Abstract
A novel ribozyme that accelerates the ligation of β-nicotinamide mononucleotide (β-NMN)-activated RNA fragments was isolated and characterized. This artificial ligase ribozyme (YFL ribozyme) was isolated by a “design and selection” strategy, in which a modular catalytic unit was generated on a rationally designed modular scaffold RNA. Biochemical analyses of the YFL ribozyme revealed that it catalyzes RNA ligation in a template-dependent manner, and its activity is highly dependent on its architecture, which consists of a modular scaffold and a catalytic unit. As the design and selection strategy was used for generation of DSL ribozyme, isolation of the YFL ribozyme indicated the versatility of this strategy for generation of functional RNAs with modular architectures. The catalytic unit of the YFL ribozyme accepts not only β-NMN but also inorganic pyrophosphate and adenosine monophosphate as leaving groups for RNA ligation. This versatility of the YFL ribozyme provides novel insight into the possible roles of β-NMN (or NADH) in the RNA world.
Keywords: RNA, ribozyme, in vitro selection, modular design, RNA motif
INTRODUCTION
Ribonucleic acid (RNA) is one of the most important biopolymers in extant biosystems as well as in the evolutionary origins of life (Cech 1993; Eddy 2001; Dworkin et al. 2003; Makarova and Kramerov 2007). Among the non-protein-coding RNAs that play a variety of roles in the cell, those with specific functions, such as catalysis (RNA enzymes or ribozymes) and molecular recognition (RNA antibodies or aptamers), have attracted a great deal of attention as promising tools in regulating or sensing higher-order biosystems, including gene circuits and signal transduction cascades (Isaacs et al. 2006). Several “riboregulators” and “ribosensors” have been developed by employing known ribozymes and aptamers (Yen et al. 2004; Win and Smolke 2007).
The tailor-made generation of novel ribozymes and aptamers with desired structures and functions is a major strategy to expand the utility of these functional RNAs (Saito and Inoue 2007). Since 1990, in vitro selection with long random RNA libraries has been employed widely to isolate novel functional RNAs (Chen et al. 2007; Joyce 2007; Stoltenburg et al. 2007). While in vitro selection is extremely powerful, it still has a number of limitations, for example, in elucidation of the structures of selected RNAs; as no structural constraints are programmed into the starting RNA library, elucidation of their secondary and tertiary structures is as laborious and time consuming as that of natural RNAs. Rational RNA design, in which all structural information is programmable, is an attractive alternative to avoid this laborious structural analysis (Westhof et al. 1996). Such rational design of structural RNA has become possible due to the remarkable progress in RNA structural biology and computational RNA modeling (Jaeger et al. 2001; Ikawa et al. 2002; Chworos et al. 2004; Yingling and Shapiro 2007). However, designing modular units responsible for sophisticated functions, such as catalysis and molecular recognition, is still a challenging task.
To combine the advantages of the two strategies and compensate for their shortcomings, a hybrid strategy designated “design and selection” has been proposed (Ikawa et al. 2004). In this strategy, a self-folding RNA is rationally designed as a structural scaffold of functional RNAs, and a novel functional unit, the structure of which cannot be designed, is then installed on the scaffold RNA by in vitro selection. The resulting RNAs should exhibit their functions depending on their modular structures, which would facilitate structural analyses and structure-based engineering. Initial experiments applying this hybrid strategy successfully generated the DSL (designed and selected ligase) ribozyme (Ikawa et al. 2004).
The second step to establish the design and selection strategy is to generate additional ribozymes. Therefore, we applied this strategy to isolate novel ribozymes capable of ligating RNA substrates activated by β-nicotinamide mononucleotide (β-NMN). β-Nicotinamide is considered a molecular fossil of the hypothetical “RNA World.” Recreation of the RNA World is an important issue in research regarding the origin of life and early evolution (Szostak et al. 2001; Joyce 2002; Chen et al. 2005; Joyce and Orgel 2006; Müller 2006), and a number of artificial ribozymes that are highly relevant to the RNA World hypothesis have been isolated (Lohse and Szostak 1996; Jenne and Famulok 1998; Sengle et al. 2001; Tsukiji et al. 2003; Fusz et al. 2005). As generation of β-NMN-dependent ligase ribozymes is attractive from both technological and scientific viewpoints, we have isolated a β-NMN-dependent ligase ribozyme designated “YFL.” Biochemical analysis indicated that the YFL ribozyme utilizes both β-NMN and inorganic pyrophosphate (ipp) as leaving groups for the ligation reaction. As greater chemical versatility has been suggested to be an important characteristic of ribozymes at the early stages of the RNA World (Breaker and Joyce 1995), the YFL ribozyme may provide new insight into the evolution of RNA ligation/polymerization systems in the RNA World (Ellington et al. 2009).
RESULTS
Design and preparation of RNA library
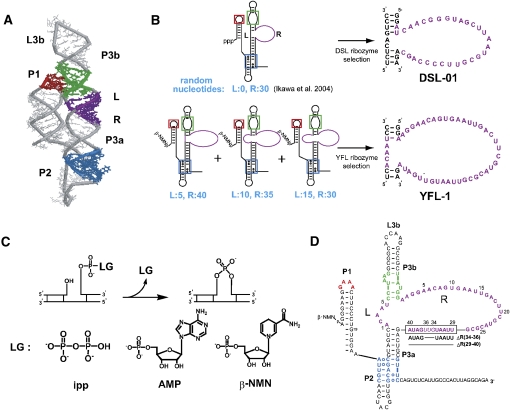
A highly structured library has been designed and constructed based on the designed type-B self-folding RNA (Fig. 1A; Ikawa et al. 2002), and the class DSL ribozyme was isolated from this library (Ikawa et al. 2004). The catalytic module of the DSL ribozyme was isolated from 30 random nucleotides inserted in the particular region (R region) of the type-B RNA (Fig. 1B). As the insertion region was rationally determined by three-dimensional (3D) molecular modeling, isolation of the DSL ribozyme confirmed the adequacy of rational design of the structured library.
FIGURE 1.
Construction scheme, structure, and reaction of the class YFL and related ribozymes. (A) 3D structure of the scaffold RNA. Colored regions correspond to those in the secondary structures shown in B and D. (B) Secondary structures of the RNA libraries and isolated sequences. For construction of the three sublibraries for YFL ribozyme selection, 5, 10, or 15 and 40, 35, or 30 random nucleotides were introduced into the L and R regions, respectively, which originally form five base pairs. (C) Leaving groups of the ligation reaction by ligase ribozymes. (Left) Inorganic pyrophosphate (ipp) used by several ligase ribozymes including the DSL. (Middle) Adenosine monophosphate (AMP) used by three ligase ribozymes isolated by Hager and Szostak (1997). (Right) β-Nicotinamide mononucleotide (β-NMN) used by the YFL-1 ribozyme. (D) Secondary structure of the YFL-1 ribozyme. Colored regions indicate structural motifs, (red) GAAA loop, (green) receptor motif for GAAA loop, and (blue) triple-helical scaffold, used in the molecular design of the scaffold. ΔR(35–37) and ΔR(29–40) indicate truncated sequences in analysis of the catalytic unit.
