Abstract
The condensin complex is the chief molecular machine of mitotic chromosome condensation. Nucleolar concentration of condensin in mitosis was previously shown to correlate with proficiency of rDNA condensation and segregation. To uncover the mechanisms facilitating this targeting we conducted a screen for mutants that impair mitotic condensin congression to the nucleolus. Mutants in the cdc14, esp1 and cdc5 genes, which encode FEAR-network components, showed the most prominent defects in mitotic condensin localization. We established that Cdc14p activity released by the FEAR pathway was required for proper condensin-to-rDNA targeting in anaphase. The MEN pathway was dispensable for condensin-to-rDNA targeting, however MEN-mediated release of Cdc14p later in anaphase allowed for proper, albeit delayed, condensin targeting to rDNA and successful segregation of nucleolus in the slk19 FEAR mutant. Although condensin was physically dislodged from rDNA in the cdc14 mutant, it was properly assembled, phosphorylated and chromatin-bound, suggesting that condensin was mis-targeted but active. This study identifies a novel pathway promoting condensin targeting to a specific chromosomal address, the rDNA locus.
Keywords: condensin, rDNA, nucleolus, chromatin, Cdc14, chromosome segregation, anaphase
INTRODUCTION
Proliferating cells must execute precise transmission of genetic information during cell division to ensure genetic integrity of species and cell lineages. An assembly of multiple protein factors controls this essential process, partially through maintaining the evolutionarily conserved and highly ordered three-dimensional organization of chromatin. One particular level of this high-order chromatin structure is mitotic chromosome condensation, which establishes the inter-domain contacts within the single chromatid, allowing its successful segregation in anaphase.1 Recent studies established that the essential role of chromosome condensation is not in making chromosomes merely “shorter and thicker,” but instead is in compartmentalization of sister chromatids to facilitate anaphase.2–4
Mitotic chromosome structure is intimately dependent on activity of the condensin complex,1 even though additional/alternative systems evidently contribute to complete mitotic chromosome compaction in higher eukaryotes.3,4 In S. cerevisiae these elusive additional mechanisms, which can largely compensate for the loss of condensin function in higher cells, are either absent or inactive. Therefore, loss of condensin activity in S. cerevisiae leads to completely impaired chromosome condensation.2,5 Budding yeast also lack condensin paralogues that form an alternative condensin complex in vertebrates.6 Thus, condensin function in S. cerevisiae is more transparent to molecular analysis, especially taking into account the relative simplicity of yeast genome organization.
Previously, we demonstrated that the condensin complex exists in S. cerevisiae and that its composition is conserved, as compared to X. laevis and human cells. S. cerevisiae condensin includes two SMC proteins: Smc2p and Smc4p 2,7 as well as three additional non-SMC subunits: Brn1p, Ycs4p and Ycs5p/Ycg1p.2,8,9 All five condensin subunits were shown to be essential for cell viability, indicating that S. cerevisiae condensin acts as an integral complex in vivo. Genetic analysis demonstrated that mutant alleles of genes encoding condensin subunits phenocopy the originally described smc2 mutant allele with respect to their role in chromosome condensation.2,5,8,9
Localization of condensin in yeast cells with GFP tagging of Smc4p and chromatin immunoprecipitation analysis (ChIP) revealed that rDNA chromatin is the primary target of condensin binding throughout the cell cycle.2 Moreover, around the metaphase-to-anaphase transition condensin undergoes an even more extreme compartmentalization to the nucleolar region.2 This process is likely homologous to the state of maximal mitotic chromosome condensation in human and fission yeast cells. Thus, the mitotic concentration of condensin in the nucleolar region was validated as a non-invasive in vivo assay for rDNA chromatin condensation in S. cerevisiae.10 Genetic analysis of conditional ts-mutants in condensin subunits confirmed that rDNA is the main target of condensin activity. Chromosomes carrying rDNA were sensitive to impaired condensin function and were lost at a high rate in the smc2 and smc4 mutants.2 Furthermore, rDNA did not segregate in condensin mutants at a non-permissive temperature, even as other chromosomes proceeded through anaphase.2 In addition to the block in rDNA segregation under non-permissive conditions, a delay in segregation of long chromosomal arms (ADE8 and MET16 loci) and YACs was also observed (see Fig. 5 in ref. 2), indicating that long chromosomal arms and chromatin saturated with repeats could bear additional sites for condensin activity. Existence of some essential secondary sites for condensin binding was corroborated by the fact that S. cerevisiae strains lacking the chromosomal rDNA array (complemented by an episomal rDNA)11 still had an essential requirement for condensin, even though Smc4p-GFP was no longer seen concentrated in the nucleolus in mitosis.2
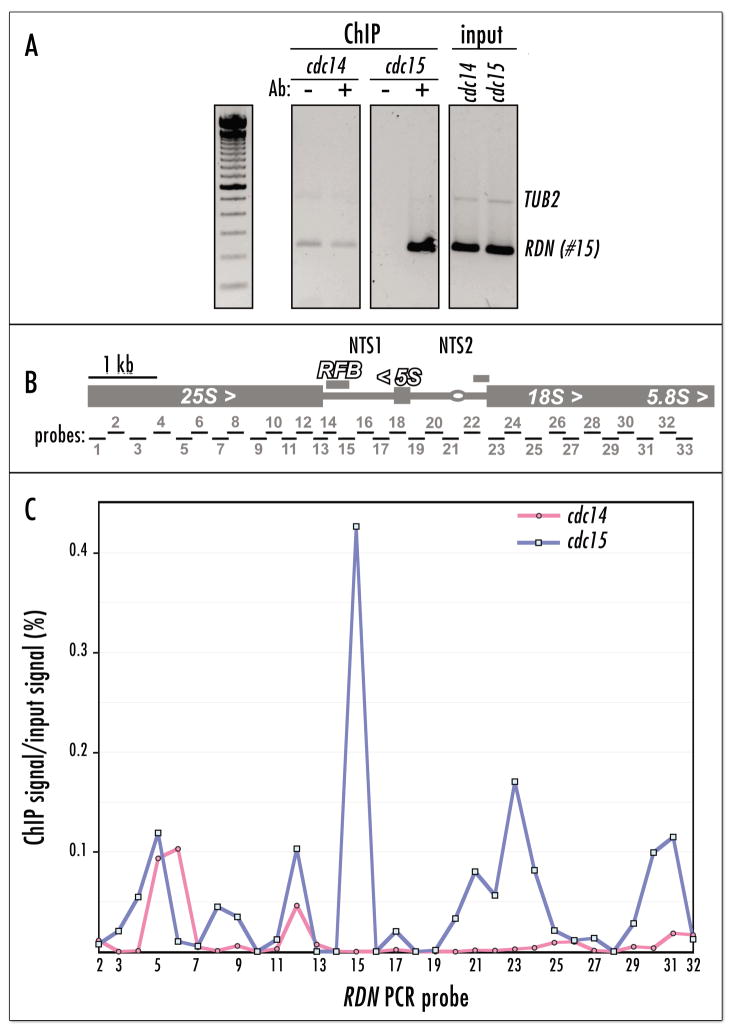
Figure 5.
