Abstract
Connexin43 (Cx43) is a membrane-spanning protein that forms channels that bridge the gap between adjacent cells and this allows for the intercellular exchange of information. Cx43 is regulated by phosphorylation and by interacting proteins. “Mode-1” interaction with 14-3-3 requires phosphorylation of Ser373 on Cx43 (Park et al. 2006). Akt phosphorylates and targets a number of proteins to interactions with 14-3-3. Here we demonstrate that Akt phosphorylates Cx43 on Ser373 and Ser369; antibodies recognizing Akt-phosphorylated sites or phospho-Ser “mode-1” 14-3-3-binding sites recognize a protein from EGF-treated cells that migrates as Cx43, and GST-14-3-3 binds to Cx43 phosphorylated endogenously in EGF-treated cells. Confocal microscopy supports the co-localization of Cx43 with Akt and with 14-3-3 at the outer edges of gap junctional plaques. These data suggest that Akt could target Cx43 to an interaction with 14-3-3 that may play a role in the forward trafficking of Cx43 multimers and/or their incorporation into existing gap junctional plaques.
Keywords: Connexin43, Akt, phosphorylation, 14-3-3, “mode-1” binding, protein interactions
INTRODUCTION
Connexin43 (Cx43, Gja1) hexamers (connexons) dock across the plasma membrane with connexons in neighboring cells to form membrane-spanning channels. These channels provide for an exchange of information between cells that is important to homeostasis, synchronization of contraction in the heart and uterus, and for the regulation of cellular responses, such as wound repair. Cx43 is regulated by phosphorylation and by interactions with other cellular proteins (Giepmans 2004; Herve et al. 2004; Lampe and Lau 2004; Warn-Cramer and Lau 2004). A number of proteins have been identified as Cx43-interacting proteins, including the tight and adherens junction proteins ZO-1 and occludin (Giepmans and Moolenaar 1998; Jin et al. 2004; Laing et al. 2001; Nagasawa et al. 2006; Toyofuku et al. 1998), the cytoskeletal proteins tubulin, β-catenin, and drebrin (Ai et al. 2000; Butkevich et al. 2004; Giepmans et al. 2001b; Hertig et al. 1996), the growth factor NOV (Fu et al. 2004; Gellhaus et al. 2004), the src kinase (Giepmans et al. 2001a; Kanemitsu et al. 1997; Lin et al. 2001; Toyofuku et al. 2001), the CIP 85 (connexin43 interacting protein; 85 kDa) and CIP 75 proteins, which have roles in Cx43 proteolytic degradation (Jin et al. 2000; Lan et al. 2005; Li et al. (Submitted)), and the CIP150 protein, which appears to have a role in Cx43 phosphorylation and the formation of gap junctions (Akiyama et al. 2005). Many of these proteins do not appear to be directly involved in the regulation of channel gating.
14-3-3 θ was identified as one of the proteins interacting with Cx43 in a yeast two-hybrid screen of a mouse embryonic library (Jin 1998; Jin et al. 2000). More specifically, the NH2-terminal half of 14-3-3 θ (residues 1-124) interacted with the C-terminal (CT) cytoplasmic domain of Cx43 (residues Ala222-Ile382) in the yeast system. This region in 14-3-3 contains several of the basic amino acids in the substrate binding pocket that coordinate the phospho-Ser/Thr residue in 14-3-3 substrates (Park et al. 2006; Wang et al. 1998). This region also contains the dimerization region for 14-3-3 (Chaudhri et al. 2003; Fu et al. 2000) and dimerization helps stabilize binding interactions with 14-3-3 substrates, in part by blocking the phosphorylation of 14-3-3 (Shen et al. 2003). In subsequent studies, in silico and in vitro, we demonstrated a “mode-1” interaction of 14-3-3 θ with phospho-Ser373 of Cx43. A second potential “mode-1” binding site at Ser244 of Cx43 did not support an interaction with 14-3-3 θ in the absence of phosphorylation at Ser373 (Park et al. 2006). Other isoforms of 14-3-3 are known to interact with phosphorylated “mode-1” binding sites (Yaffe et al. 1997) and 14-3-3 ζ also interacted with Cx43 in in vitro binding studies (Park et al. 2006).
14-3-3 proteins are small acidic proteins (isoelectric point ~4.6) of ~29-30 kDa that have been shown to interact with more than 200 different proteins (Bridges and Moorhead 2004; Pozuelo Rubio et al. 2004). They are highly conserved proteins that are found in primitive eukaryotes, but not in prokaryotes (Bridges and Moorhead 2004). There are seven different mammalian isoforms of 14-3-3 that are ubiquitously expressed (Bridges and Moorhead 2004). These isoforms can form hetero- or homo-dimers. They interact as dimers with other proteins (Shen et al. 2003), primarily, although not exclusively, through binding to a phospho-Ser/Thr site in a “mode 1” (R-S-X-pS/pT-X-P) or “mode 2” (R-X-X-X-pS/pT-X-P) motif on the target protein (Yaffe et al. 1997). Such interactions with 14-3-3 have been shown to mediate diverse functions for different proteins, including changes in protein conformation, the masking of specific molecular sites, and serving as a scaffold to assemble protein complexes (Bridges and Moorhead 2004).