In this study, we modified the original library to improve both sequence and structural diversity. The new library consists of three sublibraries with a total of 45 random nucleotides allotted to the two regions (L and R regions in Fig. 1B). Ligase ribozymes using inorganic pyrophosphate (ipp) (Fig. 1C, left) or adenosine monophosphate (AMP) (Fig. 1C, middle) as a leaving group have been reported (Hager and Szostak 1997; McGinness and Joyce 2002; Joyce 2007). To isolate the second catalytic modules for the type-B RNA, we used β-NMN as a new leaving group for RNA ligation (Fig. 1C, right). The reduced form of β-NMN was appended to the 5′-ends of the library RNAs by transcription with an 80-fold excess of NADH (8 mM) over ATP (0.1 mM). In the presence of a trace amount of [γ-32P]ATP, parallel transcription with or without NADH indicated that >90% of RNA had β-NMN at the 5′-end as determined from incorporation of 32P into γ-phosphate (Huang 2003). Note that the incorporation efficiency of β-NMN was also supported by the observation that the DSL-01 ribozyme transcribed under these conditions showed no detectable activity (see Fig. 5D).
FIGURE 5.
Effects of helical contexts in the P1 region and leaving groups on the reaction of the YFL-1 ribozyme. (A) Structures and sequences of the 5′-end of the (upper left) parental YFL-1 and mutants. These P1s possess either (upper) the original A1–U18 pair or (lower) the G1–C18 pair. (B) Activities of the YFL-1 variants with the A1–U18 pair and different leaving groups: (filled circles) reduced form of β-NMN, (filled squares) oxidized form of β-NMN, (triangles) inorganic pyrophosphate, and (filled triangles) AMP. (C) Activities of the YFL-1 variants with the G1–C18 pair and different leaving groups: (filled circles) oxidized form of β-NMN and (open circles) inorganic pyrophosphate. (D) Activities of the DSL ribozyme with different leaving groups: (filled circles) inorganic pyrophosphate and (filled squares) the reduced form of β-NMN.
Three sublibraries, each of which had 600 pmol of RNA with 4.3, 6.0, and 3.0 × 1013 different sequences, respectively, were mixed together to yield the starting RNA library with 1800 pmol of RNA with 1.3 × 1014 different sequences.
Isolation of the class YFL ligase ribozyme
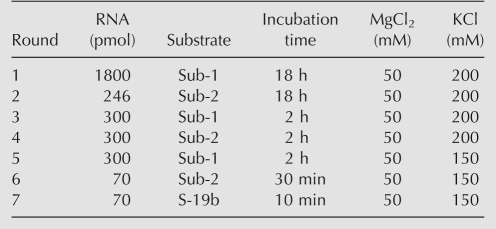
Seven rounds of in vitro selection were carried out. Two sets of substrate RNA and the corresponding P1 region, which were employed for isolation of the DSL ribozyme (Ikawa et al. 2004), were alternatively employed during the first six selection cycles to isolate catalytic modules joining the two RNA termini without base specificity (Table 1). As the seventh-round RNA pool showed detectable activity, ligated products were cloned and sequenced. Thirty-eight randomly picked clones had the correct ligation junctions, and all of their L and R regions were 5 and 40 nucleotides (nt) in length, respectively (Fig. 2A).
TABLE 1.
Conditions of the initial in vitro selection
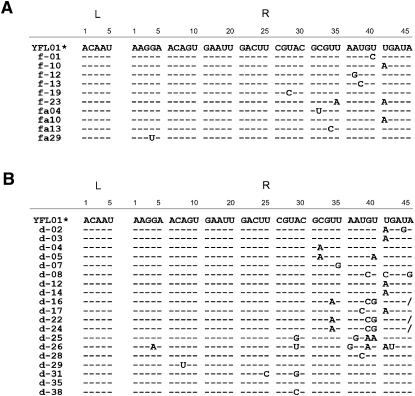
FIGURE 2.
Sequence comparison of the YFL-1 and its variants isolated from the first and second selections. (A) Sequences of 38 clones from the first selection. (Asterisks) The YFL-1 sequences were found in 28 independent clones. (B) Sequences of 26 clones from the second selection. (Asterisks) The YFL-1 sequences were found in seven independent clones. (Dashes) The bases that are the same as in YFL-1. Slashes in position R40 of clones d-16, 22, and 24 indicate single-nucleotide deletions.
The L and R regions of 28 clones had an identical sequence, and the remaining 10 clones were variants (Fig. 2A). Therefore, the isolated sequences constitute one sequence family designated the class YFL ribozyme. The activity of the most abundant sequence (YFL-1) (Fig. 1B,D) was examined in the reaction buffer used in the selection (30 mM Tris-HCl at pH 7.5, 50 mM MgCl2, 200 mM KCl). With 1 μM ribozyme and 1 μM fluorescently labeled substrate, ligated product was detectable after 5 min of incubation at 37°C (Fig. 3A). Ligation reaction was detectable in a control experiment using an isolated P1 region without the catalytic unit, but product yield was <0.04% after 96-h reaction (Supplemental Fig. S1). The apparent second-order rate constant (kapp) and the product yield after 18-h reaction were 0.20 (h−1 μM−1) and 33%, respectively (Fig. 3B; see also Supplemental Fig. S2). Under conditions of substrate excess (1 μM ribozyme, 4 μM substrate), the pseudo-first-order rate constant (k obs) and the product yield after 18-h reaction were 0.14 (h−1) and 40%, respectively (Fig. 3B).
FIGURE 3.
Activity of the YFL-1 ribozyme primed by the reduced form of β-NMN. (A) Time course of the reaction between 1 μM YFL-1 ribozyme and 1 μM 5′-FAM-labeled substrate. The reaction was performed with 30 mM Tris-HCl (pH 7.5), 50 mM MgCl2, and 200 mM KCl at 37°C. (B) Activity of the YFL-1 ribozyme. Reactions were carried out with (filled circles) equivalent amounts of ribozyme and substrate or (open squares) an excess of substrate. (C) Reactions of YFL-1 with modified substrates. The modified substrate had 2′-methoxycytidine, 2′-deoxycytidine, or 3′-deoxycytidine at the 3′-terminus. Reactions were carried out for 6 h.
Catalytic module of the YFL ribozyme
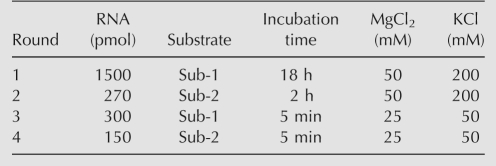
To gain insight into the catalytic module of the YFL ribozyme, a second selection with a degenerate library was carried out (Ekland and Bartel 1995). In the second library derived from the YFL-1 sequence, the L region (5 bases) was randomized, whereas the R region (40 bases) was partially mutagenized with 21% degeneracy. After four rounds of selection (Table 2), 26 clones were isolated and sequenced (Fig. 2B). In the sequence comparison of the clones from the first and second selections, the L region was invariable, and the positions 1–28 of the R region were highly conserved. However, the positions 29–40 of the R region showed considerable variation; especially, G34, U35, and U36 constituted a mutational hotspot. Several clones were assayed for catalytic activity, but none showed distinctly higher activity than the YFL-1 ribozyme (Supplemental Fig. S3).
TABLE 2.
