepin as described in A. RNAs were detected with random prime labeled probes directed against the open reading frames (ORF) of the indicated genes; (left panel) the probes against CYH2 were either directed against the entire ORF or (right panel) exclusively against its intron. (Asterisks) 3′-Extended transcripts. 18S rRNA and U3 snoRNA were detected with end-labeled oligonucleotides. (C) High-resolution Northern analysis of ACT1 and ASC1 3′ ends. Total RNA from a wild-type strain treated with cordycepin as described in A was treated with RNase H in the presence of oligonucleotides complementary to the 3′ end of the ORF of the (left panel) ACT1 or (right panel) ASC1 genes, respectively. Below each gel a scheme indicates the relative location of the oligonucleotide used to target RNAse H and the distribution of observed 3′ ends (I to V for ACT1; and I to IV for ASC1).
Moreover, we observed a pattern of ACT1 mRNA in the presence of cordycepin that was reminiscent of poly(A) site selection defects previously associated with mutants in the 3′ end formation machinery (Mandart and Parker 1995; Dichtl et al. 2002b; Kyburz et al. 2003). To analyze the 3′ ends of ACT1 , we targeted the RNA with an oligonucleotide directed against the open reading frame (ORF) and RNase H (Fig. 2C). This resulted in 5′ truncation of the mRNA and allowed the size separation on high-resolution gels of transcripts with at least five different 3′ ends (Dichtl et al. 2002b). In the absence of cordycepin site I was used predominately, but addition of the drug triggered the disappearance of this species and the accumulation of longer transcripts, which mostly use site V. Notably, this effect was not observed with Δado1 cells (data not shown), suggesting that CoTP may be the active compound mediating this effect. Interestingly, a similar phenotype was observed with ASC1 mRNA. This transcript also displayed heterogeneous 3′ ends in wild-type strains, and we could resolve at least four different RNA species following RNase H treatment as described above (Fig. 2C). In the absence of cordycepin the different transcripts were roughly equal in abundance, but addition of the drug resulted in exclusive usage of the most distal 3′ end after 2 h. These results suggested that cordycepin reduced the efficiency of 3′ end formation at a step prior to poly(A) tail synthesis by poly(A) polymerase.
The pap1-1 mutation neutralizes the effects of cordycepin on gene expression
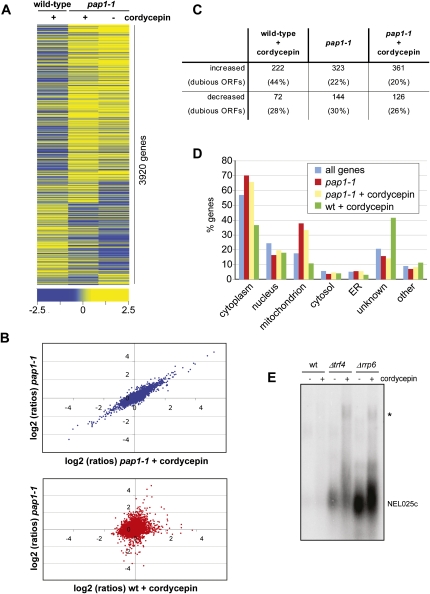
Our results indicated complex effects of cordycepin on RNA metabolism of yeast, and we were intrigued by its mitigating effects on growth of poly(A) polymerase mutants. To extend our experiments, we determined by microarray analyses changes in RNA levels that were induced within 1 h by 20 μg/mL cordycepin in wild-type and pap1-1 strains. The obtained data, depicted as a heat map in Figure 3A and as a scatter plot in Figure 3B, revealed a high correlation of signals obtained with the pap1-1 strain in the absence or presence of the drug, respectively. This suggested that the pap1-1 mutation largely neutralized the negative effects of cordycepin on gene expression. In contrast, the effects of cordycepin treatment of wild-type and the effects of the pap1-1 mutation were not correlated (Fig. 3A,B), suggesting that the two conditions had distinct effects on RNA levels.
FIGURE 3.
The pap1-1 mutation neutralizes the effects of cordycepin. Gene expression profiling was done with wild-type and pap1-1 strains following a shift for 1 h to medium lacking or containing 20 μg/mL cordycepin, respectively. All experiments were performed in triplicate. (A) Heat map of gene expression changes of cordycepin treated wild-type and nontreated and treated pap1-1 mutant strains relative to the untreated wild-type control. Rows represent genes, and columns represent average values obtained from three independent measurements for the indicated condition; only those 3920 features were represented that gave three values each for all three conditions. Clustering was done with Spearman Row correlation (substance and row) using Acuity 4.0. The color code indicates the distribution of log2 ratios. (B) Scatterplot of gene expression changes (log2 ratios). (Upper graph) Plots signals obtained with the pap1-1 strain in the absence (y-axis) and the presence (x-axis) of cordycepin. (Lower graph) Signals obtained with the pap1-1 strain in the absence of cordycepin (y-axis) are plotted against signals obtained with cordycepin-treated wild-type (x-axis). (C) The table displays the numbers of RNA transcripts that changed at least twofold (log2 value ±1) relative to untreated wild type in the respective conditions. The numbers of transcripts that were derived from dubious open reading frames are indicated in percent in brackets. (D) Transcripts that were increased under the indicated conditions were subjected to Gene Ontology analysis using the SGD slim mapper (http://www.yeastgenome.org/). The number of genes in the individual compartments is presented in percent of all analyzed genes. Also indicated is the background distribution of all genes. (E) Northern blot analysis of total RNA obtained from wild-type and mutant strains following growth in the absence and presence of 20 μg/mL cordycepin. The CUT NEL025c was detected using a random primed labeled probe. (Asterisk) Indicates a transcript that accumulates due to Pap1 dependent 3′ end formation at a downstream gene (Wyers et al. 2005).