In intracellular signaling pathways, 14-3-3 proteins are known to function as modular units for the assembly of molecular complexes, through binding interactions with phopho-Ser/Thr sites on their substrates in a manner that is similar to the interactions of SH2 (src homology 2) and PTB (phospho-tyrosine binding) domains in binding to phospho-Tyr sites on their binding partners (Muslin et al. 1996; Wilker and Yaffe 2004). In a recent X-ray crystallography study of the structure of 14-3-3 coordinated with the plant membrane H+-ATPase Ottman et al. (2007) present a 3-D reconstruction of the hexameric enzyme complexed with 14-3-3 that is reminiscent of 3-D models of connexin hexamers (Muller et al. 2002; Sosinsky and Nicholson 2005). They propose a model wherein an inactive H+-ATPase dimer interacts with a second dimer through an interaction of a 14-3-3 monomer with a C-terminal peptide on one subunit of one H+-ATPase dimer and the other 14-3-3 monomer interacts with the same region in a subunit of another H+-ATPase dimer. This mechanism allows the assembly of the hexameric H+-ATPase complex with three 14-3-3 dimers and results in an active enzyme through induced conformational changes. This represents a novel model of a role for 14-3-3 in the regulation of other proteins.
14-3-3 binding motifs are unique to Cx43 and not found in other connexins (Park et al. 2006). The Ser373 “mode-1”14-3-3-binding sequence is conserved across species, supporting an important function for the interaction of Cx43 with 14-3-3. Akt/PKB frequently mediates 14-3-3 interactions by phosphorylating target proteins (Kovacina et al. 2003). In this study, we have examined whether Akt also phosphorylates Cx43. We found that Cx43 is a substrate for Akt in vitro and appears to be a substrate in vivo, since a motif antibody to Aktphosphorylated substrates recognizes Cx43 that is phosphorylated in vivo in epidermal growth factor (EGF)-treated cells with activated Akt. Furthermore, confocal microscopy indicates that both Akt and 14-3-3 θ co-localize with Cx43. An interaction between Cx43 and 14-3-3, each of which is considered to have a scaffolding function, may play an important role in regulating the assembly of Cx43 multimers or in the assembly of a Cx43-associated protein complex or nexus (Duffy et al. 2002) at the plasma membrane, a complex that may change in response to different intracellular signals.
METHODS
Yeast Two Hybrid Screen for Cx43-Interacting Proteins
Ayeast two-hybrid screen was carried out to identify proteins that interacted with the cytoplasmic CT domain of Cx43 (Jin 1998; Jin et al. 2000). A “bait” plasmid was constructed from the LexA fusion vector pBTM116 (Bartel et al. 1993) and the cDNA encoding the CT domain of rat Cx43 (residues 222-382). A mouse embryonic library (days 9.5-10.5) constructed by Hollenberg et al. (1995) in the plasmid pVP16 was used as the “fish” vector with the L40 yeast reporter strain that is Trp-, Leu-, and His-prototrophic (Vojtek et al. 1993). From the twelve million yeast transformants that were screened, fifteen cDNAs encoded protein domains that were determined to be positive for an interaction with the CT of Cx43 in a directed two-hybrid assay.
Recombinant Fusion Proteins
Polymerase chain reaction (PCR) was used to generate plasmids to express glutathione-S-transferase (GST) fusion proteins of the cytoplasmic CT domains of rat Cx43 (residues 236-382) and mouse Cx45 (residues 265-396) as described previously (Loo et al. 1995; Park et al. 2006). PCR products were sequenced to confirm their fidelity and subcloned into the BamHI and EcoRI sites of the pGEX-KG expression vector (provided by Dr. Jack Dixon, University of Michigan). The cDNA for the Cx43 CT with a Ser373Ala mutation was prepared using primers: 5′-GCAGCCCGCGCCAGCGCCAGGCCTCGGC-3′ and 5′-GCCGAGGCCTGGCGCTGGCGCTGGCGCGGCTGC-3′ as described previously (Park et al. 2006) and a truncated GST-Cx43 CT mutant, encoding residues 236-368 of Cx43, was prepared with primers to substitute a stop codon for the serine codon at position 369, primers: 5′-TATGGATCCGTTAAGGATCGCGTCAAGGG-3′ and 5′-GTCTAGAATTCCTTAGCTGGCTCTGC TGGAAGG-3′.
A GST fusion protein containing the cDNA for mouse 14-3-3 θ (provided by Dr. Ellen Freed, Onyx Pharmaceuticals; Emeryville, CA, USA) was prepared by PCR in a similar manner. A His6x-tagged 14-3-3 θ (His-14-3-3 θ) construct was also prepared to incorporate the BamHI and EcoRI restriction sites onto the 5′ and 3′ ends, respectively, of the cDNA for full-length 14-3-3 θ for subsequent subcloning into a bacterial expression vector, pTrcHis-A (Invitrogen; Carlsbad, CA, USA). All PCR products were sequenced to confirm their fidelity. The recombinant fusion proteins were expressed in BL21 cells following the induction of protein expression with IPTG (isopropyl-β-D-galactoside). Fusion proteins were isolated and purified on glutathione agarose beads (Sigma, St. Louis, MO, USA) or Ni-NTA spin Columns (QIAGen, Valencia, CA, USA) for the GST- or His-tagged proteins, respectively (Park et al. 2006). Blue native gel electrophoresis (Cline and Mori 2001; Schagger et al. 1994) was used to characterize the migration of the 14-3-3 θ fusion proteins relative to protein standards.