Conditions of in vitro selection with the doped YFL-1 library
As in vitro selected ligase ribozymes often form nonnative 2′–5′ linkages (Ekland et al. 1995; Landweber and Pokrovskaya 1999; Rogers and Joyce 1999), the regioselectivity of the YFL-1 ribozyme was examined using substrates with modified 3′-termini (Fig. 3C; Hager and Szostak 1997). The YFL-1 ribozyme exhibited modest or detectable activity with substrate having a 2′-methoxyribose or a 2′-deoxyribose at the 3′-end, respectively. However, no product was observed in a reaction with a substrate with a 3′-deoxy terminus. These observations strongly suggest that the YFL-1 ribozyme joins RNA with a 3′–5′ linkage. These results also suggested the importance of the oxygen atom at the ribose C2′ position of the 3′-terminus of the substrate in formation of the 3′–5′ linkage.
Metal ion dependence of the YFL ribozyme
Metal cations play diverse roles in folding and/or reaction processes in almost all ribozymes (Pan et al. 1994; Suga et al. 1998; Fedor 2002; DeRose 2003). The YFL-1 ribozyme was evolved in the presence of 50 mM Mg2+ and 200 mM K+. The YFL-1 ribozyme was found to be active without K+ but showed no detectable activity without Mg2+ (Fig. 4A). The metal ion specificity of the YFL-1 ribozyme was then investigated in the presence of different metal ions. Among eight divalent cations and one trivalent cation examined, only Mg2+ supported the ligation reaction (Fig. 4B).
FIGURE 4.
Metal ion dependence of the activity of the YFL-1. (A) Time courses of the ligation reaction by YFL-1 (filled circles) with 200 mM K+ and 50 mM Mg2+, (filled squares) with 50 mM Mg2+ and no K+, and (open circles) with 200 mM K+ and no Mg2+. (B) Reactions of YFL-1 with different divalent metal ions and Co(NH3)3+. Reactions were carried out for 2 h in the presence of 50 mM divalent metal ion or Co(NH3)3+. (C) Time courses of the ligation reaction of YFL-1 with (open circles) 10 mM, (filled squares) 25 mM, (filled circles) 50 mM, (open squares) 100 mM, and (open triangles) 150 mM Mg2+ ions and 200 mM KCl.
To further investigate the effects of Mg2+ ions, we examined the ligation reaction in the presence of 10, 25, 50, 100, and 150 mM Mg2+. The final yield of ribozyme reaction increased with increasing Mg2+ concentration. Reactivity increased markedly between 10 and 50 mM Mg2+ (Fig. 4C), indicating that YFL-1 requires ≥50 mM Mg2+ for stable folding and/or to coordinate catalytic metal ions.
Effects of leaving group
The class YFL ribozyme was isolated from the pool of RNA with the reduced form of β-NMN (β-NMN-red) at the 5′-end. To examine the importance of the chemical structure of β-NMN-red in the catalytic mechanism of the ribozyme, β-NMN-red was replaced with its oxidized form (β-NMN-ox), inorganic pyrophosphate (ipp), or adenosine monophosphate (AMP) (Fig. 5A, top). The kapp values of the β-NMN-ox, ipp, and AMP variants were 0.16 h−1 μM−1, 0.052 h−1 μM−1, and 0.035 h−1 μM−1, respectively (Fig. 5B). A control experiment using an isolated P1 region showed that the P1 region RNA with an ipp leaving group was several-fold more reactive than that with a β-NMN-red leaving group (Supplemental Fig. S1). The calculated final yields of the ligation products of the β-NMN-ox variant (49%) and the ipp variant (59%) were similar to that of the YFL-1 ribozyme (44%). On the other hand, the calculated final product yield of the AMP variant was 24% (see also Fig. 5B). We also examined the effects of the leaving group in the reaction of DSL-01. Its catalytic unit was evolved under selection conditions to use the ipp leaving group (Fig. 1B). The k app value of DSL-01 with the ipp leaving group (0.16 h−1 μM−1) was similar to that of YFL-1 with the β-NMN-red leaving group (0.20 h−1 μM−1) (Fig. 5D). However, the DSL-01 with the β-NMN-red leaving group showed no detectable activity (Fig. 5D). These results indicated distinct properties of catalytic units between class YFL and DSL ribozymes (Fig. 5D).
Base specificity in the ribozyme-catalyzed ligation
Template-directed RNA ligation is the key reaction in the hypothetical RNA World (Joyce 1987; McGinness and Joyce 2003). However, only a few ribozymes have been shown to catalyze this reaction because ligase ribozymes evolved in vitro often require particular nucleotides at the reaction site (McGinness and Joyce 2003; Müller 2006).
Nucleobase preference at the 5′-end of the ribozyme was examined using the β-NMN-ox and ipp variants. The original A1–U18 pair of the P1 region was replaced with a G–C pair (Fig. 5A, bottom). Two variants still showed substantial activities, and the k app values for the β-NMN-ox and ipp variants were 0.045 h−1 μM−1 and 0.032 h−1 μM−1, respectively. On the other hand, the product yield of the ipp variant after 24-h reaction (19%) was less than that of the β-NMN-ox variant (39%) (Fig. 5C).
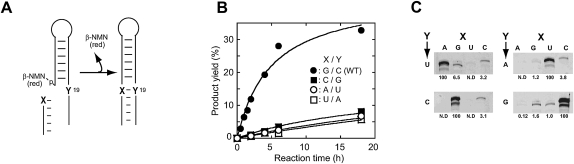
To determine the preference at the 3′-end of the substrate, four substrates with different bases at the 3′-termini (indicated as “X” in Fig. 6A) and the corresponding single mutations were also introduced at position 19 of the ribozyme (indicated as “Y” in Fig. 6A). All four combinations of variant ribozymes and substrates were active, although the original combination was the most active with regard to both reaction rate and product yield (Fig. 6B). Further experiments with all possible ribozymes and substrate combinations indicated that Watson–Crick base-pairing between the 3′-end of the substrate and position 19 of the ribozyme is required for ligation (Fig. 6C). Taken together, these data demonstrated that the YFL-1 ribozyme catalyzes RNA–RNA ligation in a template-directed manner but shows a preference for the original substrate–P1 combination used in its generation.
FIGURE 6.
(A) Ligation reaction by the YFL-1 variants with different nucleotide combinations between substrate 3′-end (position 1) and position 19. (X) The nucleotide at the 3′-end of the substrate (position-1); (Y) the nucleotide at position 19. (B) Activities of the YFL-1 variants with complementary nucleotides between positions −1 and 19: (filled circles) G–C, (filled squares) C–G, (open circles) A–U, and (open squares) U–A. (C) Relative activities of possible nucleotide combinations between positions −1 and 19. Reactions were carried out for 6 h.
Structural organization of the YFL ribozyme
The class YFL ribozyme was constructed by installing a catalytic unit into the designed self-folding RNA, the structure of which is rigidly fixed by two tertiary interactions (Ikawa et al. 2002). Therefore, the two interactions would also serve as modular units to establish a structural scaffold that would enable the catalytic unit to interact physically with the ligation site.
To determine whether the scaffold structure was maintained even after evolution of the catalytic unit, derivatives lacking one of the tertiary interactions were constructed and tested (Fig. 7A). When the interaction between P1 and P3b was disrupted by replacing the P1 GAAA loop with a GUGA loop (Costa and Michel 1997; Ikawa et al. 1999), the resulting variant (P1 GUGA) showed little activity (Fig. 7B). The ΔP2 variant lacking the triple helical scaffold, the motif rigidly connecting the three helices, also showed marked reduction of the activity (Fig. 7B). Therefore, the tertiary structure of the scaffold supports the ribozyme activity.