Condensin is physically de-localized from rDNA in the cdc14 cells. (A) Chromatin IP analysis of cdc14-1 BRN1:HA and cdc15-1 BRN1:HA strains arrested at 37°C for 2.5 hr. The RFB region, PCR probe #15 in (B), a known hot-spot of condensin localization to the rDNA repeat,2,10 is used to assess the relative enrichment for condensin binding (Brn1p-HA ChIP). The TUB2 gene probe, which contains no binding sites for condensin (unpublished), is a negative control for duplex PCR. (B) Positions of PCR probes used for quantitative PCR analysis of ChIP in (C). PCR primer sequences were exactly as in reference 36. (C) Quantitative PCR analysis of Brn1p-HA ChIP from cdc14-1 and cdc15-1 strains arrested at 37°C for 2.5 hr. The quantitative PCR values from cycle 32 (see Materials and Methods) are plotted as ratios of precipitated DNA to total DNA (in %).
Thus, studies on chromosome partition in condensin mutants and the demonstration that mitotic chromatin compaction per se is not a prominent process in S. cerevisiae have established that the primary biological role of condensin (and probably chromosome condensation driven by it) is to facilitate separation of sister chromatids in anaphase. However, the nature of condensin preference for rDNA chromatin remains unknown. It is unlikely to be a simple direct pathway.12 For example, it was shown that both SUMO deconjugation machinery10 and sister chromatid cohesion establishment (Strunnikov A, unpublished) are required for condensin targeting to rDNA in mitosis. To expand our knowledge about the mechanism of condensin targeting to rDNA we undertook a screen for conditional mutants that impair targeting of condensin to rDNA in mitosis. This screen resulted in identification of a novel pathway, which is required for condensin targeting to rDNA, but probably does not regulate the enzymatic activity of condensin. We demonstrated that activity of Cdc14p in early anaphase is responsible for proper condensin binding to rDNA chromatin in mitosis.
MATERIALS AND METHODS
All yeast genetic techniques were according to standard protocols.32,33 The genotypes of yeast strains are shown in Table 1. Yeast cell cycle arrest and release experiments were conducted as described.7,34 The quality of arrest was monitored microscopically and by FACS (not shown). For Figures 1 and 3 arrests were 4 hr at 37°C in SD-uracil media; for Figures 4A, B and Figure 5, 2.5 hr in YPD; and for Figure 4C, 2 hr in YPD.
Table 1.
Yeast Strains
| Strain name | Relevant genotype | Original source |
|---|---|---|
| 3cAS451 | MATa ade2 leu2 lys2 trp1 ura3 cdc14–1 pep4-Δ::HIS3 BRN1:6His:3HA::URA3 | this work |
| 1cAS452 | MATa leu2 trp1 ura3 cdc15–1 pep4–Δ::HIS3 BRN1:6His:3HA::URA3 | this work |
| 3aAS453 | MATa ade2 leu2 trp1 ura3 cdc14–1 YCS5:6His:3HA::URA3 | this work |
| 6cAS453 | MATa leu2 lys2 ura3 YCS5:6His:3HA::URA3 | this work |
| 640-1AS432 | MATα ade2 his3 leu2 trp1 ura3 cdc14–1 SMC4:GFP::URA3 | this work |
| 640-3AS426 | MATα his3 leu2 ura3 cdc15–1 SMC4:GFP::URA3 | this work |
| 640-8AS491 | MATa ade2 his3 trp1 lys2 leu2 met15 ura3 esp1-1 SMC4:GFP::URA3 | this work |
| 640-K291-9C | MATa his3 ura3 cdc5–1 SMC4:GFP12::URA3 | L. H. Johnston |
| 640-WY46 | MATa ade2 his3 leu2 trp1 ura3 tem1–Δ::pGAL:UPL-NET1::TRP1 bar1 SMC4:GFP12::URA3 | W. Shou |
| 640-WY69 | MATa ade2 his3 leu2 trp1 ura3 bar1 net1–Δ::his5sp SMC4:GFP12::URA3 | W. Shou |
| 640-BY4741 | MATa his3 leu2 met15 ura3 SMC4:GFP::URA3 | ATCC |
| 640-4002451-BY4741 | MATa his3 leu2 met15 ura3 slk19–Δ::kanMX SMC4:GFP::URA3 | ATCC |
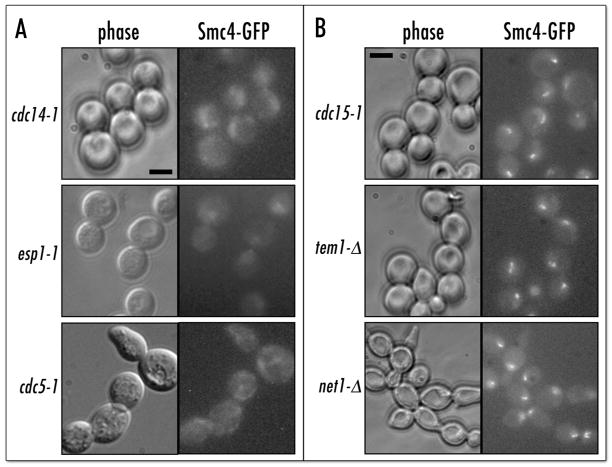
Figure 1.
Condensin is delocalized in FEAR but not MEN mutants. (A) Smc4p-GFP signal is delocalized in FEAR mutants at 37°C. The esp1–1 strain (Table 1) was kept in mitosis with nocodazole. Scale bar 5 μm. (B) MEN mutants properly localize condensin to rDNA. Arrest conditions: cdc15-1 (37°C), tem1-Δ(3-hr shut-off of pGAL:NET1 at 23°C), net1-Δ (mitotic delay at 23°C). Strain genotypes in Table 1.