Cell Culture
Rat-1 fibroblasts were cultured in Dulbecco's Modified Eagle's Medium (DMEM; Sigma) with 10% fetal calf serum (FCS; Hyclone, Logan, UT, USA) at 37°C in a humidified incubator with 5% CO2. For some studies, the cells were transferred to a low serum medium overnight (0.5% FCS) prior to a treatment with EGF (100 ng/mL).
Preparation of Cell Lysates and Western Blotting
For immunoprecipitation studies, Rat-1 cells were lysed in 0.5% NP-40 buffer with 50-mM Tris-HCl (pH 7.4), 100-mM NaCl, and protease and phosphatase inhbitors (100 μg/mL aprotinin, 50 μg/mL leupeptin, 1 μg/mL pepstatin, 1-mM PMSF (phenylmethylsulfonyl fluoride), 10 mM NaF, and 50 μM Na3VO4). Other antibodies used for Western blotting included: a rabbit peptide antibody to the C-terminus of Cx43 (residues 368-382), a rabbit antibody to phospho-Ser473 of activated Akt (Cell Signaling, Danvers, MA, USA), a goat antibody specific for the Akt-3 isoform (M14, Santa Cruz Biotechnology, Santa Cruz, CA, USA), a mouse antibody to a phospho-Ser “mode-1” 14-3-3-binding site motif (4E2, Cell Signaling), and a rabbit antibody to phosphorylated Ser/Thr sites on Akt substrates (110B7, Cell Signaling). Secondary antibodies coupled to horseradish peroxidase (HRP) were obtained from Santa Cruz Biotechnology. The immunoblots were developed with an enhanced chemiluminescence (ECL, Piscataway, NJ, USA) kit from Amersham.
GST Pull-Downs
Ten micrograms of the GST-CxCT fusion proteins (or GST alone) were incubated with His-tagged 14-3-3 θ or with a whole-cell lysate (WCL) obtained from EGF-treated Rat-1 cells (30 min treatment with 100 ng/mL recombinant human EGF, Upstate Biotechnology, Lake Placid, NY, USA). The cells were lysed in a binding buffer (50 mM Tris-HCl (pH 7.5), 150-mM NaCl, 0.1% NP-40, 20% glycerol, and 1-mM PMSF) with added phosphatase inhibitors. Following incubation with the WCL and mixing for one hr at 4°C, the GST-CxCT fusion proteins were isolated with 10 μl of glutathione agarose beads. The beads were washed three times with 500 μL of binding buffer and the bound proteins were eluted with SDS-PAGE sample buffer and heating the samples. Proteins were resolved on 10% polyacrylamide NuPAGE gels (Invitrogen) and electrotransferred to Immobilon-P membranes (Milli-pore, Billerica, MA, USA) for Western blotting with anti-GST (B14, mouse monoclonal from Santa Cruz Biotechnology), anti-Cx43 (P2D12, mouse monoclonal to peptide residues 360-382 of Cx43 from the Fred Hutchinson Cancer Center, Seattle) or anti-14-3-3 θ (C-17, rabbit polyclonal from Santa Cruz Biotechnology).
Phosphorylation of Cx43 by Akt
GST fusion proteins were phosphorylated with purified Akt-1 and γ -[32P]-ATP in in vitro reactions using a kit obtained from Cell Signaling. GST alone or reactions without Akt served as negative controls and the GST-GSK3 fusion protein, provided with the kit, served as a positive control. Reactions were carried out at 30 °C for 30 min and were stopped by the addition of SDS sample buffer and heating the samples. The proteins were resolved by SDS-PAGE on 12% polyacrylamide gels and the gels were then stained, dried, and exposed to film at -80°C.
Immunofluorescence Microscopy
Rat-1 cells were plated onto glass coverslips and grown for 1 to 2 days. Cells were treated with or without EGF (100 ng/mL) and then rinsed with phosphate buffered saline (PBS) and fixed in methanol at -20°C. The cells were permeabilized with 0.2% Triton X-100 in PBS at room temperature, and then rinsed with PBS and blocked with 1% bovine serum albumin (BSA) in PBS. The cover slips were incubated with a primary antibody (mouse monoclonal anti-Cx43, P4G9, Fred Hutchinson Cancer Center); rabbit anti-14-3-3 θ (Santa Cruz Biotechnology), or goat anti-Akt-3 (Santa Cruz Biotechnology) and subsequently incubated with a secondary antibody coupled to AlexaFluor dye, either Alexa488 (green) or Alexa594 (red). The cover slips were mounted on glass slides with a phenylenediamine/glycerol solution (Biomeda, Foster City, CA, USA) and the cells viewed with a Zeiss Axioplan Universal microscope (Thornwood, NY, USA) equipped with epifluorescence. Confocal images were collected on a Leica (Bannockburn, IL, USA) TCS SP5 scanning laser confocal microscope.