FIGURE 7.
Structural requirements of the YFL-1 ribozyme. (A) Mutations at the (upper) P2 region and (lower) the loop–receptor interactions between P1 and P3. Mutations were introduced in the P2 region, (red) GAAA loop in P1, and/or (green) receptor motif in P3 in Figure 1D. (B) Activities of the YFL-1 variants with mutations in the P1 loop or P2 region. (C) Activities of the YFL-1 variants with mutations in the P2 region shown in Figure 6A, upper. (D) Activities of the YFL-1 variants with mutations in loop–receptor motifs shown in Figure 6A, lower.
The importance of the two tertiary interactions in the YFL ribozyme was further confirmed by compensatory mutations (Fig. 7A). Base substitutions at the triple helical scaffold resulted in more severe reduction of activity than ΔP2 mutation (Fig. 7C). However, the compensatory mutations restoring base-pairing resulted in recovery of the activity (Fig. 7C). The interaction between GAAA at P1 and its receptor at P3b was also replaced with another loop–receptor combination. The mutants with the GGAA loop at P1 or R(GGAA) receptor at P3b were almost inactive because of disruption of P1–P3b interaction. However, a double mutant with the GGAA loop and its specific receptor [R(GGAA)] showed significant restoration of the activity (Fig. 7D; Geary et al. 2008). The class DSL ribozyme also possesses modular organization identical to that of the class YFL ribozyme (Fig. 1). However, the activity of the DSL-01 ribozyme was markedly reduced when its GAAA–receptor interaction between P1 and P3b was replaced with different GNRA–receptor sets (Ikawa et al. 2004). Thus, it remains doubtful whether the GNRA–receptor interaction between P1 and P3b can be manipulated as a modular unit in the class DSL ribozyme. In contrast to the DSL ribozyme, the YFL ribozyme is the first ribozyme with clear structural modularity in the GAAA–receptor unit as it is replaceable as a different set of modular units without severe loss of activity (Fig. 7D).
Miniaturization of the YFL ribozyme
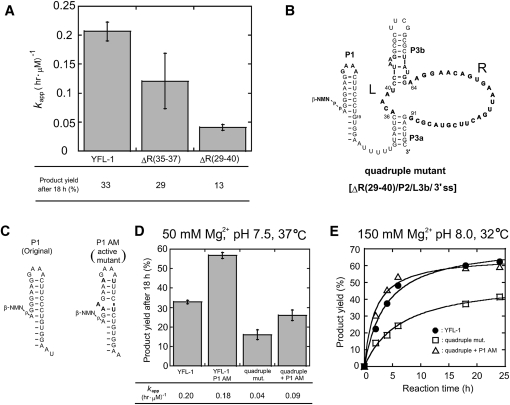
The results of comparative analysis revealed the conserved and variable regions in the catalytic unit of the YFL-1 ribozyme. As the variable region is expected to be nonessential, it was truncated (Fig. 8A). The positions R(34–36) represented a mutational hotspot. Positions R(29–40) were a moderately variable region including the mutational hot spot R(34–36) (Fig. 2). The two truncated mutants, ΔR(34–36) and ΔR(29–40), both retained distinct activities, although they were less active than the full-length YFL-1 (Fig. 8B).
FIGURE 8.
Truncation of the YFL-1 ribozyme. (A) Effects of truncation of the variable region in the catalytic units. ΔR(35–37) and ΔR(29–40) indicate truncated variants, the sequences of which are shown in Figure 1D. (B) Secondary structure of the quadruple mutant with truncations at the tip of P3b, the single-stranded region at the 3′-terminus, P2, and positions R29–40 of the catalytic unit. (C) Secondary structures of (left) the original P1 and (right) its AM variant. (D) Activities of YFL-1, the quadruple mutant, the P1 AM variant, and the double mutant. Reactions were performed with 30 mM Tris-HCl (pH 7.5), 50 mM Mg2+, 200 mM K+ at 37°C. (E) Time courses of the activities of (filled circles) the YFL-1, (open squares) quadruple mutant, and (open triangles) the double mutant derived from the quadruple mutant and the P1 AM variant. Reactions were performed with 30 mM Tris-HCl (pH 8.0), 150 mM Mg2+, 200 mM K+ at 32°C.
Some transfer RNAs and hammerhead ribozymes fold much more rapidly and uniformly than large RNAs, including the group I intron and RNaseP RNAs (Schroeder et al. 2004). As the folding pathways of such smaller RNAs are much simpler than those of larger RNAs, they may escape the kinetic traps caused by misfolded or partially folded structures often observed in large RNAs (Thirumalai and Woodson 2000; Woodson 2000; Treiber and Williamson 2001; Misra et al. 2003). Therefore, miniaturizing given RNA structures is a useful way to make artificial progenitors to generate advanced variants with improved functions (Green and Szostak 1992). Based on a double mutant of ΔP2 and ΔR(29–40), we further truncated the tip of P3b and the single-stranded region at the 3′-terminus. The resulting quadruple mutant retained distinct activity. Under the standard assay conditions, the ligation rate (k app) and product yield (18 h) were both 4.6-fold and 2.0-fold smaller than those of the parental YFL-1 ribozyme, respectively (Fig. 8D).
By screening reaction conditions (Supplemental Table S1), we found conditions (150 mM Mg2+ at pH 8.0, 32°C) under which final product yields of the YFL-1 and the quadruple mutant were improved to 73% and 52%, respectively (Fig. 8E). Under these conditions, the k app value of the mutant (0.15 h−1 μM−1) was also close to that of the parental ribozyme (0.26 h−1 μM−1) (Supplemental Table S2).
As a possible mutation that would further improve the quadruple mutant, we introduced a set of mutations identified by Voytek and Joyce designated P1 active mutations and abbreviated as “P1-AM” (Voytek and Joyce 2007). P1-AM was identified during the directed evolution of the DSL ribozyme. P1-AM derivatives of the YFL-1 and quadruple mutant both showed approximately twofold increases in product yield (18 h), while the degree of improvement in the k app value was less marked (Fig. 8C,D). Under the reaction conditions that gave improved product yield (150 mM Mg2+ at pH 8.0, 32°C), introduction of P1-AM significantly improved product yield (18 h) of the ribozyme from 37% to 59% (Fig. 8E).
DISCUSSION
β-NMN-dependent ligase ribozymes in the “RNA World”
Ligase ribozymes are the most promising progenitors of replicase ribozymes, which are central to the hypothetical RNA world (McGinness and Joyce 2003; Müller 2006). Among artificial ligases and their derivatives generated by in vitro selection, an evolved form of the class I ligase ribozyme polymerized up to 20 nt in a template-directed manner (Zaher and Unrau 2007; Müller and Bartel 2008).
To further improve ribozyme-catalyzed RNA ligation and polymerization, recognition and activation of the substrate nucleotide is a crucial issue. However, the chemical structure of RNA predicts that recognition of anionic moieties, such as triphosphate in nucleoside triphosphate (NTP), would be more challenging than that of cationic or neutral moieties (Sassanfar and Szostak 1993; Sazani et al. 2004). Therefore, pyrophosphate in NTP may not be the most suitable leaving group for RNA-based RNA ligation or polymerization. The chemical nature of catalytic RNA may result in a preference for a leaving group with a “handle” moiety that can yield strong and specific recognition. This is supported by the observation that natural group I intron ribozymes use guanine as the handle of the leaving group. The guanine base moiety is recognized by the conserved binding pocket in the catalytic core of the group I introns (Cech 1990; Stahley and Strobel 2006; Vicens and Cech 2006). The group I introns are readily converted to ligase ribozymes that perform template-directed primer extension with GpN substrate, although the group I intron ligase failed to show primer extension with GpG (Bartel et al. 1991).