Moreover, the microarray data suggested that cordycepin treatment of the wild type resulted in more transcripts that were increased rather than decreased (Fig. 3C). This was surprising since we observed a loss of cellular poly(A) associated with cordycepin treatment (Fig. 2A). These observations may indicate that global levels of transcript bodies (which were detected in the microarray) and of poly(A) tails [which were detected in the poly(A) analysis] were differentially affected by the drug. Interestingly, RNAs derived from dubious open reading frames (dubious ORFs) were over-represented in the category of increased transcripts (Fig. 3C, 44%). A Gene Ontology (GO) analysis revealed, furthermore, that cytoplasmic components were decreased and unknown components were approximately twofold increased under this condition (Fig. 3D). Unknown components are largely constituted by dubious ORFs that also include cryptic unstable transcripts (CUTs) (Wyers et al. 2005). CUTs are rapidly degraded by the nuclear exosome in a reaction that is strongly stimulated by the noncanonical TRAMP poly(A) polymerase complex (Wyers et al. 2005; Houseley et al. 2006). To test whether cordycepin interfered with this pathway, we probed by Northern blot for the abundance of the well-characterized CUT NEL025c in wild-type, exosome (Δrrp6), and TRAMP (Δtrf4) mutant strains. The CUT was barely detectable in wild type (Fig. 3E). In contrast, NEL025c was stabilized in Δrrp6 and Δtrf4 strains, as expected, and cordycepin enhanced this effect, resulting in a striking accumulation of extended transcripts. Taken together, these observations are consistent with the idea that the increase of transcript levels following cordycepin treatment can partially be explained by elevated levels of CUTs and extended CUT containing RNAs.
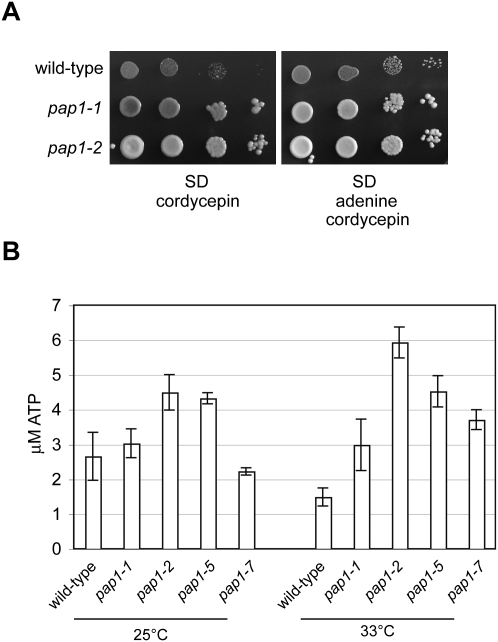
Most interestingly, we observed a highly significant up-regulation of ATP synthesis pathways in pap1-1 strains including, among others, mRNAs encoding proteins involved in nucleotide precursor generation (P-value = 2.69 E-35), mitochondrial electron transport (P-value = 3.17 E-25), and oxidative phosphorylation (P-value = 1.43 E-40). Moreover, mitochondrial components were strongly over-represented in the GO term analysis (Fig. 3D). This may indicate that less effective Pap1 stimulated the cell to increase ATP production, suggesting that the pap1-1 mutant had an increased ratio of ATP/CoTP in the presence of cordycepin. Since this ratio may determine cordycepin toxicity (see above), enhanced ATP synthesis may underlie the observed cordycepin resistance of pap1 mutants. To further test this idea, we supplemented cordycepin-containing medium with adenine. Adenine was used instead of ATP because yeast do not have an adenosine transporter (Maser et al. 1999). Figure 4A shows that excess adenine was sufficient to suppress the growth-inhibitory effect of the drug on a wild-type strain, whereas growth of pap1 mutant strains was not noticeably affected.
FIGURE 4.
Increased ATP levels in pap1 mutants. (A) Tenfold serial dilutions of wild-type and pap1 mutant strains were spotted on SD medium containing 20 μg/mL cordycepin that was supplemented with 20 μg/mL adenine as indicated. The yeast were grown for 3 d at 25°C and photographed. (B) Wild-type and pap1 mutants strains were grown at 25°C or shifted for 1 h to 33°C as indicated and assayed for ATP content. Represented in the graph are average values for ATP content obtained from three independent yeast cultures; the error bars display standard error.
Measurement of the cellular ATP content revealed significantly increased levels in pap1-2 and pap1-5 strains at 25°C compared to wild type (Fig. 4B). Following growth at semipermissive temperature (33°C), all tested mutant strains displayed markedly (two- to fourfold) increased ATP levels compared to the wild-type control. These data support the conclusion that ATP levels were elevated in response to less active poly(A) polymerase in the tested temperature-sensitive mutants.
Genome-wide identification of cordycepin targets
The yeast gene deletion collection (Winzeler et al. 1999) is a valuable tool to explore the molecular targets of therapeutic agents and xenobiotics (Hillenmeyer et al. 2008). To identify mutants that displayed cordycepin-dependent growth phenotypes, we tested more than 4800 haploid deletion strains by spotting them as single drops on synthetic minimal plates lacking or containing 20 μg/mL cordycepin. Candidate strains were identified by visual inspection and re-examined in two consecutive rounds by dropping 10-fold serial dilutions on plates containing 40 and 20 μg/mL cordycepin, respectively. This identified 155 sensitive strains and five strains that showed increased resistance (see also Fig. 1C). The mutants were categorized as strong (57), medium (56), and weak (47) indicating the severity of the growth difference and the relative confidence in the observed phenotype. Figure 5A shows growth phenotypes of selected strains, a full list of identified mutants is supplied in Supplemental Data S1. The distribution of GO components revealed that cordycepin targets were distributed without bias in the individual cellular compartments. Significant functional categories included ATPases (P-value = 0.001) and chromatin binding (P-value = 0.003). Several subunits of the transcriptional coactivator SAGA (Spt3, Spt7, Spt8, and Spt20), the SWR1 chromatin remodeling ATPase, and its substrate, the H2A variant Htz1, were found to be cordycepin-responsive.
FIGURE 5.