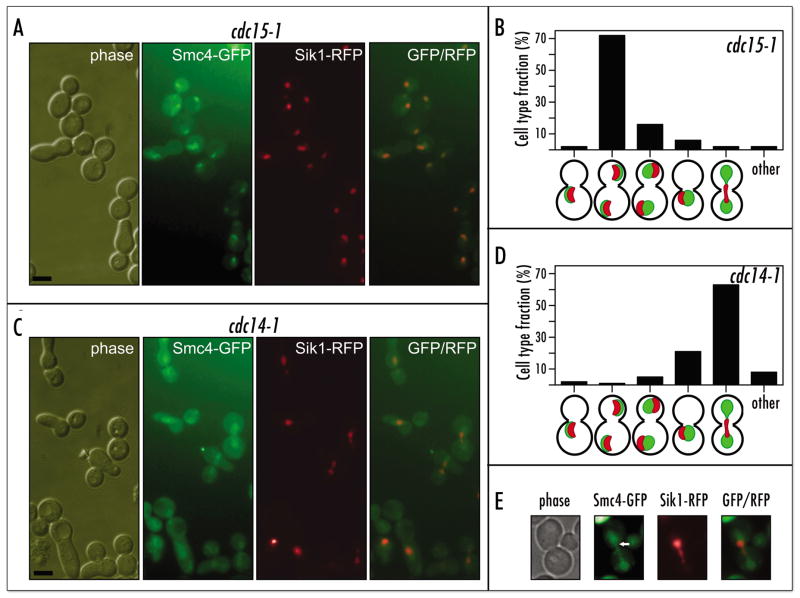
Figure 3.
Diffuse condensin distribution in the cdc14 mutant correlates with a nucleolus segregation defect. (A) Micrograph and (B) quantification of relative distribution of Smc4p-GFP and Sik1p-mRFP in the 640-3AS426/SIK1:mRFP::kanMX cells after a temperature shift. Scale bar 5 μm. (C) Micrograph and (D) quantification of relative distribution of Smc4p-GFP and Sik1p-mRFP in the 640-1AS432/SIK1:mRFP::kanMX cells after a temperature shift. Scale bar 5 μm. (E) Same as in (C), with higher magnification.
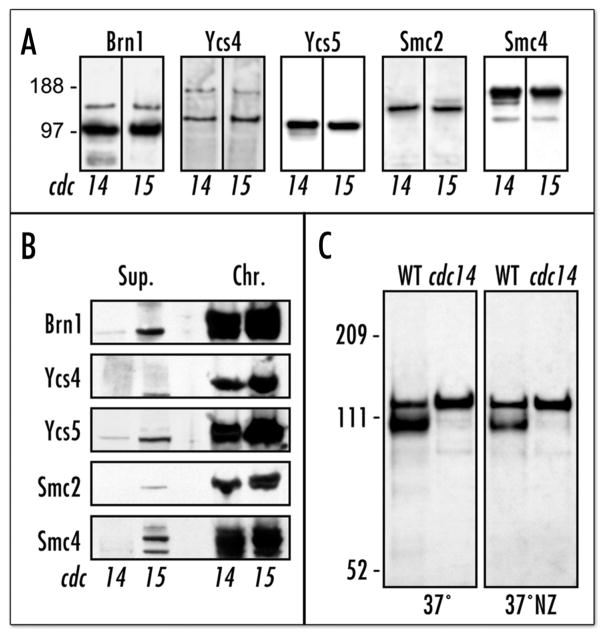
Figure 4.
Mistargeted condensin is functional. (A) Western-blot of Brn1p-HA-precipitated condensin complex, showing comparison of condensin composition in cdc14 and cdc15 mutants at non-permissive temperature. Strains: Table 1. (B) Western-blot comparing chromatin-bound and soluble condensin fractions35 in cdc14 and cdc15 mutants at non-permissive temperature. Supernatant fractions (Sup.) are equivalent to 1/5 of the chromatin (Chr.) fractions. The strains are identical to (A). (C) Western-blot (anti-HA) analysis of Ycs5p-6xHis-3HA IMAC eluates from 3aAS453 (cdc14–1) and 6cAS453 (WT) incubated at two different conditions (See Materials and Methods). The upper Ycs5p band is a phosphorylated from. Mock purification (no 6xHis tag) produces no anti-HA-reactive bands at these stringent conditions (not shown).
Microscopy was performed with the AxioVert (Zeiss) microscope with epifluorescence. The images were captured by integrated 0.2 μm-step Z-axis scanning with cooled CCD camera. The pLF640 plasmid and gene-targeting protocol used to construct the Smc4p-GFP fusion in vivo has been described.2 The SIK1:mRFP::kanMX DNA fragment was amplified by PCR with SIK1-flanking primers from the SIK1:mRFP::kanMX strain DNA25 and used to transform yeast strains described in Table 1. Replacement of SIK1 with SIK1:mRFP::kanMX was confirmed by PCR analysis. Before microscopy cells were fixed with formaldehyde (5 min, 3.7%), washed with PBS and kept on ice.
Anti-condensin antibodies have been described.2 Chromatin-binding assays (Fig. 4B) were performed according to reference 35 with slight modifications. Immunoprecipitations (Fig. 4A) were performed with the same buffer (EBX) supplemented with DNase I in capped 2-ml columns (BioRad). Cell lysates were incubated (4°C) with anti-HA antibodies (12CA5, Roche) and coupled to CNBr-Sepharose (Amersham). The antibody-bound proteins were eluted from the column-packed beads with 2% SDS (90°C). The Ycs5 protein tagged with 3xHA and 6xHis tags and the in-vivo-replacement tagging procedure has been described.2 Two strains (6cAS453 and 3aAS453) were used to partially purify and concentrate the tagged Ycs5 protein by IMAC. After cells were released from alpha-factor arrest the cultures were split in half and immediately shifted to 37°C. To one of the halves nocodazole was added within minutes after release. After 2 hr at 37°C or at 37°C with nocodazole cells were harvested and immediately resuspended/disrupted (with glass beads) in a chaotropic extraction buffer (6 M Guanidine-HCl, 50 mM Tris pH8.0, 1xPBS, 0.35 M NaCl) to prevent loss of phosphorylation during extraction. Extracts were clarified by centrifugation and incubated with Ni-NTA-Superflow resin (Quiagen) for 12 hr at 4°C. The resin suspension was passed through a 2-ml disposable BioRad column to form an open column bed. After several washes (with the extraction buffer, with a similar buffer where Guanidine-HCl was replaced by 6 M urea and with the urea buffer supplemented with 20 mM imidazole) the bound proteins were collected in 100 μl of stripping buffer (2% SDS, 20 mM EDTA, 1 mM β-mercaptoethanol). Ycs5p-HA was visualized by Western blotting after separation by PAGE (100V, 2 hr) in the 3–8% gradient Tris-acetate gel (Invitrogen). These conditions allowed for resolution of phosphorylated and non-phosphorylated forms of Ycs5p-HA, which were not resolved by other PAGE conditions (4–12% Bis-Tris/MOPS gels, 200V, 45 min) used in this work (Fig. 4A and B).