To generate new ligase ribozymes, we used the substrate RNA modestly activated by 5′–5′-pyrophosphate linkage. The phosphodiester bond in nucleoside-5′–5′-pyrophosphate is chemically less reactive than that in triphosphate (Bartel and Szostak 1993; Hager and Szostak 1997). However, the linkage can form readily under prebiotic conditions and can also be used as the reactive intermediate in ligation by the protein enzymes, DNA ligase and RNA ligase (White 1976, 1982; Orgel 1986). In this study, β-nicotinamide riboside was employed as a handle moiety in the nucleoside monophosphate leaving group. β-Nicotinamide riboside and its derivatives play key roles in redox and other processes in extant biosystems and have also been shown to be produced under prebiotic conditions (White 1976; Cleaves and Miller 2001). Therefore, in the RNA World, β-nicotinamide riboside or its monophosphate (β-NMN) may have played multiple roles as a cofactor of redox reactions and an activator of nucleotides. This model was partly supported by the isolation of RNA aptamers recognizing NAD(H) (Burgstaller and Famulok 1994; Lauhon and Szostak 1995) and NAD(H)-dependent ribozymes catalyzing redox conversion of alcohol and aldehyde (Tsukiji et al. 2003). The results of the present study provided additional evidence to support the multifunctionality of β-nicotinamide riboside in the RNA World.
A strategy merging in vitro selection and rational design
Using β-NMN as a leaving group, we have isolated new ligase ribozymes designated YFL. The YFL ribozyme shares its structural scaffold with the DSL ribozyme that joins two RNA fragments using the pyrophosphate leaving group. Before construction of the DSL ribozyme, a natural RNA structure extracted from the Tetrahymena intron was employed as a scaffold for an artificial ribozyme (Jaeger et al. 1999). The resulting class hc ligase ribozyme (302 nt) shares a large structural domain P4–P6 (160 nt) with the Tetrahymena group I ribozyme (414 nt). While it is not clear whether the regions outside the P4–P6 scaffold also have similar modularity in the class hc and Tetrahymena ribozymes, the YFL and DSL ribozymes share a common modular organization composed of the shared scaffold (Fig. 1A) and unique catalytic modules (Fig. 1). This observation strongly suggests that the design and selection strategy could be widely applicable to construction of a variety of functional RNAs with modular architectures.
In the DSL-01 and YFL-1 ribozyme selection experiments, libraries with relatively short random nucleotide sequences (30 and 45 random nucleotides, respectively) were employed, and the two ribozymes were isolated after relatively few rounds of in vitro selection (five and seven, respectively). These features were regarded as advantages of the design and selection strategy (Voytek and Joyce 2007). Based on these advantages, further evolution experiments were performed, which yielded an advanced form of the class DSL ribozyme (Voytek and Joyce 2007).
Evaluation of the YFL-1 ribozyme
Although the YFL ribozyme was evolved to catalyze RNA–RNA ligation using the 5′-terminal phosphate activated by β-NMN (5′-XppRNA, where X and p are β-nicotinamide riboside and phosphate, respectively), subsequent analyses revealed that the YFL ribozyme also utilizes ipp and AMP and that the identity of leaving groups was not a factor determining the k app values of the ribozyme. These observations suggest that the catalytic unit of YFL-1 accepts the β-NMN leaving group by providing a space for it, and this space should also accept ipp and AMP leaving groups. However, AMP should cause steric and/or electrostatic conflicts, resulting in inactive structure(s) (Fig. 5B,C). Interestingly, primer extension activity of the group I intron ligase was improved by using a 2-aminopurine (2-AP) handle through mutation of the guanine binding pocket to recognize 2-AP (Bartel et al. 1991). These observations suggest that non-Watson–Crick-type nucleobases, such as β-nicotinamide and 2-aminopurine, were also used in primordial ribozymes in the RNA World.
Under single-turnover conditions, k obs for ligation reaction of YFL-1 was 0.14 h−1, which is an order of magnitude smaller than the typical values of well-studied ligase ribozymes utilizing the pyrophosphate leaving group. Interestingly, the k obs value is similar to those of three ligase ribozymes utilizing AMP as a leaving group for RNA–RNA ligation, reaction rates of which were reported to be 0.38, 0.046, and 0.012 h−1 (Hager and Szostak 1997).
These three ribozymes were isolated from an RNA library with a modular unit. In the starting RNA library used in this selection experiment, an AMP aptamer sequence was inserted in the middle of long random sequences (Hager and Szostak 1997) as a possible recognition site for the AMP leaving group, and the random sequences provided other elements needed for the ligase ribozymes. The three ligases all showed strict requirements for AMP and were hardly active with pyrophosphate, suggesting that the designed library provided substantial advantage to sequences selectively recognizing the AMP leaving group. In contrast, the starting library for the YFL ribozyme selection was programmed with no advantage for strong recognition of the β-NMN leaving group. Although further experiments are required, the differences in modular design of the two RNA libraries may have been a determinant of the ribozymes exhibiting similar reaction rates but distinct selectivities against leaving group structures. Although the degrees of rate acceleration by the YFL ribozyme and AMP-dependent ligases are similar, the lack of marked specificity for leaving groups in the YFL ribozyme reaction may have been advantageous for molecular evolution in the RNA World, where primitive RNA ligation/polymerization systems must have evolved to advanced systems utilizing ipp leaving groups (Ellington et al. 2009).
Unfortunately, the catalytic activity of the YFL ribozyme is still insufficient as a progenitor for generating β-NMN-dependent RNA polymerases. Therefore, we are currently planning further in vitro evolution combined with rational modular engineering to improve its catalytic efficiency.
MATERIALS AND METHODS
Oligonucleotides
DNA and RNA oligonucleotides were purchased from Hokkaido System Science and Dharmacon, Inc., respectively. The standard and variant FAM (5-carboxyfluorescein)-labeled oligonucleotides used in the activity assays were purchased from IDT, Inc. and JBioS, respectively.