Identification of cordycepin-responsive haploid deletion mutants. (A) The growth phenotypes of strains identified by drop-test screening were analyzed on medium containing or lacking 40 μg/mL cordycepin as indicated. Strains were grown for 3 d at 30°C and photographed. (B) Pools of all viable haploid deletion mutants from the systematic deletion library were grown in complete liquid medium with or without 20 μg/mL cordycepin for 12 or 18 generations (Experiments 1 and 2, respectively) by serial dilutions. Cells were then collected, the genomic DNA was extracted, and the barcode sequence tags associated with each gene deletion were amplified by PCR and hybridized on barcode DNA arrays as described (Decourty et al. 2008). The figure plots for each experiment (Experiment 1 on the x-axis and Experiment 2 on the y-axis) the log2 of the ratio for each deletion mutant of the barcode signal between the cultures without and with cordycepin (computed as in Decourty et al. 2008). Negative log2 (ratios) indicate strains more sensitive to cordycepin than the average population of mutants. Mutants in PEX genes, SWR1 complex genes, and SKI 2, 3, and 8 genes are color-coded as indicated. Arrows and labels point to mutants with strong phenotypes. (C) Cordycepin-sensitive growth phenotypes of pex mutants. The indicated strains were analyzed as described in A. (D) Graph depicting the overlap of mutants identified in drop-test screening versus functional profiling (after 12 generations). Also indicated are the numbers of multi-drug-resistance genes that were identified with the individual procedures or which are shared, respectively. In brackets the number of strains are indicated that are identified in the drop-test screening that could not be analyzed by functional profiling due to slow growth of the mutants.
A limitation of evaluating drug-dependent growth by drop-test is that weak differences are difficult to detect. We decided therefore to employ a quantitative approach to measure the relative fitness of mutant strains in the absence and presence of the drug by chemical-genetic profiling (Ooi et al. 2006). In these experiments, all mutants of the deletion collection were competitively grown in a single culture, and the relative abundance of individual strains was measured after zero, 12, and 18 generations in medium lacking or containing 20 μg/mL cordycepin. Considering only signals of at least twofold or higher, we identified 205 cordycepin responsive mutants after 12 generations and 496 mutants after 18 generations (complete data sets are available in Supplemental Data S2). However, due to severe growth defects, this procedure will not resolve signals for around 300 to 400 strains (Decourty et al. 2008). The strong increase of identified strains from 12 to 18 generations suggested that even small growth differences could be resolved with this approach. The scatterplot shown in Figure 5B revealed a strong correlation of signals obtained after 12 and 18 generations, demonstrating that the smaller data set was almost completely included in the larger one underscoring the significance of the obtained results.
We could identify several groups of genes that were significantly enriched and that are highlighted in Figure 5B. This included 15 PEX genes (P-value = 2.42 E-10), which are required for the biogenesis and proliferation of peroxisomes (Kiel et al. 2006). Since cordycepin caused reduced growth of pex mutants (Fig. 5C; data not shown), we suggest a role for these organelles in suppressing toxicity of the drug. Interestingly, the catabolism of purines was associated with peroxisomes in mammals (Wanders and Waterham 2006). If this holds true also for yeast, cordycepin levels may be increased in pex mutants due to decreased nucleotide turn-over providing a plausible explanation for the increased sensitivity to the drug.
Most interestingly, chromatin remodeling complexes (P-value = 1.03 E-06) and the SWR1 complex (P-value = 8.90 E-07) were significantly enriched in these profiling experiments, and the same GO terms were also identified with the drop-test procedure. Similarly, the cordycepin-sensitive growth of mutants lacking SAGA components Spt3 and Spt8 was reproduced in both approaches. We were surprised to identify in this screen strains lacking the Pan2 and Pan3 proteins as cordycepin-sensitive. Pan2/3 form the Pab1p-dependent poly(A) nuclease that acts in the shortening of mRNA poly(A) tails (Boeck et al. 1996; Brown et al. 1996) and that may act in the message-specific maturation of poly(A) length (Brown and Sachs 1998). In contrast, we found that the absence of Pop2, which is associated with the major yeast deadenylase Ccr4 (Daugeron et al. 2001; Tucker et al. 2002), suppressed cordycepin toxicity (Fig. 1C). Thus, these cordycepin-dependent phenotypes may reflect partially antagonistic effects of Pan2/3 and Ccr4/Pop2 deadenylase complexes, respectively, on gene expression in yeast.
Finally, we compared the results obtained with the drop-test and functional profiling approaches, respectively. Figure 5D shows that a mere 37 mutant strains were identified with both procedures. Since only four of those overlapping mutants corresponded to multi-drug-resistance genes (Parsons et al. 2006), this group of mutants appeared particularly responsive to cordycepin. The limited overlap is mainly due to fact that a large fraction of mutants identified in the drop-test (85 of 107 strains) escaped the analysis by functional profiling due to the slow growth phenotype. Interestingly, we found that a published drop-test screen with mycophenolic acid (Desmoucelles et al. 2002) also overlapped only little with results of a profiling experiment that we performed with the same drug (S. Holbein, L. Decourty, A. Jacquier, and B. Dichtl, unpubl.). These observations indicate that the outcomes of drug-sensitivity screening of the yeast deletion collection may be strongly dependent on the employed approach.
Cordycepin-responsive strains are connected to poly(A) metabolism
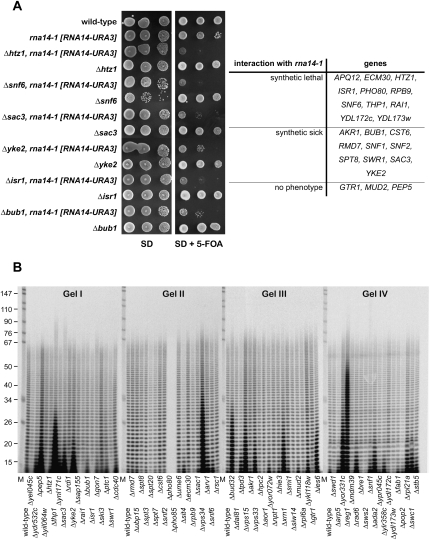
Since pre-mRNA 3′ end formation is targeted by cordycepin, drug mediated growth phenotypes may indicate a functional link to poly(A) metabolism. We therefore tested whether cordycepin-induced growth phenotypes can be duplicated by combining selected mutations with the temperature-sensitive rna14-1 allele, which is defective in cleavage and polyadenylation of pre-mRNA (Minvielle-Sebastia et al. 1994). Out of 24 double mutants tested, 11 were synthetic lethal, 10 were synthetic sick, and three did not exhibit a phenotype (Fig. 6A). These results suggested a functional association of cordycepin-responsive mutants and pre-mRNA processing.