ChIP assays for Figure 5A were performed using the following protocol. Cells from 50-ml cultures were harvested and cross-linked in 1% formaldehyde for 30 min at room temperature. After cross-linking, cells were washed twice in Tris-Buffered Saline (TBS, pH 7.4). Cells were suspended in 400 μl of Lysis Buffer (50 mM HEPES-OH pH 7.5, 0.75 M NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% Na Deoxycholate) with Complete protease inhibitors (Roche). An equal amount of glass beads was added and the cells were lysed in a mini bead beater for 10 minutes at 4°C. The lysate was collected and sonicated to an average fragment size of less than 1 kb. The lysate was collected by centrifugation for 5 min in a microfuge at 4°C. For the Brn1p-HA ChIP experiment, the lysate was divided in two halves. The anti-HA antibody (Roche) (0.5 μg) was added to one half of the sample, while the other half was used as the “no antibody” control. The samples were then incubated on a mini-rotator overnight at 4°C. 15 μl of Protein G beads suspension (Pierce) was added to each sample and sample was incubated on a mini-rotator for 3 hours at room temperature. After the beads settled down the buffer was removed. The beads were washed five times using 1 ml of lysis buffer (0.75 M NaCl) with protease inhibitors, and washed three times using the lysis buffer (0.5 M NaCl). The beads were then washed once using wash solution 3 (10 mM Tris-HCl pH 8.0, 0.25 M LiCl, 0.5% NP-40, 0.5% Na Deoxycholate, 5 mM EDTA) and TE (pH 8.0), respectively, and were resuspended in 20 μl of TE (pH 8.0). Samples were incubated at 65°C for 6 hours to reverse cross-links and placed at 95°C for 20 min. For input DNA, 100 μl TE with 1% SDS was added to 20 μl of lysate. After reversing cross-links at 65°C for 6 hours, 100 μg Proteinase K was added and the samples were incubated at 37°C for 2 hr. Samples were extracted once with phenol: chloroform:isoamyl-alcohol and once with chloroform. 200 μl ethanol, 10 μl Na acetate and 5 mg glycogen were added and sample was precipitated at −20°C overnight. Precipitated and washed DNA was dissolved in 100 μl of TE. After PCR analysis (1 cycle of 95°C for 5 min, followed by 25 cycles of 95°C for 1 min, 55°C for 30 sec, and 72°C for 1 min) samples were run on 1.5 % agarose gel at 125 V for 45 min. For duplex PCR reactions, TUB2 (which does not bind condensin in vivo) PCR product was used as a negative control for ChIP and an internal control for input DNA. The linear ranges of PCR amplification were estimated by serial dilutions of immunoprecipitated DNA.
The quantitative ChIP protocol was essentially the same as described above. Quantitative PCR reactions were performed by the Real-time PCR System (Stratagene MX3000P). PCR reactions (50 μl) contained 1 μl template DNA (ChIP or Input), 25 μl 2x SYBR Green master mix (Stratagene), and 50 nM primers. PCR parameters were 1 cycle of 95°C for 10 min, followed by 35 cycles of 95°C for 1 min, 55°C for 30 sec, and 72°C for 1 min. Primer concentrations and PCR cycle parameters was optimized to eliminate formation of primer dimers. Primers for rDNA sequences were the same as used in reference 36. Primers #1 and #33 were routinely excluded from the quantitative PCR set to allow analysis of all samples for a given strain in a single 96-well PCR plate. No notable condensin binding was found to these two sites in a separate experiment (not shown). PCR products were quantified using the MX3000P software (Stratagene) in the cycle corresponding to the linear range of amplification (cycles 32 and 33). The enrichment ratio was determined by calculating the ratio of rDNA-ChIP to rDNA-input (total chromatin) and normalizing the data to the “no antibody” control background. All the rDNA-ChIP to rDNA-input ratios were calculated as: 2{Ct (Input) − Ct (ChIP)} divided with {dilution rate (ChIP)/dilution rate (Input)}. Ct values of all PCR products were determined by the MX3000P software.
RESULTS
Our original finding that the essential biological function of condensin is to segregate tandem repeat-containing sister chromatids2 allowed us to develop several new approaches to study molecular mechanics and regulation of condensin in budding yeast. One of these approaches was to monitor the nucleolar accumulation of Smc4p-GFP in mitosis as an in-vivo assay for chromosome condensation.10 We undertook a systematic screening of conditional mutants that arrested in mitosis to identify the candidate genes encoding potential condensin regulators and/or rDNA-targeting factors. This collection also included some mutants defective in mitotic rDNA segregation (e.g. cdc14–1). We replaced the SMC4 gene with a fully functional SMC4-GFP fusion in more than forty conditional mutants and monitored localization of Smc4p-GFP after a three-hour conditional shift (37°C in most cases). In some cases we used synchronous release from a G1 arrest into non-permissive conditions in the presence of nocodazole (e.g. for esp1–1). After completion of the initial screening we focused our attention on the cdc14–1 mutant, as it showed the most striking defect in condensin localization to nucleolus in mitosis (Fig. 1A), especially as compared to the cdc15-1 mutant, which arrests at a similar point in late anaphase (Fig. 1B). The cdc14 mutant is known to have decondensed rDNA, while in cdc15 rDNA is properly condensed.13
We first tested whether the enzymatic activity of Cdc14p was required for proper condensin localization to nucleolar chromatin. It is an important consideration, as Cdc14p, similarly to condensin, is present in the nucleolus, but in the enzymatically inactive state (RENT complex).14,15 Moreover, condensin mis-localization in the cdc14-1 mutant could not be reversed by returning the cells to the permissive temperature (data not shown). This potentially indicates that either the Cdc14p activity per se is not required for condensin concentration at the rDNA locus, or that condensin relocalization from rDNA to other sites is itself irreversible (at least in late anaphase). To address this question we ectopically expressed the mutant cdc14–Δ(1–374) allele which encoded protein defective in phosphatase activity16 in the cdc14–1 cells. This phosphatase-deficient Cdc14p did not rescue the cdc14–1 defects in condensin localization (data not shown). Thus, mutations in CDC14 specifically impair the condensin-to-rDNA-targeting pathway, revealing a new role for this multifunctional phosphatase.