DNA oligonucleotides
For library construction
Pool R-1: 5′-AACCTAAGGCACCAAGGCCGCTATGG-N30-GACTGCGTTCCAGTCTCATTGCCCACTTAGGCAGA-3′ (N30 indicates 30 random nucleotides);
Pool R-2: 5′-AACCTAAGGCACCAAGGCCGCTATGG-N35-GACTGCGTTCCAGTCTCATTGCCCACTTAGGCAGA-3′ (N35 indicates 35 random nucleotides);
Pool R-3: 5′-AACCTAAGGCACCAAGGCCGCTATGG-N40-GACTGCGTTCCAGTCTCATTGCCCACTTAGGCAGA-3′ (N40 indicates 40 random nucleotides);
Pool L-1: 5′-AACCTAACCTTGGTGCCGCCTTAGG-N15-GACTACGCTGCAACCGTAGTTGCAATTTCCACAGGGAAGTTTCCTTCCCTAATAGTGAGTCGTATTACTTC-3′ (N15 indicates 15 random nucleotides);
Pool L-2: 5′-AACCTAACCTTGGTGCCGCCTTAGG-N10-GACTACGCTGCAACCGTAGTTGCAATTTCCACAGGGAAGTTTCCTTCCCTAATAGTGAGTCGTATTACTTC-3′ (N10 indicates 10 random nucleotides);
Pool L-3: 5′-AACCATAACTTGGTGCCGCCTTAGG-N5-GACTACGCTGCAACCGTAGTTGCAATTTCCACAGGGAAGTTTCCTTCCCTAATAGTGAGTCGTATTACTTC-3′ (N5 indicates 5 random nucleotides);
Fw-R: 5′-CAGTCATGACGAGAGTAACCTAAGGCACCAAGGCCGCTAT-3′;
Fw- L: 5′-GAAGTAATACGACTCACTATTAGGGAAGGAAACTTCCCTGTGGAAATT-3′, where the T7φ2.5 promoter sequence is shown in italics;
Rv-R: 5′-TCTGCCTAAGTGGGCAATGTGACTGGAACTC-3′;
Rv-1: 5′-CAGTCACGACCGAGACAACCTAACCTTGGTGCCGCCTTAG-3′; and
YFL-1 dope antisense: 5′-AACCATAACTTGGTGCCGCCTTAGGaaggaacagtgaattgacttcgtacgcgttaatgttgataGACTGCGTTCCAGTCTCATTGCCCACTTAGGCAGA-3′, where lowercase letters correspond to degenerate bases comprised of 79% of the original bases and 21% of three other bases.
For in vitro selection
Rv-S: 5′-TCTGCCTAAGTGGGCAATGAGACTGG-3′;
Rv-M: 5′-TCTGCCTAAGTGGGCAATGAGACTGGAAC-3′;
Rv-LL: 5′-TCTGCCTAAGTGGGCAATGAGACTGGAACGCAG-3′;
RT-1: 5′-CGTACACGTACTCACGCGTATACAGTC-3′;
RT-2: 5′-ACTTCCGAGCTGTAGAGTTAGCAGCGA-3′;
RT-3: 5′-CTTGACGTCAGCCTGGAAAACCCT-3′;
Fw-L: 5′-GAAGTAATACGACTCACTATTAGGGAAGGAAACTTCCCTGTGGAAATTG-3′;
Fw-2: 5′-GAAGTAATACGACTCACTATTAGGGAAGGAAACTTCCCTGTGTCAATTG-3′; and
Fw-3: 5′-GAAGTAATACGACTCACTATTAGGTCAGGAAACTGACCTCGTGGAATTGCAACTA-3′.
5′-Biotin-modified RNA oligonucleotides for in vitro selection
Sub-1: 5′ biotin-CGUACACGUACTCACGCGUAUACAGUCCAC-3′;
Sub-2: 5′ biotin-ACUUCCGAGCUGUAGAGUUAGCAGCGACAC-3′; and
Sub-19b: 5′ biotin-UACUUGACGUCAGCCUGGAAAACCCUCG-3′.
FAM-labeled oligonucleotide for standard activity assays
5′-FAM-CGTACACGTACTCACGCGTATACA-rGrUrCrCrArC-3′, where A, C, G, and T indicate deoxynucleotides, and rA, rC, rG, and rU indicate ribonucleotides.
Library construction
Template DNAs for three sublibraries were constructed as follows. The 3′-half fragments of the sublibraries were prepared by PCR. Three pool-R oligonucleotides (pool R-1, pool R-2, and pool R-3) including 30, 35, and 40 nt of random sequence, respectively, were amplified by PCR with ExTaq DNA polymerase (Takara) using a sense primer (Fw-R) and an antisense primer (Rv-R). The 5′-half fragments of the sublibraries were also prepared in the same manner. Three pool-L oligonucleotides (pool L-1, pool L-2, and pool L-3) including 15, 10, and 5 nt of random sequence, respectively, were amplified by PCR using a sense primer (Fw-L) and an antisense primer (Rv-1). Half of each PCR product was digested with BanI (New England Biolabs), and the remaining half was digested with StyI (New England Biolabs). The R-1, R-2, and R-3 PCR fragments were ligated with L-1, L-2, and L-3 fragments, respectively, using T4 DNA ligase (Takara).
The DNA library for doped selection was also constructed in a similar manner using YFL-1 dope AS and pooled L-3 oligonucleotides as the PCR template to generate 3′ and 5′ fragments, respectively.
The resulting ligated DNAs were used as templates for in vitro transcription with T7 RNA polymerase under standard reaction conditions except the nucleotide composition consisted of 1 mM GTP, UTP, and CTP; 0.1 mM ATP; and 50 mM β-NADH (Sigma). The transcripts of the sublibraries were mixed and used for the in vitro selection experiments.
In vitro selection
The purified RNAs were dissolved in H2O, and then denatured for 5 min at 80°C, followed by pre-incubation for 3 min at 37°C. RNA folding was initiated by adding 5× reaction buffer. After incubation for 10 min at 37°C, substrate RNA was added to start the ligation reaction. The final concentrations of the ribozyme and substrate were both 1 μM. The reaction was carried out with 30 mM Tris-HCl (pH 7.5), 50 mM MgCl2, and 200 mM KCl at 37°C. Three 5′-biotinylated substrate RNAs (Sub-1, Sub-2, and S-19b) were used. Sub-1 was used in rounds 1, 3, and 5; Sub-2 was used in rounds 2, 4, and 6; and S-19b was used in round 7.
The reaction was stopped by ethanol precipitation. The ligated RNAs were captured on streptavidin paramagnetic particles (Promega) and hybridized with a primer complementary to the 3′-region of the pooled RNAs. Reverse transcription was performed with ReverTra Ace MMLV RNaseH-minus reverse transcriptase (Toyobo) with four primers: Rv-S in rounds 1 and 4; Rv-M in rounds 2 and 5; Rv-R in rounds 3 and 6; and Rv-LL in round 7. The resulting cDNAs were eluted with 150 mM KOH, followed by neutralization with 150 mM HCl. The resulting cDNAs were subjected for selective PCR with the Rv primer and the selective primers complementary to the sequences of the substrate RNAs (RT-1 in rounds 1, 3, and 5; RT-2 in rounds 2, 4, and 6; RT-3 in round 7). The PCR products were electrophoresed on 2% agarose gels, and the desired products were isolated using Nucleospin ExtractII (Macherey-Nagel). PCR to regenerate templates as well as replace P1 sequence was carried out with the primer including the sequence of the T7 ϕ2.5 promoter (Huang 2003) followed by the sequence for one of the three P1 regions (Fw-L in rounds 2 and 4; Fw-2 in rounds 1, 3, and 5; and Fw-3 in round 6). The resulting DNAs were used as templates for the following round.
The selective PCR products from the round 7 pool were cloned into the pGEM-T vector (Promega). Individual plasmid clones were isolated and sequenced using an automated DNA sequencer (CEQ8000XL; Beckman Coulter).
Preparation of YFL-1 ribozyme mutants
Plasmids encoding YFL-1 ribozyme mutants with base substitutions and/or deletions in the P2 region, catalytic units, and/or P3 region were constructed by PCR-based mutagenesis with plasmid pYFL-1 carrying the parental YFL-1 sequence as a template. To introduce mutation at the P1 region, the template DNAs for in vitro transcription were prepared directly by PCR with appropriate sense primers for a series of mutants with the T7 ϕ2.5 promoter and P1 mutations, Rv-S, and pYFL-1. Template DNAs with a truncated 3′-region were also generated directly by PCR with appropriate antisense primers, Fw-L, and pYFL-1.