FIGURE 6.
Cordycepin-responsive mutants are linked to poly(A) metabolism. (A) Mutations causing cordycepin sensitivity genetically interact with rna14-1. The temperature-sensitive rna14-1 strain, carrying a URA3-marked plasmid containing the wild-type RNA14 gene, was used for disruption of genes causing cordycepin sensitivity. Double mutants were challenged with 5-flouroorotic acid (5-FOA) to force loss of the URA3-marked plasmid exhibiting the phenotype of the double mutation. Shown are serial dilutions of indicated single and double mutant strains on synthetic (SD) medium lacking or containing 5-FOA. Growth phenotypes were categorized as synthetic lethal (no growth), synthetic sick (reduced growth), or normal (no phenotype) when compared to the rna14-1 mutant alone. (B) Cellular poly(A) length distribution in cordycepin-responsive strains. Total RNA was obtained from indicated mutant strains and analyzed as described in the legend of Figure 2A, except that poly(A) tails were resolved on 10% sequencing gels.
To corroborate these findings, we asked whether poly(A) metabolism was affected in cordycepin-responsive strains and analyzed the cellular poly(A) length distribution of selected mutants. Figure 6B shows that wild-type strains exhibited an even distribution of poly(A) ranging from about 10 to 70 adenylate residues (As). While the majority of tested strains were similar to wild type, some mutants showed marked accumulation of short (up to 20 As), intermediate (up to 35 As), and long (up to 50 As) A-tracts. The maximum poly(A) length of most strains was within the expected range of ∼70 As. However, Δthp1 and Δsac3 mutants (Gel I) showed some hyperadenylation, a phenotype that was previously associated with defective mRNA export (Hilleren and Parker 2001). Somewhat curiously, Δpho85 and Δpho80 RNAs did not give any poly(A) labeling (Gel II). We recently showed that polyphosphate levels were elevated in these mutants, resulting in the inhibition of the in vitro labeling reaction and pointing to a possible role for this metabolite in regulating poly(A) polymerase (Holbein et al. 2008). Short poly(A) tails were stabilized, for example, in the Δxrn1 strain, that lacks 5′–3′ exonuclease activity required for cytoplasmic mRNA turn-over (Gel III). This is consistent with reports showing that the absence of Xrn1 stabilized mRNAs, which were decapped and maintained residual 10 to 15 As following deadenylation (Muhlrad et al. 1994). Other mutants with short A-tracts included Δsac3, Δyke2 (Gel I), Δvps33, Δies3 (Gel III), and Δrad6, Δada2, Δrpl21a, Δswc1 (Gel IV). Intermediate poly(A) tracts were observed in the Δvps34 mutant (Gel II). Vps34 is a phosphatidylinositol 3-kinase that forms a complex with Vps15 (Stack et al. 1993); interestingly, the Δvps15 mutant was also identified in our screening and also displayed aberrant poly(A) length distribution (Gel III). These observations suggested a role for the Vps34–Vps15 complex or the signaling molecule phosphatidylinositol 3-phosphate, respectively, in poly(A) metabolism. Other mutants that had accumulated A-tracts of intermediate length included Δpep5, Δthp1 (Gel I), Δbud32 (Gel III), and Δyor331c (Gel IV). The Δreg1 strain accumulated long poly(A) tails (Gel IV). Interestingly, Reg1 is a regulatory subunit for the type-1 protein phosphatase Glc7p that is involved in glucose repression (Tu and Carlson 1995). The poly(A) phenotype observed here may reflect a previously described role for Reg1 in controlling mRNA translation in response to nutrient status (Ashe et al. 2000) and may point to a requirement for Reg1-Glc7 in the maintenance of poly(A) tail length homeostasis.
DISCUSSION
We present a comprehensive study of the effects of cordycepin in the yeast S. cerevisiae that was initiated to increase the understanding of the molecular modes of action of this drug, on the one hand, and, on the other hand, to exploit its properties for the identification of novel genes that are functionally connected to poly(A) metabolism.
It is well established that cordycepin can terminate RNA synthesis when incorporated by RNA polymerases and poly(A) polymerase (Horowitz et al. 1976; Muller et al. 1977). While this conclusion is supported strongly by in vitro experiments, the effects of the drug in vivo remained less well characterized. Consistent with previous reports (Lecoq et al. 2001; Naula et al. 2003), our analyses support the idea that CoTP rather than cordycepin is mediating the toxicity of the drug in yeast. Since CoTP is an analog of ATP, this nucleotide may interfere with numerous cellular processes that require ATP binding or hydrolysis. While this remains a viable possibility, our analysis of suppressors of cordycepin toxicity identified mutants in three major activities: (1) nucleotide synthesis pathways that are involved in the conversion of cordycepin to CoTP; (2) poly(A) polymerase; and (3) cytoplasmic mRNA turn-over reactions. The latter observation suggested that mRNA stabilization suppressed toxicity, supporting the idea that it is, indeed, the inhibition of RNA synthesis that was limiting cell growth. Consistently, we observed strongly reduced steady-state mRNA levels following cordycepin treatment. This reduction of mRNA levels may result from the inhibition of transcript synthesis by RNA polymerase II, the abortion of poly(A) addition by poly(A) polymerase, or the inhibition of other reactions that are necessary for stable mRNA accumulation.
Unexpectedly, we observed cordycepin mediated phenotypes that cannot be explained by the RNA chain termination potential of the drug. Particularly striking were rapid effects on the 3′ ends of ACT1 and ASC1 mRNAs. For ACT1, it was suggested that a cluster of alternative poly(A) sites guides 3′ end formation and that the efficiency of poly(A) site recognition may determine the relative usage of these sites (Mandart and Parker 1995). Consistent with this idea, several mutants in 3′ end factors interfered with ACT1 3′ ends (Mandart and Parker 1995; Dichtl et al. 2002b; Kyburz et al. 2003). The duplication of these phenotypes by cordycepin in the wild type was unexpected since the preferred usage of site I was shifted to the more distal site V, resulting in longer transcripts (Fig. 2C). Since shorter RNAs would be predicted to result from chain termination, we conclude that the defect at ACT1 did not result from cordycepin-induced termination of RNA polymerase II transcription. The accumulation of mRNAs with more distal 3′ ends also argued against chain termination during poly(A) synthesis by Pap1. The latter conclusion was furthermore underscored by the positive effect that cordycepin had on growth of pap1 mutants (see below). Indeed, it was maybe one of the most surprising results of this study that the drug did not enhance the growth defect of pap1 mutants. Since the ACT1 defect was observed only minutes following exposure of the yeasts to the drug, and since the phenotype was suppressed in Δado1 cells, it was probably directly mediated by CoTP. We also observed heterogeneous 3′ ends for ASC1 mRNA, and also in this case cordycepin treatment affected their relative abundance in favor of the more distal 3′ ends. Therefore, similar mechanisms may mediate the effects of cordycepin on 3′ end formation on ACT1 and ASC1.