Two other mutants analyzed in the original screen showed similarly de-localized condensin: esp1-1 and cdc5-1 both displayed diffuse nuclear Smc4p-GFP signals. Interestingly, both the ESP1 and CDC5 genes are linked with CDC14 in two mitotic pathways. ESP1 and CDC5 genes are components of FEAR network,17–19 while CDC5 is also required for MEN pathway.20,21 As both the FEAR and MEN pathways involve Cdc5p-controlled release of Cdc14p activity from a sequestered (nucleolar) form, we assumed that the Cdc14 protein was the main effector controlling condensin targeting. It seemed unlikely, however, that the MEN pathway would mediate condensin targeting, as condensin concentrates in the rDNA locus between metaphase and anaphase, long before exit from mitosis. Indeed analysis of Smc4p-GFP localization in the cdc15-1 (included in the original screening), as well as in the tem1-Δ and net1-Δ MEN mutants demonstrated that condensin was properly concentrated in the nucleolus (Fig. 1B).
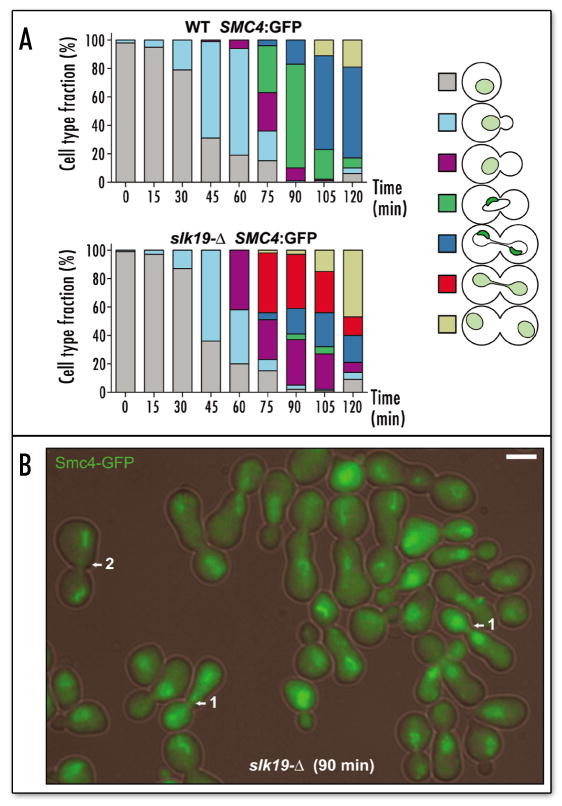
The FEAR network, on the other hand, facilitates the earliest known release of Cdc14p activity in the cell cycle and thus could be responsible for the Cdc14p-controlled step in condensin targeting. We tested whether components of FEAR network were involved in condensin targeting. With the esp1–1 mutant it was difficult to assess the dynamics of condensin targeting, as esp1–1 cells arrest in mitosis only for a short period of time and then proceed to exit mitosis with unsegregated sister chromatids.22,23 Thus, we focused our attention primarily on the slk19–Δ mutant. The slk19–Δ cells had a notable delay in mitosis and allowed monitoring of condensin targeting with higher precision. Microscopic analysis of asynchronous slk19–Δ population revealed that high proportion of cells were delayed in late anaphase with properly targeted condensin, however no cells had singular nucleolus-concentrated condensin in large-budded cells (not shown), morphology characteristic for metaphase-to-anaphase transition (Fig. 4E in ref. 2). This observation was consistent with the hypothesis that failure to release the Cdc14 protein early in mitosis results in improper condensin localization, however subsequent release of Cdc14 by MEN allows eventual proper targeting of condensin and successful segregation of rDNA chromatids. In order to verify this hypothesis we monitored condensin localization to nucleolus following synchronous release from a G1 arrest. We followed the budding pattern and bud size in conjunction with the position and concentration of the Smc4p-GFP signal (Fig. 2). The wild-type (Slk19+) cells proceeded synchronously though the cell cycle (Fig. 2A) and demonstrated nucleolar concentration of Smc4p-GFP at the metaphase-to-anaphase transition and during anaphase, as expected. The slk19–Δ cells proceeded through anaphase with a slight delay, but without displaying cells with a singular concentrated condensin signal. Instead, the slk19–Δ cells had a new class of cells, virtually absent in the wild-type population. These slk19–Δ cells were apparently undergoing anaphase with condensin diffusely distributed in the nucleus (red, Fig. 2A; arrow 1, Fig 2B), instead of condensin being concentrated in the nucleolus. However, this cell class was diminished by the end of the time-course, giving way to cells with two separate concentrated Smc4p-GFP signals (dark blue, Fig. 2A; arrow 2, Fig 2B). Thus slk19–Δ and FEAR network are required for the early anaphase condensin concentration at the nucleolus, but are dispensable in the mid-to-late anaphase, the time when Cdc14p is released by the MEN network.
Figure 2.
FEAR mutant slk19 is defective in anaphase concentration of condensin. (A) Condensin-to-nucleolus concentration in the synchronous cell cycle. Parallel analysis of 640-BY4741 (WT) and 640-4002451-BY4741 (slk19-Δ) strains released from alpha-factor arrest allowed comparative monitoring of Smc4p-GFP dynamics in the nucleus. Diffuse condensin, light green nucleus; nucleolus-targeted condensin, crescent-shaped dark-green subnuclear structure. (B) Micrograph of the 90-min point in the time-course analysis of the slk19-Δ strain. Arrow 1, unique class of cells undergoing anaphase with mistargeted condensin; Arrow 2, mid-to-late anaphase recapture of condensin by nucleolus. Scale bar 5 μm.
The putative mechanism of condensin de-localization in cdc14p mutants could be either dislodging of condensin from its proper location in rDNA chromatin or diffusion of nucleolar chromatin itself with condensin, presumably inactive, but still bound. The latter seemed less likely as the nucleolus was reported to be relatively compact, albeit enlarged, in the arrested cdc14 mutant cells.24 Nevertheless, we monitored the position of nucleolus with an alternative fluorescent marker to exclude the possibility that rDNA chromatin was stretched out in the cdc14–1 cells to give the appearance of improperly targeted condensin. We utilized the recently described Sik1p-mRFP fusion25 as a constitutive nucleolar marker.