Ligation activity assays
The ribozymes were dissolved in H2O, then denatured by incubation for 5 min at 80°C, followed by additional incubation for 3 min at 37°C. RNA folding was initiated by adding 5× reaction buffer at 37°C. After folding for 10 min, the ligation reaction was initiated by adding the substrate the 5′-end of which was labeled with 5-carboxyfluorescein (FAM). The standard reaction conditions were as follows. Final ribozyme and substrate concentrations were both 1 μM. The reactions were carried out at 37°C in the presence of 30 mM Tris-HCl (pH 7.5), 50 mM MgCl2, and 200 mM KCl. At various time points, an aliquot was treated with a half volume of stop solution consisting of 80% formamide and 100 mM EDTA. Samples were separated on 12% polyacrylamide denaturing gels and quantified with a Pharos FX FluoroImager (Bio-Rad). Rate constants were derived from at least two independent experiments. The bimolecular ligation reaction between the YFL ribozyme and substrate can be described by the following equation (Atkins 1998):
 |
where t is time and k app is the apparent second-order rate constant. Under reaction conditions with equivalent molar concentrations of the ribozyme and substrate, the equation can be transformed as follows:
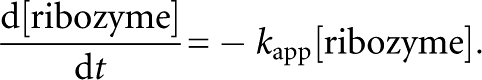 |
When the initial concentrations of ribozyme and substrate are both 1 μM, the above differential equation can be transformed to the following equation (Atkins 1998):
where t is time, k is the apparent second-order rate constant (k app), and Fa is the calculated final yield. The pseudo-first-order rate constant with excess substrate was determined as described previously (Ikawa et al. 2005).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Drs. Shigeyoshi Matsumura and Kazuo Harada for critical reading of the manuscript. We also thank the anonymous reviewers for their helpful comments. This work was partly supported by Grants-in-Aid for Young Scientists (A) (No.18685020 to Y.I.), Exploratory Research (No.19657071 to Y.I.), and the Global COE Program, “Science for Future Molecular Systems” (to H.F.) from the MEXT, Japan.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1461309.
REFERENCES
- Atkins P.W. Physical chemistry. 6th ed. Oxford University Press; Oxford, UK: 1998. Chapter 25-3. [Google Scholar]
- Bartel D.P., Szostak J.W. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- Bartel D.P., Doudna J.A., Usman N., Szostak J.W. Template-directed primer extension catalyzed by the Tetrahymena ribozyme. Mol. Cell. Biol. 1991;11:3390–3394. doi: 10.1128/mcb.11.6.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker R.R., Joyce G.F. Self-incorporation of coenzymes by ribozymes. J. Mol. Evol. 1995;40:551–558. doi: 10.1007/BF00160500. [DOI] [PubMed] [Google Scholar]
- Burgstaller P., Famulok M. Isolation of RNA aptamers for biological cofactors by in vitro selection. Angew. Chem. Int. Ed. Engl. 1994;33:1084–1087. [Google Scholar]
- Cech T.R. Self-splicing of group I introns. Annu. Rev. Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Cech T.R. The efficiency and versatility of catalytic RNA: Implications for an RNA world. Gene. 1993;135:33–36. doi: 10.1016/0378-1119(93)90046-6. [DOI] [PubMed] [Google Scholar]
- Chen I.A., Salehi-Ashtiani K., Szostak J.W. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 2005;127:13213–13219. doi: 10.1021/ja051784p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li N., Ellington A.D. Ribozyme catalysis of metabolism in the RNA world. Chem. Biodivers. 2007;4:633–655. doi: 10.1002/cbdv.200790055. [DOI] [PubMed] [Google Scholar]
- Chworos A., Severcan I., Koyfman A.Y., Weinkam P., Oroudjev E., Hansma H.G., Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- Cleaves H.J., Miller S.L. The nicotinamide biosynthetic pathway is a by-product of the RNA world. J. Mol. Evol. 2001;52:73–77. doi: 10.1007/s002390010135. [DOI] [PubMed] [Google Scholar]
- Costa M., Michel F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose V.J. Metal ion binding to catalytic RNA molecules. Curr. Opin. Struct. Biol. 2003;13:317–324. doi: 10.1016/s0959-440x(03)00077-0. [DOI] [PubMed] [Google Scholar]
- Dworkin J.P., Lazcano A., Miller S.L. The roads to and from the RNA world. J. Theor. Biol. 2003;222:127–134. doi: 10.1016/s0022-5193(03)00020-1. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. Noncoding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Ekland E.H., Bartel D.P. The secondary structure and sequence optimization of an RNA ligase ribozyme. Nucleic Acids Res. 1995;23:3231–3238. doi: 10.1093/nar/23.16.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland E.H., Szostak J.W., Bartel D.P. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science. 1995;269:364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- Ellington A.D., Chen X., Robertson M., Syrett A. Evolution origins and directed evolution of RNA. Int. J. Biochem. Cell Biol. 2009;41:254–265. doi: 10.1016/j.biocel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Fedor M.J. The role of metal ions in RNA catalysis. Curr. Opin. Struct. Biol. 2002;12:289–295. doi: 10.1016/s0959-440x(02)00324-x. [DOI] [PubMed] [Google Scholar]
- Fusz S., Eisenführ A., Srivatsan S.G., Heckel A., Famulok M. A ribozyme for the aldol reaction. Chem. Biol. 2005;12:941–950. doi: 10.1016/j.chembiol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Geary C., Baudrey S., Jaeger L. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res. 2008;36:1138–1152. doi: 10.1093/nar/gkm1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R., Szostak J.W. Selection of a ribozyme that functions as a superior template in a self-copying reaction. Science. 1992;258:1910–1915. doi: 10.1126/science.1470913. [DOI] [PubMed] [Google Scholar]
- Hager A.J., Szostak J.W. Isolation of novel ribozymes that ligate AMP-activated RNA substrates. Chem. Biol. 1997;4:607–617. doi: 10.1016/s1074-5521(97)90246-5. [DOI] [PubMed] [Google Scholar]
- Huang F. Efficient incorporation of CoA, NAD, and FAD into RNA by in vitro transcription. Nucleic Acids Res. 2003;31:e8. doi: 10.1093/nar/gng008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa Y., Naito D., Aono N., Shiraishi H., Inoue T. A conserved motif in group IC3 introns is a new class of GNRA receptor. Nucleic Acids Res. 1999;27:1859–1865. doi: 10.1093/nar/27.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa Y., Fukada K., Watanabe S., Shiraishi H., Inoue T. Design, construction, and analysis of a novel class of self-folding RNA. Structure. 2002;10:527–534. doi: 10.1016/s0969-2126(02)00739-6. [DOI] [PubMed] [Google Scholar]
- Ikawa Y., Tsuda K., Matsumura S., Inoue T. De novo synthesis and development of an RNA enzyme. Proc. Natl. Acad. Sci. 2004;101:13750–13755. doi: 10.1073/pnas.0405886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa Y., Matsumoto J., Horie S., Inoue T. Redesign of an artificial ligase ribozyme based on the analysis of its structural elements. RNA Biol. 2005;2:137–142. doi: 10.4161/rna.2.4.2302. [DOI] [PubMed] [Google Scholar]