Another unexpected observation was the appearance of extended CYH2 mRNAs in the presence of cordycepin. Transcripts with discrete length appeared within minutes of drug addition and extended several kilobases beyond the normal 3′ end of the pre-mRNA, strongly suggesting that RNA chain termination was not the major consequence of cordycepin treatment. Instead, cordycepin apparently interfered with efficient 3′ end formation at the CYH2 poly(A) site as was observed with ACT1 and ASC1 mRNAs. In contrast to the latter transcripts, which harbor clusters of poly(A) sites in their 3′ UTR, extended CYH2 RNAs may result from 3′ end formation at the next available poly(A) sites, which lie far downstream from the regular site. How cordycepin reduces 3′ end formation efficiency remains unclear, but involved pathways may include the coupling of 3′ end formation to transcription (Lykke-Andersen and Jensen 2007). Consistent with this idea, we found that mutations in kinases and phosphatases that act on the CTD are hypersensitive to cordycepin (Fig. 1A).
Furthermore, our gene expression profiling experiments revealed that cordycepin treatment caused an increase of transcripts that were derived from dubious ORFs. This included the well-studied CUT NEL025c, which is degraded by exosome and TRAMP mediated mechanisms (Wyers et al. 2005; Houseley et al. 2006). A satisfying explanation for the observed effects on NEL025c could be that TRAMP mediated polyadenylation was inhibited following cordycepin incorporation causing less efficient turn-over of the RNA. Alternatively, it seems possible that exosome-mediated degradation was impaired when the substrate RNA lacked a 3′ hydroxyl moiety. However, the major effect of cordycepin on NEL025c expression was the appearance of extended transcripts, which were likely to result from 3′ end formation at sites downstream from the Nrd1-dependent terminator that normally guides 3′ end formation of this transcript (Fig. 3E; Arigo et al. 2006; Thiebaut et al. 2006). Downstream termination on this transcription unit has been shown to include a Pap1 dependent site, providing another possible explanation for the accumulation of these and other RNAs in the presence of the drug. Since the Nrd1 complex is targeted to transcription units by serine 5 phosphorylated CTD (Gudipati et al. 2008; Vasiljeva et al. 2008), it is tempting to speculate that cordycepin treatment reduced the coupling of 3′ end formation to transcription also at NEL025c, as we suggested above for ACT1, ASC1, and CYH2 mRNAs.
Overall, our data support the idea that the observed biochemical effects of cordycepin occurred largely independently from RNA chain termination. This unexpected conclusion may indicate that the nuclear concentration of CoTP that built up under the conditions employed here was sufficiently high to reduce the efficiency of 3′ end formation, but too low to cause significant chain termination either during transcription or polyadenylation. It will be interesting to test whether the same conclusion may apply to other organisms, be it prokaryotes or metazoans. However, our work implies that the impact of cordycepin on RNA synthesis has to be interpreted with caution and that it cannot generally be reduced to the chain termination potential of the drug. While the target of cordycepin action remains to be identified, the ameliorating effects of cordycepin on growth of pap1 mutants (Fig. 1B) suggested an epistatic relationship between polyadenylation and cordycepin action. Such an epistatic effect is consistent with the idea that mutation of poly(A) polymerase and the presence of cordycepin interfere with the same pathway (Schuldiner et al. 2005; Segre et al. 2005).
Moreover, our analyses revealed that inactivation of poly(A) polymerase provoked higher cellular ATP levels in temperature-sensitive mutant strains. Our data are also consistent with the idea that the CoTP:ATP ratio is an important determinant for cordycepin toxicity, providing a plausible explanation of how poly(A) polymerase mutants suppressed cordycepin toxicity. Interestingly, a recent report revealed an ATP-sensing mechanism to connect RNA abundance to respiration (Amiott and Jaehning 2006). Our results suggest a reverse flow of information and raise the interesting possibility that the “efficiency” of gene expression (in this case, of polyadenylation) is somehow sensed and functionally linked to the energy status of the cell. However, further analyses are required to untangle this intriguing interplay of ATP homeostasis and 3′ end formation.
One of our motivations to analyze the effects of cordycepin was to establish a correlation of drug mediated phenotypes and functional classes of genes. Direct analysis of mutants involved in RNA synthesis and turn-over pathways clearly established cordycepin-responsive phenotypes. Our screening approaches using the deletion collection of non-essential genes identified genome-wide targets for the drug, and only a rather limited number of those genes is known to be involved in RNA synthesis and processing. This may reflect that genes involved in those processes often are essential for cell viability and were therefore not represented in the mutant collection that we screened. Nevertheless, we were able to link several functional categories of mutants with high significance to cordycepin action. This included chromatin associated proteins like the SAGA and SWR1 complexes, but the molecular reasons for cordycepin-responsive growth of the respective mutants remain to be determined.