We investigated whether Smc4p-GFP was properly colocalized with Sik1p-mRFP to rDNA during anaphase arrest mediated by the cdc14-1 mutation. The cdc14–1 and cdc15–1 mutants expressing Smc4p-GFP were shifted to 37°C and condensin localization was compared to Sik1p localization (Fig. 3). In the arrested cdc15-1 cells, used as a control, the Smc4p-GFP fusion was fully colocalized with Sik1p-mRFP at two polar sites in mother and daughter cells, respectively (Fig. 3A and B). In the majority of arrested cdc14-1 cells, the nucleolar Sik1p-RFP signal was found undivided, in the mid-zone of anaphase cells (Fig. 3C and D). This localization was consistent with the reported failure of nucleolar segregation in cdc14 mutants24 and was reminiscent of condensin-mutant induced rDNA non-segregation.2 However, the Smc4p-GFP fusion did not colocalize with Sik1p-RFP (Fig. 3C and D). These results strongly suggest that condensin is not enriched in the rDNA at the cdc14-1-mediated anaphase arrest. Moreover, in some cells, at higher magnification condensin appeared to be excluded, at least partially, from the nucleolar area (Fig. 3E).
Thus, while it is evident that condensin is more uniformly distributed throughout the nucleus in the cdc14-1-arrested cells, the mechanism of condensin depletion/exclusion from the nucleolus is not clear. It could involve disassembly of the condensin complex, release of condensin from chromatin, relocalization of condensin to alternative chromosomal sites or inactivation through inadequate posttranslational modification.
In order to assess the degree and mechanism of condensin’s inability to target rDNA in the cdc14-1 cells, we first compared the integrity and chromatin association of condensin in the cdc14 and cdc15 mutants. Condensin immunoprecipitation via the HA-tagged Brn1p demonstrated that condensin was fully assembled in both mutants at 37°C and that there were no significant variations in the relative subunit amounts (Fig 4A). Moreover, the condensin complex was also fully associated with chromatin in both mutants at the restrictive temperature, suggesting that condensin was redistributed to other genomic sites in the arrested cdc14 cells (Fig 4B). Finally we tested whether condensin phosphorylation was perturbed in the arrested cdc14 cells. Indeed, targeting of condensin to rDNA may be coupled to its activation via post-translational modification, as it is the case in X. laevis.26 Recently an important role of Ycs5p phosphorylation for condensin activity in yeast mitosis has been substantiated.12 The anaphase-specific increase in the phosphorylation level of Ycs5p was attributed to activity of Ipl1p kinase.12 Cdc14p, in its turn, controls some aspects of Ipl1p localization.27 To test whether this post-translational modification of condensin is dependent on Cdc14p we assessed phosphorylation of Ycs5p in the cdc14-arrested cells. Ycs5p-6xHis-3HA was purified for following strains and conditions: asynchronous wild-type at 37°C, cdc14 arrested at 37°C, wild-type arrested at 37°C with nocodazole and, finally, cdc14 released from G1 into 37°C in the presence of nocodazole (Fig. 4C). Nocodazole treatment resulted in reduction of unphosphorylated Ycs5p in wild-type cells but had little effect on the Ycs5p-6xHis protein purified from cdc14 cells, as Ycs5p was likely fully phosphorylated both at the late-anaphase cdc14-specific arrest and at the nocodazole-mediated arrest. Thus, condensin appears to be functional, albeit mis-targeted, in the arrested cdc14 cells.
As the amount of condensin in the cell appeared to be equivalent in the cdc14 and cdc15 mutants we applied the chromatin immunoprecipitation approach to determine the relative amounts of condensin bound to rDNA chromatin. First we assessed the occupancy of the RFB region in the rDNA (Fig. 5B), which was shown to be a hot-spot for condensin binding.2,10 Comparison of the RFB occupancy by Brn1p-HA in the cdc14 and cdc15 mutants at the non-permissive temperature indicated that virtually no condensin was bound to RFB in the cdc14 mutant (Fig. 5A). To verify this with a more rigorous method we combined Brn1p-HA chromatin immunoprecipitation with quantitative real-time PCR analysis. This approach allowed us to delineate both qualitative and quantitative changes in condensin binding across the rDNA repeat in the arrested cdc14 cells. As Figure 5C shows, the cdc14 rDNA has only two condensin-binding peaks, out of six observed in the cdc15 cells. These results conclusively confirmed that condensin complex was mis-localized in the cdc14-arrested cells. The nearly complete loss of condensin from rDNA could explain the rDNA de-condensation (see ref. 13) and rDNA non-segregation (Fig. 3 and ref. 24) phenotype of the cdc14 mutants.
DISCUSSION
While the composition, in vitro enzymatic activity and general molecular architecture of condensin have been elucidated to a great degree,1 the molecular mechanism of the chromatin condensation reaction in vivo remains a mystery. Solving the two following problems is necessary to understand the molecular nature of condensin activity in vivo. First, as condensin undergoes cell cycle dependent changes in localization and activity levels, its function should be regulated in a cell cycle-dependent fashion. However, data from different systems suggest that either condensin regulation is not conserved in evolution or all regulatory mechanisms have not yet been found. Second, the existing data on condensin supercoiling activity with naked DNA28 cannot be extrapolated to accommodate a chromatin fiber as a substrate. Thus, while accepting the paradigm that the basic activity of condensin is to change the superhelical state of chromatin, we have yet to identify a natural substrate for condensin activity in vivo and to understand the mechanism of affinity between condensin and chromatin in order to elucidate the molecular mechanism of chromosome condensation.
As condensin is not a sequence-specific DNA-binding complex, there are two possible alternative scenarios for specific recognition of chromatin motifs by condensin: either condensin recognizes a specific chromatin structure (such as repeats, positioned nucleosomes, histone modifications) or some auxiliary proteins (facilitators) attract condensin and, possibly, anchor it at a specific place. In either case, loss of some of the factors involved in determining the specificity of condensin binding should recapitulate the condensin mutant phenotype. Conversely, mutations in proteins that directly regulate condensin activity, e.g., through post-translational modification, should also manifest a condensin mutant-like phenotype.