- Isaacs F.J., Dwyer D.J., Collins J.J. RNA synthetic biology. Nat. Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Wright M.C., Joyce G.F. A complex ligase ribozyme evolved in vitro from a group I ribozyme domain. Proc. Natl. Acad. Sci. 1999;96:14712–14717. doi: 10.1073/pnas.96.26.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L., Westhof E., Leontis N.B. TectoRNA: Modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne A., Famulok M. A novel ribozyme with ester transferase activity. Chem. Biol. 1998;5:23–34. doi: 10.1016/s1074-5521(98)90084-9. [DOI] [PubMed] [Google Scholar]
- Joyce G.F. Nonenzymatic template-directed synthesis of informational macromolecules. Cold Spring Harb. Symp. Quant. Biol. 1987;52:41–51. doi: 10.1101/sqb.1987.052.01.008. [DOI] [PubMed] [Google Scholar]
- Joyce G.F. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- Joyce G.F. Forty years of in vitro evolution. Angew. Chem. Int. Ed. 2007;46:6420–6436. doi: 10.1002/anie.200701369. [DOI] [PubMed] [Google Scholar]
- Joyce G.F., Orgel L.E. Progress toward understanding the origin of the RNA world. In: Gesteland R.F., et al., editors. The RNA world. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 23–56. [Google Scholar]
- Landweber L.F., Pokrovskaya I.D. Emergence of a dual-catalytic RNA with metal-specific cleavage and ligase activities: The spandrels of RNA evolution. Proc. Natl. Acad. Sci. 1999;96:173–178. doi: 10.1073/pnas.96.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauhon C.T., Szostak J.W. RNA aptamers that bind flavin and nicotinamide redox cofactors. J. Am. Chem. Soc. 1995;117:1246–1257. doi: 10.1021/ja00109a008. [DOI] [PubMed] [Google Scholar]
- Lohse P.A., Szostak J.W. Ribozyme-catalyzed amino acid transfer reactions. Nature. 1996;381:442–444. doi: 10.1038/381442a0. [DOI] [PubMed] [Google Scholar]
- Makarova J.A., Kramerov D.A. Noncoding RNAs. Biochemistry. 2007;72:1161–1178. doi: 10.1134/s0006297907110016. [DOI] [PubMed] [Google Scholar]
- McGinness K.E., Joyce G.F. RNA-catalyzed RNA ligation on an external RNA template. Chem. Biol. 2002;9:297–307. doi: 10.1016/s1074-5521(02)00110-2. [DOI] [PubMed] [Google Scholar]
- McGinness K.E., Joyce G.F. In search of an RNA replicase ribozyme. Chem. Biol. 2003;10:5–14. doi: 10.1016/s1074-5521(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Misra V.K., Shiman R., Draper D.E. A thermodynamic framework for the magnesium-dependent folding of RNA. Biopolymers. 2003;69:118–136. doi: 10.1002/bip.10353. [DOI] [PubMed] [Google Scholar]
- Müller U.F. Re-creating an RNA world. Cell. Mol. Life Sci. 2006;63:1278–1293. doi: 10.1007/s00018-006-6047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U.F., Bartel D.P. Improved polymerase ribozyme efficiency on hydrophobic assemblies. RNA. 2008;14:552–562. doi: 10.1261/rna.494508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel L.E. RNA catalysis and the origins of life. J. Theor. Biol. 1986;123:127–149. doi: 10.1016/s0022-5193(86)80149-7. [DOI] [PubMed] [Google Scholar]
- Pan T., Dichtl B., Uhlenbeck O.C. Properties of an in vitro selected Pb2+ cleavage motif. Biochemistry. 1994;33:9561–9565. doi: 10.1021/bi00198a023. [DOI] [PubMed] [Google Scholar]
- Rogers J., Joyce G.F. A ribozyme that lacks cytidine. Nature. 1999;402:323–325. doi: 10.1038/46335. [DOI] [PubMed] [Google Scholar]
- Saito H., Inoue T. RNA and RNP as new molecular parts in synthetic biology. J. Biotechnol. 2007;132:1–7. doi: 10.1016/j.jbiotec.2007.07.952. [DOI] [PubMed] [Google Scholar]
- Sassanfar M., Szostak J.W. An RNA motif that binds ATP. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- Sazani P.L., Larralde R., Szostak J.W. A small aptamer with strong and specific recognition of the triphosphate of ATP. J. Am. Chem. Soc. 2004;126:8370–8371. doi: 10.1021/ja049171k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R., Barta A., Semrad K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- Sengle G., Eisenführ A., Arora P.S., Nowick J.S., Famulok M. Novel RNA catalysts for the Michael reaction. Chem. Biol. 2001;8:459–473. doi: 10.1016/s1074-5521(01)00026-6. [DOI] [PubMed] [Google Scholar]
- Stahley M.R., Strobel S.A. RNA splicing: Group I intron crystal structures reveal the basis of splice site selection and metal ion catalysis. Curr. Opin. Struct. Biol. 2006;16:319–326. doi: 10.1016/j.sbi.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Stoltenburg R., Reinemann C., Strehlitz B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Suga H., Cowan J.A., Szostak J.W. Unusual metal ion catalysis in an acyl-transferase ribozyme. Biochemistry. 1998;37:10118–10125. doi: 10.1021/bi980432a. [DOI] [PubMed] [Google Scholar]
- Szostak J.W., Bartel D.P., Luisi P.L. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- Thirumalai D., Woodson S.A. Maximizing RNA folding rates: A balancing act. RNA. 2000;6:790–794. doi: 10.1017/s1355838200000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber D.K., Williamson J.R. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- Tsukiji S., Pattnaik S.B., Suga H. An alcohol dehydrogenase ribozyme. Nat. Struct. Biol. 2003;10:713–717. doi: 10.1038/nsb964. [DOI] [PubMed] [Google Scholar]
- Vicens Q., Cech T.R. Atomic level architecture of group I introns revealed. Trends Biochem. Sci. 2006;31:41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Voytek S.B., Joyce G.F. Emergence of a fast-reacting ribozyme that is capable of undergoing continuous evolution. Proc. Natl. Acad. Sci. 2007;104:15288–15293. doi: 10.1073/pnas.0707490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Masquida B., Jaeger L. RNA tectonics: Toward RNA design. Fold. Des. 1996;1:R78–R88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- White H.B., III Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- White H.B., III . Evolution of coenzymes and the origin of pyridine nucleotides. In: Everse J., et al., editors. The pyridine nucleotide coenzymes. Academic Press; New York: 1982. pp. 1–17. [Google Scholar]
- Win M.N., Smolke C.D. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson S.A. Recent insights on RNA folding mechanisms from catalytic RNA. Cell. Mol. Life Sci. 2000;57:796–808. doi: 10.1007/s000180050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen L., Svendsen J., Lee J.S., Gray J.T., Magnier M., Baba T., D'Amato R.J., Mulligan R.C. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- Yingling Y.G., Shapiro B.A. Computational design of an RNA hexagonal nanoring and an RNA nanotube. Nano Lett. 2007;7:2328–2334. doi: 10.1021/nl070984r. [DOI] [PubMed] [Google Scholar]
- Zaher H.S., Unrau P.J. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA. 2007;13:1017–1026. doi: 10.1261/rna.548807. [DOI] [PMC free article] [PubMed] [Google Scholar]