Our analyses demonstrated that cordycepin meditated growth phenotypes can be useful indicators to identify genes that are involved in poly(A) metabolism. Several mutant strains were identified that showed aberrant poly(A) length distribution profiles (Fig. 6B). While the mechanistic relation of cordycepin-responsive growth and poly(A) distribution remains unclear for certain mutants, we can rationalize the situation in some cases. For example, Δxrn1 strains stabilize decapped mRNAs that retain residual short poly(A) tails (Muhlrad et al. 1994). The apparent accumulation of short poly(A) tracts can therefore reflect defective mRNA turn-over, which we found to suppress the toxic inhibition of RNA synthesis by cordycepin (Fig. 1C). In addition to mRNA turn-over enzymes, translation efficiency is thought to affect mRNA stability (Meyer et al. 2004). In cordycepin-hypersensitive Δreg1 strains, the accumulation of long poly(A) tracts may reflect a synergism between deregulated translation (Ashe et al. 2000) and reduced mRNA synthesis in the presence of cordycepin. Other cordycepin phenotypes may have resulted from enhancement of nuclear deficiencies that have an impact on poly(A) metabolism. For example, Thp1 and Sac3 form a nuclear-pore-associated complex that is required for efficient mRNA export (Fischer et al. 2002). Nuclear poly(A) accumulation may underlie the disturbed poly(A) profiles observed here in Δthp1 and Δsac3 strains, and less efficient gene expression in these strains likely synergized with the toxic effects of cordycepin. Moreover, it seems possible that loss of control of adenylation or deadenylation activities may contribute to poly(A) tail phenotypes in certain mutants. Non-essential components involved in poly(A) metabolism may primarily be involved in fine-tuning and regulatory aspects of gene expression. In this respect, it will be interesting to test, for example, which role signal-transduction pathways and signaling molecules like the Vps34/Vps15 and its product phosphatidylinositol 3-phosphate play in the control of global poly(A) levels during growth and development or in response to changing environmental conditions. It seems likely that these and other cordycepin-responsive pathways identified in this work may help to uncover novel regulatory layers of poly(A) metabolism.
MATERIALS AND METHODS
Growth and construction of yeast strains
The S. cerevisiae strains used in this study are listed in Table 1. Yeast were grown in rich YPD medium (2% glucose, 2% bacto-tryptone, 1% yeast extract) or synthetic complete or synthetic drop-out media (2% glucose, 0.67% yeast nitrogen base, 1× amino acids). Cordycepin (Sigma-Aldrich) was supplemented at a concentration of 20 μg/mL unless stated otherwise. For synthetic interaction analyses, kanMX4-marked alleles were amplified from haploid mutant strains (Winzeler et al. 1999) and transferred into the rna14-1 mutant background. Loss of URA3-marked plasmids was forced on SD plates containing 1 mg/mL 5-FOA (Zymo Research).
TABLE 1.
Yeast strains used in this study
Drug sensitivity screening
For drop-test screening, all haploid mutants of the yeast gene deletion collection (Euroscarf) (Winzeler et al. 1999) were grown in 96-well microtiter plates and spotted on agar plates as single drops. Growth was examined daily for a 5-d period by comparing the drops on SD and SD cordycepin (20 μg/mL; Sigma-Aldrich) medium. Candidate strains were re-examined in two consecutive rounds by analyzing growth of 10-fold serial dilutions on SD supplemented with 40 μg/mL and 20 μg/mL cordycepin, respectively. The verified strains were then categorized as strong, medium, and weakly responsive to cordycepin, reflecting the relative confidence in the observed phenotype (Supplemental Data S1).
Chemical-genetic profiling was done essentially as described (Decourty et al. 2008). All haploid mutants of the yeast gene deletion collection were pooled and cultured overnight in SD medium at 30°C. At an OD600 of 1.0, the culture was diluted 20-fold in SD medium lacking and containing 20 μg/mL cordycepin, respectively, and the OD600 was monitored every hour. After zero, 12, and 18 generations, 10 mL of cell culture was harvested. Genomic DNA extraction, labeling of the probes, hybridization to arrays, and analysis of the data was done as described (Decourty et al. 2008). Data are available in Supplemental Data S2.
RNA analyses
RNA extractions were done with a hot-phenol method as previously described (Holbein et al. 2008). Northern blot analysis was done with RNAs resolved either on 1.2% denaturing formaldeyde/agarose gels or with high-resolution 6% polyacrylamide/8.3 M urea gels (Dichtl et al. 2002b). ASC1, ACT1, ADH1, PGK1, and CYH2 mRNAs were detected with 32P random prime labeled PCR products (primer sequences are available upon request) corresponding to parts of the open reading frames. For CYH2, a PCR product was also used that encompassed intronic sequences; the CUT NEL025c was detected with a labeled PCR product amplified with forward GATTAACAGTTAGATCCAG and reverse GGACTTTCAGATCAGTCAGTG primers. 18S rRNA (ACGAAAAATCAAATAC), U3 snoRNA (AGGACATTTCTATAGGAATCG), and tRNALEU 3 (GAACTCTTGCATCTTACGATAGC) were detected with 5′ end labeled oligonucleotides. RNase H analyses (Dichtl et al. 2002b) and poly(A) tail labeling (Holbein et al. 2008) were done as previously described. Northern filters and dried gels were exposed to BAS-MS imaging plates (Fuji) and developed on a FLA-7000 PhosphorImager (Fuji).
ATP measurements
ATP quantification was done as described (Freimoser et al. 2006). One OD600 equivalent of yeast cells was pelleted, the supernatant was discarded, 50 μL of 1 M sulfuric acid were added, and the suspension was neutralized with 50 μL of 2 M NaOH and 100 μL of Tris-malate buffer (1 M, pH 7.5, 6% neutral red solution [0.1% neutral red in 70% ethanol]). Cell debris was pelleted by centrifugation, 20 μL of the supernatant was added to 80 μL of Tris buffer (20 mM at pH 8, 2 mM EDTA), and 4 μL of phosphoenol pyruvate were added (2.5 mM PEP at pH 8, 0.125 M MgSO4, 0.312 M K2SO4). Five microliters of this sample were added to 45 μL of luciferase buffer (10 mM Tris-H2SO4 at pH 7.4, 3.5 mM MgSO4). Following addition of 50 μL of luciferase solution (Roche ATP Bioluminescence Assay kit CLS II), relative light units emitted were measured in a luminometer (Lumat LB 9507; Berthold Technologies).