14-3-3 Overlays
Cellular proteins from EGF-treated Rat-1 cells were solubilized with hot SDS sample buffer and heating of the samples. The proteins were resolved by SDS-PAGE on a 12 % polyacrylamide gel, and electrotransferred to Immobilon-P membranes. The GST-Cx43CT fusion protein was also run on the gel. The membranes were blocked with 2% bovine milk in Tris-buffered saline (TBS), 50-mM Tris-HCl (pH 7.8), 150-mM NaCl, and 1-mM MgSO4 and incubated overnight at 4 °C with His-14-3-3 θ (20 μg/mL) in the binding buffer that was used for the GST pull-downs (described above). His-14-3-3 θ was used in the overlay assay and binding was detected with an anti-His-tag monoclonal antibody (Santa Cruz Biotechnology, sc-8036). The blots were developed with the ECL plus kit from Amersham.
RESULTS
14-3-3 θ Interacts with Cx43, But Not with Cx45
In vitro binding studies using recombinant proteins and molecular modeling studies of rat/mouse 14-3-3 θ docked with the phospho-Ser373 peptide of Cx43 (Park et al. 2006) supported the interaction of the CT domain of Cx43 with 14-3-3 θ, an interaction originally identified in a yeast two-hybrid assay (Jin 1998). The recombinant 14-3-3 θ fusion proteins (GST- or His-tagged) formed homo-dimers as would be expected for mammalian 14-3-3 θ (Jones et al. 1995), with the dimers migrating at ~95 and ~69 kDa for the GST- and His-tagged proteins, respectively (Fig. 1(a)). GST-14-3-3 θ also formed higher molecular weight complexes, consistent with the ability of the GST moiety to self associate (Ji et al. 1992; Parker et al. 1990). His-tagged 14-3-3 θ interacted with GST-Cx43CT in GST pull-downs, but not with GST alone or GST-Cx43CT with a Ser373Ala mutation (GST-Cx43CT S373A), confirming the requirement for the Ser373 site of Cx43 for the interaction (Fig. 1 (b), and Park et al. 2006). Modeling studies had also supported the fit of the rat Cx43 phospho-Ser373 peptide (R-A-S-pS-R-PR) in the binding pocket of rat/mouse 14-3-3 θ, with the Arg56 and Arg60 residues in the binding pocket of 14-3-3 θ contributing to coordination of the phosphate group (Park et al. 2006). These residues were present in the NH2-terminal half of 14-3-3 θ that interacted with Cx43 in the yeast two-hybrid assay.
Figure 1.
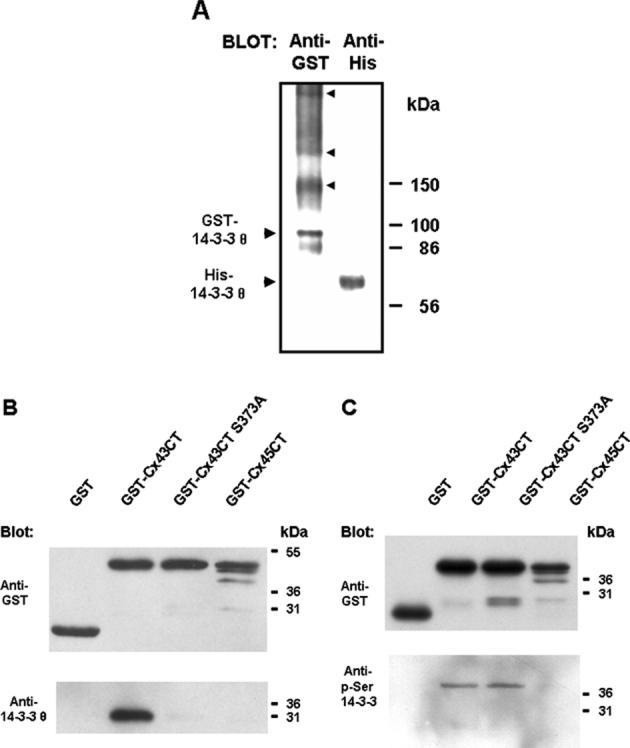
Specificity of the interaction of Cx43 with 14-3-3. (a) Recombinant His-tagged 14-3-3 θ migrated as a dimer on blue native gel electrophoresis as would be expected for the mammalian protein (large arrowheads mark the position of the dimer). GST-14-3-3 θ also migrated as a dimer and as higher molecular weight complexes (marked by smaller arrowheads). (b) His-tagged 14-3-3 θ interacted with and was pulled-down GST-Cx43CT, but not GST alone, GST-Cx43CT with a Ser373Ala mutation (GST-Cx43CT S373A), or GSTCx45CT. (c, lower blot) The GST-Cx43CT and GST-Cx43CT S373A proteins were both recognized by an antibody to phospho-Ser “mode-1” 14-3-3-binding sites. The protein load for the GST proteins is shown in the upper blot in (c).