Based on these assumptions combined with our previous findings that the rDNA repeats is the primary binding target for condensin in vivo2 and that condensin undergoes relocalization to the rDNA chromatin at the metaphase-to-anaphase transition we designed some specific tests for condensin activity and chromatin-binding in S. cerevisiae.2,10 Using these genetic and cell biology assays (condensin targeting to the nucleolus and mis-segregation of the rDNA-containing chromosomes) we conducted screening of candidate mutants for defects in chromosome condensation regulation and/or condensin targeting to chromatin. This approach previously resulted in identification of at least two pathways essential for condensin targeting to rDNA in mitosis: the sister chromatid cohesion (SCC) pathway (Strunnikov A, unpublished) and the SUMO/Smt3p-deconjugation pathway.10
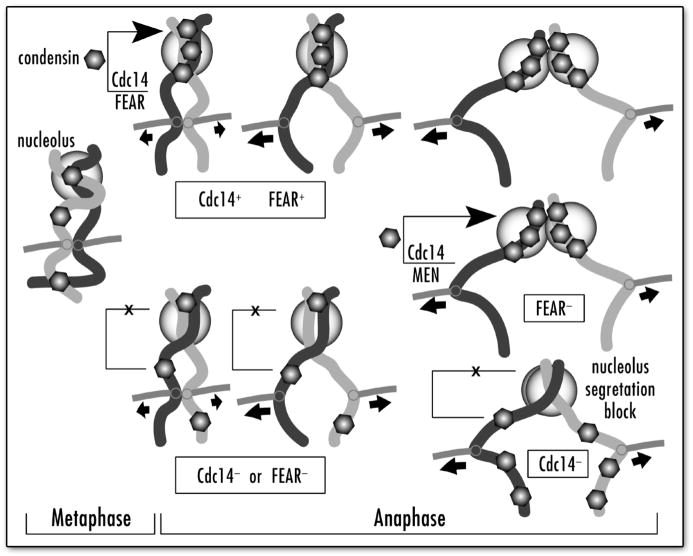
Here, using the Smc4p-GFP rDNA-targeting assay we identified the third pathway essential for condensin targeting to rDNA. We demonstrated (Figs. 1, 2 and 3) that the Cdc14p activity specifically released by the FEAR,15 but not the MEN29 network, is required for proper condensin targeting to nucleolar chromatin in mitosis (Fig. 6). This finding allowed us for the first time to determine the specific point at which condensin is concentrated in the nucleolus in the normal cell cycle. While previously we concluded that this concentration is an attribute of both metaphase and anaphase,2 the role of the FEAR/Cdc14p pathway in this process indicates that maximal condensin accumulation in the nucleolus probably happens in early anaphase, when FEAR is activated. We also found that late-anaphase release of Cdc14p (by MEN) can compensate the condensin-targeting defect when the FEAR pathway is inactivated, thus allowing mutants in FEAR genes to survive (Fig. 2).
Figure 6.
Interplay between Cdc14p, FEAR and MEN pathways in condensin localization to nucleolus in anaphase. Apparent chromosomal condensin distribution and corresponding success of rDNA/nucleolus segregation are shown for normal cell cycle, FEAR-deficient and Cdc14p deficient cells. Also shown is a supplementary role of MEN in condensin targeting when FEAR is inactivated.
The exact protein target of the Cdc14p phosphatase activity that mediates the early-anaphase condensin concentration in the nucleolus/rDNA chromatin is not yet known. We prefer the hypothesis that some nucleolar protein controlled by Cdc14p is required for condensin targeting to rDNA. The alternative explanation, that Cdc14p directly acts on condensin seems unlikely, as we show (Fig. 4) that condensin is fully assembled, properly phosphorylated and apparently chromatin-bound in the cdc14 mutant. Moreover, as the non-rDNA chromosomes segregate properly in the cdc14 mutant, with condensin still bound to chromatin (Fig. 4B), it seems quite probable that condensin is still active but mis-targeted in the cdc14 cells at the non-permissive temperature. Our analysis of the double cdc14-1 and smc2-8 mutant alleles shows that such cells are viable at 23°C, but display blocked segregation of all chromosomes, not just the rDNA-containing, at the non-permissive temperature (data not shown). This suggests that the mutant condensin, which is re-localized due to the cdc14 mutation, impairs segregation of the non-rDNA sister chromatids in this double mutant. Such a feature, i.e. the existence of active but mis-targeted condensin, makes the mutants in the CDC14 pathway unique, in comparison to the smt4 and SCC mutants (also defective in condensin targeting), as in these mutants segregation of all chromosomes is impaired. Identification of the non-rDNA condensin-binding sites and analysis of their occupancy by condensin at the cdc14 arrest would lend conclusive proof to the theory that Cdc14p is involved in nucleolar targeting but not in activation of the condensin complex per se. Moreover, characterization of such a unique enzymatic pathway, which is responsible for specific targeting of condensin to a well-characterized chromatin domain, should allow comprehensive investigation of the mechanisms attracting condensin to rDNA in mitosis.
As a result of our work, we come very close to understanding the mechanism that prevents nucleolar segregation in the cdc14 mutants.24,30,31 Using a highly sensitive and accurate quantitative ChIP approach we demonstrated that condensin is largely absent from the rDNA repeats at the cdc14-mediated arrest (Fig. 5). This inevitably leads to inability of sister chromatids (in the rDNA region) to segregate (Fig. 6), i.e. recapitulation of the classic condensin-mutant phenotype.2 Loss of condensin from rDNA is not, however, a feature of late anaphase, as in the cdc15 mutant condensin was properly bound to the rDNA repeats, as was determined by microscopic analysis (Figs. 1 and 3) and by quantitative ChIP (Fig. 5). Thus, the cdc14 mutant’s failure to segregate sister rDNA regions and the similar phenotype of condensin mutants2 are both mediated by the inability of condensin to perform its function. However the mechanisms in the two cases are significantly distinct: in case of cdc14 condensin does not bind the rDNA repeats, while in several tested condensin mutants the complex still occupies the rDNA sites (data not shown), yet presumably is inactive.