Microarray analyses
Wild-type S228C and pap1-1 were grown in SD medium at 25°C to OD600 0.3, and growth was continued for 1 h in the presence or absence of 20 μg/mL cordycepin. All subsequent steps will be described elsewhere (Halbeisen and Gerber 2009). Shortly, RNA was extracted from cell pellets using a hot-phenol method and further purification with the RNeasy Micro Kit (QIAGEN). Ten micrograms of total RNA were used for reverse transcription using superscript II (Invitrogen) with oligo(dT) and pdN9 primers and Ambion spike control. Following RNA hydrolysis and neutralization, DNA was concentrated to 9 μL using the Microcon-30 concentrator (Amicon). NHS-Cy3 and NHS-Cy5 dyes (Amersham) were coupled to the DNA and purified using the Qia-quick PCR purification kit (QIAGEN). The Cy3- and Cy5-labeled cDNA samples were mixed and concentrated in Microcon-30 concentrators (Amicon) to a final volume of 10.5 μL, and labeling efficiency was measured by NanoDrop using the microarray application.
The concentrated labeled DNA was mixed with MWG formamide buffer (Ocimun Biosolutions), and poly(A) RNA was added to a final concentration of 0.5 mg/mL. The mix was incubated for 10 min at 80°C, loaded on Nexterion slides (carrying 70-mer oligo probes for 10,944 features), and hybridized overnight at 42°C. Following washes in 2× SSC, 0.2% SDS and 2× SSC, and 0.2× SSC, the arrays were immersed in 100% ethanol to fix the cDNA, dried by centrifugation, and scanned with Axon instruments scanner 4000. Scanning parameters were adjusted to similar fluorescent intensities in both channels, and data were collected with GENEPIX 3.0 (Axon Instruments). Spots with abnormal morphology were identified by eye and excluded from further analysis. Arrays were normalized by the Stanford Microarray Database (SMD) (Gollub et al. 2003). The calculated log2 ratios were imported into Acuity 4.0 (Axon Instruments) and filtered for regression correlation of >0.5 (filters for large variations in the ratios of pixels within each spot) and a signal-to-noise ratio of >2.5 (signal over background). Average log2 ratios were calculated from the triple experiments for each gene spot. RNAs were considered as increased when the log2 ratio was >1 and decreased when log2 was <−1. Further analysis of the data was carried out with the Saccharomyces Genome Database GO term finder (http://www.yeastgenome.org/). Microarray data were deposited at GEO (http://www.ncbi.nlm.nih.gov/geo; GEO accession: GSE14619), and processed data are also available as Supplemental Data S3.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We are grateful to Georges Martin for the generous gift of recombinant Pap1 and Walter Keller and Scott Butler for the pap1 mutant strains. We thank Regula Halbeisen, André Gerber, and Stefan Zoller for assisting with microarray analyses; Marko Jovanovic for help with the heat-map; and André Halbach for discussion and comments on the manuscript. This work was supported by the University of Zürich, the Swiss National Science Foundation (grant PP00A-102941 to B.D.), and the EU (grant LSHG-CT-2005-518280 to B.D.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1458909.
REFERENCES
- Ahn Y.J., Park S.J., Lee S.G., Shin S.C., Choi D.H. Cordycepin: Selective growth inhibitor derived from liquid culture of Cordyceps militaris against Clostridium spp. J. Agric. Food Chem. 2000;48:2744–2748. doi: 10.1021/jf990862n. [DOI] [PubMed] [Google Scholar]
- Amberg D.C., Goldstein A.L., Cole C.N. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Amiott E.A., Jaehning J.A. Mitochondrial transcription is regulated via an ATP “sensing” mechanism that couples RNA abundance to respiration. Mol. Cell. 2006;22:329–338. doi: 10.1016/j.molcel.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Arigo J.T., Eyler D.E., Carroll K.L., Corden J.L. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Ashe M.P., De Long S.K., Sachs A.B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch J.C., Perrin F., Lacroute F. Yeast temperature-sensitive mutants specifically impaired in processing of poly(A)-containing RNAs. Mol. Gen. Genet. 1978;165:123–127. doi: 10.1007/BF00269900. [DOI] [PubMed] [Google Scholar]
- Boeck R., Tarun S., Jr, Rieger M., Deardorff J.A., Muller-Auer S., Sachs A.B. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- Brown C.E., Sachs A.B. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Tarun S.Z., Jr, Boeck R., Sachs A.B. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae . Mol. Cell. Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.T., Bai X., Johnson A.W. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K.G., Manson W., Spring F.S., Hutchinson S.A. Cordycepin, a metabolic product isolated from cultures of Cordyceps militaris (Linn.) Link. Nature. 1950;166:949. doi: 10.1038/166949a0. [DOI] [PubMed] [Google Scholar]
- Daugeron M.C., Mauxion F., Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decourty L., Saveanu C., Zemam K., Hantraye F., Frachon E., Rousselle J.C., Fromont-Racine M., Jacquier A. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl. Acad. Sci. 2008;105:5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmoucelles C., Pinson B., Saint-Marc C., Daignan-Fornier B. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem. 2002;277:27036–27044. doi: 10.1074/jbc.M111433200. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Blank D., Ohnacker M., Friedlein A., Roeder D., Langen H., Keller W. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell. 2002a;10:1139–1150. doi: 10.1016/s1097-2765(02)00707-4. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Blank D., Sadowski M., Hubner W., Weiser S., Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 2002b;21:4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T., Strasser K., Racz A., Rodriguez-Navarro S., Oppizzi M., Ihrig P., Lechner J., Hurt E. The mRNA export machinery requires the novel Sac3p–Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimoser F.M., Hurlimann H.C., Jakob C.A., Werner T.P., Amrhein N. Systematic screening of polyphosphate (poly P) levels in yeast mutant cells reveals strong interdependence with primary metabolism. Genome Biol. 2006;7:R109. doi: 10.1186/gb-2006-7-11-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garas M., Dichtl B., Keller W. The role of the putative 3′ end processing endonuclease Ysh1p in mRNA and snoRNA synthesis. RNA. 2008;14:2671–2684. doi: 10.1261/rna.1293008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollub J., Ball C.A., Binkley G., Demeter J., Finkelstein D.B., Hebert J.M., Hernandez-Boussard T., Jin H., Kaloper M., Matese J.C., et al. The Stanford Microarray Database: Data access and quality assessment tools. Nucleic Acids Res. 2003;31:94–96. doi: 10.1093/nar/gkg078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati R.K., Villa T., Boulay J., Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat. Struct. Mol. Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- Halbeisen R.E., Gerber A.P. Stress-dependent coordination of transcriptome and translatome in yeast. PLoS Biol. 2009 doi: 10.1371/journal.pbio.1000105. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M.E., Fung E., Wildenhain J., Pierce S.E., Hoon S., Lee W., Proctor M., St Onge R.P., Tyers M., Koller D., et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P., Parker R. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′ end formation of nascent transcripts. RNA. 2001;7:753–764. doi: 10.1017/s1355838201010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbein S., Freimoser F.M., Werner T.P., Wengi A., Dichtl B. Cordycepin-hypersensitive growth links elevated polyphosphate levels to inhibition of poly(A) polymerase in Saccharomyces cerevisiae . Nucleic Acids Res. 2008;36:353–363. doi: 10.1093/nar/gkm990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz B., Goldfinger B.A., Marmur J. Effect of cordycepin triphosphate on the nuclear DNA-dependent RNA polymerases and poly(A) polymerase from the yeast, Saccharomyces cerevisiae . Arch. Biochem. Biophys. 1976;172:143–148. doi: 10.1016/0003-9861(76)90059-x. [DOI] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Kiel J.A., Veenhuis M., van der Klei I.J. PEX genes in fungal genomes: Common, rare or redundant. Traffic. 2006;7:1291–1303. doi: 10.1111/j.1600-0854.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- Konrad M. Analysis and in vivo disruption of the gene coding for adenylate kinase (ADK1) in the yeast Saccharomyces cerevisiae . J. Biol. Chem. 1988;263:19468–19474. [PubMed] [Google Scholar]
- Kyburz A., Sadowski M., Dichtl B., Keller W. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′ end formation. Nucleic Acids Res. 2003;31:3936–3945. doi: 10.1093/nar/gkg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq K., Belloc I., Desgranges C., Daignan-Fornier B. Role of adenosine kinase in Saccharomyces cerevisiae: Identification of the ADO1 gene and study of the mutant phenotypes. Yeast. 2001;18:335–342. doi: 10.1002/1097-0061(20010315)18:4<335::AID-YEA674>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S., Jensen T.H. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie. 2007;89:1177–1182. doi: 10.1016/j.biochi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Mandart E., Parker R. Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol. Cell. Biol. 1995;15:6979–6986. doi: 10.1128/mcb.15.12.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser P., Sutterlin C., Kralli A., Kaminsky R. A nucleoside transporter from Trypanosoma brucei involved in drug resistance. Science. 1999;285:242–244. doi: 10.1126/science.285.5425.242. [DOI] [PubMed] [Google Scholar]
- Meyer S., Temme C., Wahle E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker P.J., Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′ end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- Mortimer R.K., Johnston J.R. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Decker C.J., Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ → 3′ digestion of the transcript. Genes & Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muller W.E., Seibert G., Beyer R., Breter H.J., Maidhof A., Zahn R.K. Effect of cordycepin on nucleic acid metabolism in L5178Y cells and on nucleic acid-synthesizing enzyme systems. Cancer Res. 1977;37:3824–3833. [PubMed] [Google Scholar]
- Naula N., Hilti N., Schweingruber A.M., Schweingruber M.E. Cordycepin in Schizosaccharomyces pombe: Effects on the wild type and phenotypes of mutants resistant to the drug. Curr. Genet. 2003;43:400–406. doi: 10.1007/s00294-003-0413-4. [DOI] [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eukaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S.L., Pan X., Peyser B.D., Ye P., Meluh P.B., Yuan D.S., Irizarry R.A., Bader J.S., Spencer F.A., Boeke J.D. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 2006;22:56–63. doi: 10.1016/j.tig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Parsons A.B., Lopez A., Givoni I.E., Williams D.E., Gray C.A., Porter J., Chua G., Sopko R., Brost R.L., Ho C.H., et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Patel D., Butler J.S. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol. Cell. Biol. 1992;12:3297–3304. doi: 10.1128/mcb.12.7.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson R.R. Cordyceps: A traditional Chinese medicine and another fungal therapeutic biofactory? Phytochemistry. 2008;69:1469–1495. doi: 10.1016/j.phytochem.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg M.E., Masocha W., Ferella M., Petitto-Assis F., Goto H., Kristensson K., McCaffrey R., Wigzell H. Treatment of African trypanosomiasis with cordycepin and adenosine deaminase inhibitors in a mouse model. J. Infect. Dis. 2005;192:1658–1665. doi: 10.1086/496896. [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Collins S.R., Thompson N.J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J.F., et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Segre D., Deluna A., Church G.M., Kishony R. Modular epistasis in yeast metabolism. Nat. Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- Stack J.H., Herman P.K., Schu P.V., Emr S.D. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugar A.M., McCaffrey R.P. Antifungal activity of 3′-deoxyadenosine (cordycepin) Antimicrob. Agents Chemother. 1998;42:1424–1427. doi: 10.1128/aac.42.6.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut M., Kisseleva-Romanova E., Rougemaille M., Boulay J., Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: A role for the Nrd1–Nab3 pathway in genome surveillance. Mol. Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Tu J., Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae . EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M., Staples R.R., Valencia-Sanchez M.A., Muhlrad D., Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae . EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valay J.G., Simon M., Dubois M.F., Bensaude O., Facca C., Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J. Mol. Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L., Kim M., Mutschler H., Buratowski S., Meinhart A. The Nrd1–Nab3–Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders R.J., Waterham H.R. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wyers F., Rougemaille M., Badis G., Rousselle J.C., Dufour M.E., Boulay J., Regnault B., Devaux F., Namane A., Seraphin B., et al. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Zhu J.S., Halpern G.M., Jones K. The scientific rediscovery of a precious ancient Chinese herbal regimen: Cordyceps sinensis: Part II. J. Altern. Complement. Med. 1998a;4:429–457. doi: 10.1089/acm.1998.4.429. [DOI] [PubMed] [Google Scholar]
- Zhu J.S., Halpern G.M., Jones K. The scientific rediscovery of an ancient Chinese herbal medicine: Cordyceps sinensis: part I. J. Altern. Complement. Med. 1998b;4:289–303. doi: 10.1089/acm.1998.4.3-289. [DOI] [PubMed] [Google Scholar]