The GST-Cx43CT fusion protein was recognized on Western blots by a phospho-Ser “mode 1” 14-3-3-binding site antibody that recognizes the motif R/K-X-X-pS-X-P (see Fig. 1(c)), indicating bacterial phosphorylation of the recombinant protein (Deutscher and Saier 2005; Macek et al. 2007) on a 14-3-3 “mode-1” binding site(s). The GST-Cx43CT S373A mutant was also recognized by the phospho-Ser “mode-1” 14-3-3-binding site antibody (Fig. 1 (c)), suggesting that the Ser244 “mode-1” site (Park et al. 2006) may also have been bacterially phosphorylated in the recombinant protein. However, binding studies with the GST-Cx43CT S373A mutant demonstrated that if Ser244 was phosphorylated, it was not sufficient to support an interaction with 14-3-3 θ in the absence of phosphorylation at the Ser373 site (Fig. 1 (b); Park et al., 2006). The 14-3-3-binding sequence at Ser373 is conserved in rat, mouse, human, bovine and chicken Cx43, consistent with functional significance of an interaction and since 14-3-3 motifs are not found in the CT domains of other connexins (Park et al. 2006), a function specific to Cx43 is suggested. Specificity of this interaction was also supported by a lack of binding of 14-3-3 θ to the GST fusion protein that contained the CT domain of Cx45 (Fig. 1(b)). Cx45 is also a phosphoprotein and has a cytoplasmic CT domain similar in size to the Cx43 CT; however, Cx45 does not contain 14-3-3 binding sequences.
Phosphorylation of Cx43 by Akt
The amino acid sequence at Ser373 of Cx43 fits a consensus motif for phosphorylation by Akt, RX-R-X-X-pS/pT (Obata et al. 2000), as shown in Fig. 2(a). Scansite (Obenauer et al. 2003) identified the Ser373 site of Cx43 as a likely site for Akt phosphorylation, since it was within the top 2% (1.178%) of the potential Akt phosphorylation sites found in the protein data bank. We examined whether GST Cx43CT served as a substrate for Akt in in vitro reactions, using a GST-GSK3 fusion protein as a positive control. Purified Akt-1 phosphorylated GST-GSK3 and GST-Cx43CT, but not GST alone (Fig. 2(b)). Akt also phosphorylated the GST-Cx43CT S373A fusion protein (Fig. 2(b)), indicating the presence of an additional Akt target site on Cx43. A second site for Akt phosphorylation is predicted at Ser369 of Cx43 (a site within the top 4.35% of the potential Akt sites in the protein data bank, Fig. 2(a)). Although this is a less favorable site, it may be a target for Akt in vitro. The protein load was somewhat less for the GST-Cx43 protein than the S373A mutant (Fig. 2(b)). Normalization of the incorporation of 32P into the proteins to the protein load indicated that the S373A mutant was phosphorylated at ~80%, compared to the wild-type fusion protein. The close proximity of the Ser369 and Ser373 sites may not allow both of these sites to be well phosphorylated in the wild-type protein.
Figure 2.
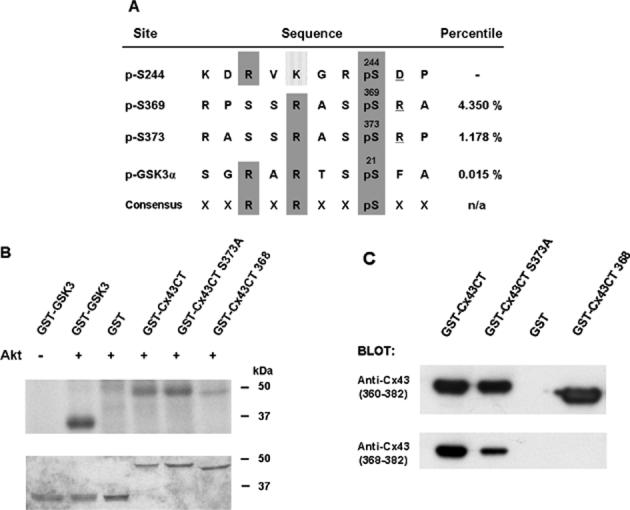
Cx43 is a substrate for Akt in vitro. (a) The alignment of potential Akt phosphorylation sites in Cx43 with a known Akt site in GSK3 and the consensus sequence for Akt phosphorylation. Scansite percentile scores, shown in the last column, represent substrates within the top percentage of potential Akt phosphorylation sites in the protein data bank. (b) Purified Akt phosphorylated GST-Cx43CT and GST-GSK3 (positive control), but not GST alone. Incorporation of radioactivity into the fusion proteins is shown in the upper part of the panel and the Coomassie stain of the gel is shown in the lower part of the panel. Akt also phosphorylated the GST-Cx43CT S373A mutant, consistent with a second target site for Akt at Ser369. Deletion of Ser368-Ile382 of Cx43 in the GST-Cx43CT mutant (GST-Cx43CT 368) significantly reduced phosphorylation by Akt. (c) The truncated mutant was recognized by a peptide antibody to residues 360-382, but not by a peptide antibody to residues 368-382 of Cx43, confirming the loss of the C-terminal amino acids in the fusion protein.