Figure 6 recapitulates our current view of condensin regulation by the FEAR/Cdc14 pathway. We propose that the main regulatory role of FEAR in condensin targeting to rDNA in early anaphase is to liberate the sequestered Cdc14p activity. Cdc14p, however, is the key player in this pathway, as we demonstrated that the enzymatically inactive Cdc14p cannot complement the condensin mis-targeting phenotype. Moreover, we suggest that the late-anaphase release of Cdc14p (by MEN), that still occurs in the FEAR mutant slk19, is able to correct the absence of Cdc14p in early anaphase by facilitating condensin targeting to rDNA later in anaphase. The cdc14 mutant, however, has a permanent defect in condensin targeting to rDNA and in condensin-dependent segregation of the rDNA sister chromatids. Irreversibility of the cdc14 rDNA non-segregation and condensin mis-targeting phenotypes indicates that condensin cannot return to the rDNA chromatin after a certain cell cycle point. While the molecular nature of this turning point is unknown, it seems likely that condensin becomes immobile at the end of anaphase, possibly due to yet uncharacterized decondensation pathway.
Relationship of the Cdc14 pathway to two other pathways controlling condensin targeting, the SCC and SUMO-deconjugation pathway remains to be established. The recent finding that the SCC function controls the pre-anaphase step in rDNA compaction12 may indicate that this pre-anaphase condensation is a prerequisite for condensin targeting at the onset of anaphase. However, as the observed FEAR-mediated condensin targeting is an anaphase event, it seems unlikely that cohesion plays a significant role at that point of the cell cycle. The Smt4p-controlled pathway10 for condensin targeting could potentially overlap with the Cdc14p-mediated pathway described here, as no SUMO/Smt3p modifications essential for mitosis have been identified yet. However, neither condensin nor Cdc14p itself are hyper-sumoated in the smt4-Δ mutant (data not shown), indicating that over-sumoation of these proteins is not responsible for condensin mis-targeting observed in the smt4-Δ nuclei.10
Acknowledgments
We would like to thank numerous colleagues who supplied the strains and gave advice for the original Smc4p-GFP screening; Ray Deshaies, who, before FEAR was discovered, has brought to our attention that some process other than the MEN should control condensin targeting via Cdc14p; L. Aragon, L. Freeman, N. Dhillon and R. Kamakaka for helpful discussions; M. Lichten, for advice on large-scale ChIP; Kei Ko, for technical help.
References
- 1.Swedlow JR, Hirano T. The making of the mitotic chromosome: modern insights into classical questions. Mol Cell. 2003;11:557–69. doi: 10.1016/s1097-2765(03)00103-5. [DOI] [PubMed] [Google Scholar]
- 2.Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol. 2000;149:811–24. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. C.elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 2002;16:729–42. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell. 2003;5:323–36. doi: 10.1016/s1534-5807(03)00199-0. [DOI] [PubMed] [Google Scholar]
- 5.Lavoie BD, Hogan E, Koshland D. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J Cell Biol. 2002;156:805–15. doi: 10.1083/jcb.200109056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirano T. Chromosome shaping by two condensins. Cell Cycle. 2004;3:26–8. [PubMed] [Google Scholar]
- 7.Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation defines a subgroup within the SMC-family. Genes Dev. 1995;9:587–99. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- 8.Lavoie BD, Tuffo KM, Oh S, Koshland D, Holm C. Mitotic chromosome condensation requires Brn1p, the yeast homologue of barren. Mol Biol Cell. 2000;11:1293–304. doi: 10.1091/mbc.11.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouspenski II, Cabello OA, Brinkley BR. Chromosome condensation factor Brn1p is required for chromatid separation in mitosis. Mol Biol Cell. 2000;11:1305–13. doi: 10.1091/mbc.11.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strunnikov AV, Aravind L, Koonin EV. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genet. 2001;158:95–107. doi: 10.1093/genetics/158.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oakes M, Aris JP, Brockenbrough JS, Wai H, Vu L, Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavoie BD, Hogan E, Koshland D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 2004;18:76–87. doi: 10.1101/gad.1150404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–30. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–23. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 15.Jensen S, Geymonat M, Johnston LH. Mitotic exit: delaying the end without FEAR. Curr Biol. 2002;12:R221–3. doi: 10.1016/s0960-9822(02)00756-x. [DOI] [PubMed] [Google Scholar]
- 16.Taylor GS, Liu Y, Baskerville C, Charbonneau H. The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J Biol Chem. 1997;272:24054–63. doi: 10.1074/jbc.272.38.24054. [DOI] [PubMed] [Google Scholar]
- 17.Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–20. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan M, Uhlmann F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat Cell Biol. 2003;5:249–54. doi: 10.1038/ncb940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geymonat M, Jensen S, Johnston LH. Mitotic exit: the Cdc14 double cross. Curr Biol. 2002;12:R482–84. doi: 10.1016/s0960-9822(02)00963-6. [DOI] [PubMed] [Google Scholar]
- 20.Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr Biol. 2001;11:784–8. doi: 10.1016/s0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- 21.Yeong FM, Lim HH, Surana U. MEN, destruction and separation: mechanistic links between mitotic exit and cytokinesis in budding yeast. BioEssays. 2002;24:659–66. doi: 10.1002/bies.10106. [DOI] [PubMed] [Google Scholar]
- 22.Tinker-Kulberg RL, Morgan DO. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 1999;13:1936–49. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 24.Granot D, Snyder M. Segregation of the nucleolus during mitosis in budding and fission yeast. Cell Motil Cytoskeleton. 1991;20:47–54. doi: 10.1002/cm.970200106. [DOI] [PubMed] [Google Scholar]
- 25.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 26.Kimura K, Hirano M, Kobayashi R, Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- 27.Pereira G, Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–4. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- 28.Bazett-Jones DP, Kimura K, Hirano T. Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol Cell. 2002;9:1183–90. doi: 10.1016/s1097-2765(02)00546-4. [DOI] [PubMed] [Google Scholar]
- 29.Jensen S, Johnston LH. Complexity of mitotic exit. Cell Cycle. 2002;1:300–3. doi: 10.4161/cc.1.5.142. [DOI] [PubMed] [Google Scholar]
- 30.Buonomo SB, Rabitsch KP, Fuchs J, Gruber S, Sullivan M, Uhlmann F, et al. Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev Cell. 2003;4:727–39. doi: 10.1016/s1534-5807(03)00129-1. [DOI] [PubMed] [Google Scholar]
- 31.Torres-Rosell J, Machin F, Jarmuz A, Aragon L. Nucleolar Segregation Lags Behind the Rest of the Genome and Requires Cdc14p Activation by the FEAR Network. Cell Cycle. 2004;3:496–502. [PubMed] [Google Scholar]
- 32.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics, a Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1990. [Google Scholar]
- 33.Brown A, Tuite M. Methods in Microbiology. San Diego, London: Academic Press; 1998. Yeast Gene Analysis; p. 502. [Google Scholar]
- 34.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–86. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–76. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]