To confirm that these were the primary target sites for Akt in Cx43, we prepared a GST-Cx43CT fusion protein with deletion of the last fifteen amino acids (deletion of residues Ser368-Ile382, GST-Cx43CT 368). The truncated mutant migrated faster than wild-type GST-Cx43CT due to the loss of amino acids and was recognized by a peptide antibody to residues 360-382, but not by a peptide antibody to residues 368-382 of Cx43 (Fig. 2(c)). This indicated that the epitope recognized by the residues 360-382 antibody was in the amino-terminal portion of the peptide between Asp360-Ser368 of Cx43. The residues 368-382 antibody appeared to react less strongly with GST-Cx43CT S373A than with GST-Cx43CT (Fig. 2(c)), suggesting that the serine at position 373 may be part of the epitope recognized by this antibody. The GST-Cx43CT 368 mutant lacks the Ser373 and Ser369 sites and was not a good substrate for Akt (Fig. 2(b)). The minimal phosphorylation observed with this mutant in vitro may have occurred at Ser244, a site that has some homology with the consensus Akt phosphorylation sequence (Fig. 2(a)), but that is not identified as an Akt target by Scansite (Obernauer et al., 2002).
Akt and 14-3-3 θ Interact with Cx43 In Vivo
We examined whether Akt also targets Cx43 in vivo. Rat-1 fibroblasts were treated with EGF to activate intracellular signaling and induce Cx43 phosphorylation. Cx43 was immunoprecipitated from whole cell lysates (WCLs) prepared with a 0.5% NP-40 lysis buffer containing protease and phosphatase inhibitors (see Methods). This mild buffer was used to enrich native protein-protein interactions. However, 14-3-3 θ did not co-precipitate with Cx43 from the WCLs at the endogenous levels of protein expression (Fig. 3(a), left-hand panel). Since the phosphorylated isoforms of Cx43 are much less soluble than non-phosphorylated Cx43 (Musil and Goodenough 1991), we examined the proteins that remained in the 0.5% NP-40-insoluble membrane pellet, after solubilizing them in hot SDS sample buffer and then heating the samples. Cx43 was detected in the 0.5% NP-40-insoluble fraction (Fig. 3(a), right panel) and a protein in this fraction that migrated as Cx43 was recognized by both an Akt phosphorylated-substrate antibody and a phospho-Ser “mode 1” 14-3-3-binding site antibody (Fig. 3 (a)), as shown by stripping and reprobing the blot. Akt-3 was the isoform that was activated in the EGF-treated Rat-1 cells as demonstrated by the phosphorylation of Ser473 of Akt on a protein that migrated as Akt-3 and was recognized by an Akt-3 specific antibody (Fig. 3(b)). The data are consistent with the possibility of Akt-mediated phosphorylation of Cx43 in vivo on a phospho-Ser “mode-1” 14-3-3-binding site. 14-3-3 θ was also found in the 0.5% NP-40 insoluble fraction (Fig. 3(a)), consistent with a potential interaction with phosphorylated Cx43.
Figure 3.
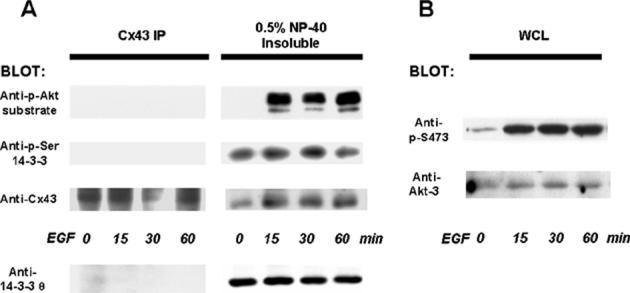
Cx43 is a substrate for Akt and for 14-3-3-binding in vivo. 14-3-3 θ was not detected in immunoprecipitates (IP) of Cx43 from EGF-treated Rat-1 cells lysed in a 0.5% NP-40 buffer, as shown by Western blotting (bottom blot on the left-hand side of (a)). Cx43 that remained in the 0.5% NP-40-insoluble fraction was solubilized with hot SDS sample buffer and was recognized on Western blots by an antibody to phospho-Ser “mode-1”14-3-3-binding sites and an antibody to phosphorylated Akt-substrates (right side of (a)), as shown by stripping and reprobing the blot. The protein bands recognized by the motif antibodies migrated in the same position as Cx43. 14-3-3 θ was also detected in the 0.5% NP-40 insoluble fraction (bottom blot on the right hand side of (a)). (b) Akt was activated in the EGF-treated cells, as shown by the phosphorylation of Ser473 of Akt on a protein in the whole cell lysate (WCL) that migrated as Akt-3 and was recognized by an Akt-3 specific antibody.
To determine whether Cx43 and Akt or Cx43 and 14-3-3 θ co-localize in vivo, we used immunofluorescence microscopy on EGF-treated Rat-1 cells. EGF induced a transient redistribution of Cx43 (stained in green) in the Rat-1 cells and an apparent co-localization with 14-3-3 θ (stained in red) as shown by the yellow-orange color in the merged images (lower panels in Fig. 4 (a)). Co-localization appeared to be diffuse and largely cytoplasmic. An apparent co-localization of Cx43 with Akt (stained in red) was also observed (upper panels in Fig. 4(a)). A more careful examination of protein co-localization by confocal microscopy provided further support for the co-localization of Cx43 with Akt and with 14-3-3 θ in the EGF-treated cells (Fig. 4(b)). Co-localization of Cx43 with 14-3-3 θ and to a lesser extent with Akt was also detected in the untreated cells, where a basal level of activated Akt was detected (see Fig. 3(b)).
Figure 4.
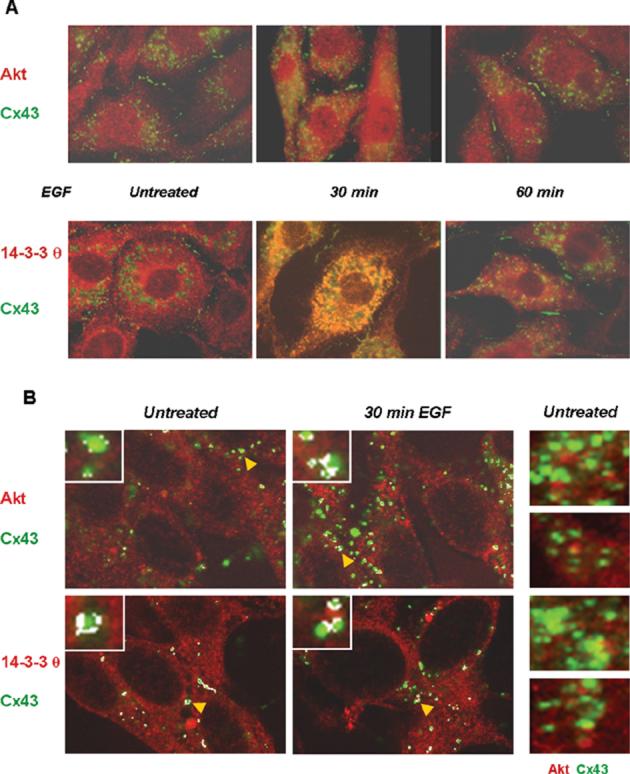
Cx43 co-localizes with Akt and with 14-3-3 θ in vivo. Untreated and EGF-treated (100 ng/mL) Rat-1 cells were immunostained for Cx43 (green) and Akt-3 (red) or 14-3-3 θ (red). (a) Apparent co-localization of the proteins was detected in the EGF-treated cells as shown by the yellow/orange color in the merged images. (b) Confocal microscopy images of the co-stained proteins. Here, the areas of most intense co-localization were highlighted by white pixelation using the confocal microscopy software. The insets in (b) show enlargements of randomly selected Cx43 gap junctional plaques. The arrowheads (yellow) point to the gap junctional plaques enlarged in the insets. Co-localization of Cx43 and Akt is also shown without the white pixilation by the yellow-orange color in the merged images shown on the right-hand side of (b) for untreated cells.
The areas of strongest co-localization were highlighted by white pixilation using the quantification option of the confocal software for the images shown on the left hand side of Fig. 4(b). The enlargements in the insets to Fig. 4(b) show co-localization of Cx43 with Akt or with 14-3-3 θ at the outer edges of gap junctional plaques. This was also apparent in the confocal images without the use of the white pixilation, as shown in the panels on the right hand side of Fig. 4(b). The yellow-orange color indicative of co-localization is found at the outer edges of the plaques. There are also some plaques where there appears to be little or no co-localization of Akt with Cx43 in the untreated cells. Co-localization of these proteins supports the possibility of Akt-mediated phosphorylation of Cx43 in the cells and the potential for an in vivo interaction of Cx43 with 14-3-3 θ.
Further evidence for the interaction of endogenously phosphorylated Cx43 with 14-3-3 θ was obtained in an overlay assay. These assays are usually carried out with radiolabeled or digoxygenin (DIG)-labeled 14-3-3 proteins, to provide the sensitivity that allows detection of a binding interaction of 14-3-3 to a denatured substrate immobilized on a membrane. We used unlabeled His-14-3-3 θ in these studies and were able to detect a binding interaction between His-14-3-3 θ and GST-Cx43CT (Fig. 5(a), lane 2 of the upper panel, marked by an arrowhead). This demonstrates that the interaction can take place on a surface, as well as in solution and that it is a direct interaction between the two proteins. His-14-3-3 θ also appeared to bind to Cx43 that was phosphorylated endogenously in EGF-treated Rat-1 cells, as shown in lane 1 (marked by an arrowhead). The blot was developed with an anti-His tag antibody using the ECL plus kit from Amersham for enhanced sensitivity, which also enhanced background. The membrane was then stripped and reprobed with an anti-Cx43 antibody using the regular ECL kit (lower panel in Fig. 5(a)). His-14-3-3 θ appeared to bind to the trailing edge of the GST-Cx43CT protein (Fig. 5(a)) on a 12% polyacrylamide SDS-PAGE gel. This may represent more phosphorylated, slower migrating forms of the fusion protein that have the appropriate phosphorylation for an interaction with 14-3-3. Although there is much less Cx43 protein in the lane containing the cell lysate, it appears to be a better substrate for an interaction with 14-3-3.
Figure 5.
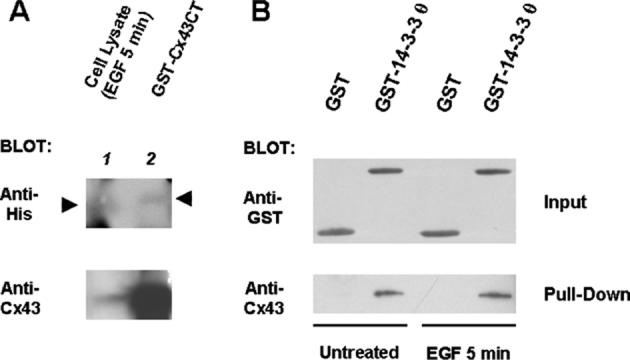
14-3-3 θ binds to Cx43 phosphorylated in vivo. (a) His-14-3-3 θ bound to the GST-Cx43CT fusion protein and to Cx43 phosphorylated endogenously in Rat-1 cells as demonstrated here in an overlay assay. Proteins were resolved on a 12% polyacrylamide SDS-PAGE gel and transferred to an Immobilon-P membrane that was probed with His-14-3-3 θ and developed with an anti-His tag antibody. (b) GST-14-3-3 θ, but not GST alone, bound to Cx43 in cell lysates from EGF-treated and untreated Rat-1 cells as shown here. The GST input proteins are shown in the upper blot and the pull-down of Cx43 in the lower blot.
Additional support for the interaction of Cx43 with 14-3-3 in vivo is provided in Fig. 5 (b). Cx43 that was phosphorylated endogenously in Rat-1 cells and then extracted from the cell membrane with a binding buffer containing 0.1% NP-40 and 20% glycerol with added protease and phosphatase inhibitors was recognized in solution and pulled down by a GST-14-3-3 θ fusion protein, but not by GST alone. Cx43 that was phosphorylated endogenously in either untreated or EGF-treated cells interacted with GST-14-3-3 θ in these GST pull-downs.
DISCUSSION
These studies demonstrate that Akt phosphorylates Cx43 on Ser369 and Ser373 and that an Akt-phosphorylated substrate motif antibody recognizes Cx43 from EGF-treated cells where Akt is activated. They also provide evidence for an interaction of full-length, endogenously phosphorylated Cx43 with 14-3-3 θ, in agreement with the interaction of the C-terminal domain of Cx43 with the NH2-terminal half of 14-3-3 θ that was identified in the yeast two-hybrid system (Jin 1998) and our previous in vitro binding studies with fusion proteins of the C-terminal domain of Cx43 (Park et al. 2006). Since a GST fusion protein containing the C-terminal domain of Cx45 (a connexin that does not have 14-3-3 binding sequences) and the CT of Cx43 with a Ser373Ala mutation did not bind to His-tagged 14-3-3 θ (Fig. 1(b)), the data support an interaction that is specific for Cx43 and that requires phosphorylation of Ser373 of Cx43. Previous studies (Park et al. 2006) demonstrated that GST Cx43CT also interacted with His-14-3-3 ζ and “mode-1” phosphorylated binding sites in 14-3-3 substrates are known to interact with other 14-3-3 isoforms (Yaffe et al. 1997).
Akt plays an important role in intracellular signaling pathways that are activated by an exposure to extracellular growth factors, as a downstream target of PI3 kinase, and regulates cell proliferation and survival, cell differentiation, and glucose homeostasis (Dummler and Hemmings, 2007). Akt is also constitutively activated in many tumor cells due to amplification of the p110α PI3K catalytic subunit or deletion/mutation of the PTEN (phosphatase and tensin homolog) gene that regulates PI3 kinase (Sansal and Sellers 2004). The identification of additional Akt substrates may provide some insight into cellular responses elicited by Akt activation. The sequences recognized by Akt and 14-3-3 overlap and Akt phosphorylates a number of proteins to promote their interactions with 14-3-3 (Kovacina et al. 2003). In our study, Akt phosphorylated Cx43 in vitro (Fig. 2 (b)) at the predicted target sites, Ser373 and Ser369 (Fig. 2(a)), and removal of these sites, in the truncated GST Cx43CT fusion protein, prevented effective phosphorylation by Akt (Fig. 2(b)). The identification of Akt-phosphorylated sites and phosphorylated “mode-1” 14-3-3-binding sites on Cx43 that was phosphorylated endogenously in EGF-treated Rat-1 cells provides further support for the interaction of these proteins in vivo (Fig. 3(a)) and lends credence to the possibility of Akt-mediated phosphorylation of Cx43 in vivo. Whether Akt-mediated phosphorylation or phosphorylation by another kinase such as PKA (Yogo et al. 2006; Giepmans 2004) promotes an interaction of Cx43 with 14-3-3 in vivo remains a subject for further study.
The validity of an interaction of Cx43 with 14-3-3 was also supported by the recognition of endogenously phosphorylated Cx43 in cell lysates from EGF-treated cells by a GST-14-3-3 fusion protein in GST pull-down studies, by an apparent binding interaction of Cx43 with 14-3-3 θ in an overlay assay (Fig. 5(b)), and by the co-localization of Cx43 with 14-3-3 θ by immunofluorescence microscopy (Fig. 4). In the overlay assay the endogenously phosphorylated Cx43 protein appeared to be a better substrate for 14-3-3 binding than the bacterially phosphorylated fusion protein. If the fusion protein was phosphorylated at the Ser369 or the Ser372 site, a PKC (protein kinase C) site (Saez et al. 1997) this would likely reduce phosphorylation at the adjacent Ser373 site. Phosphorylation of both Ser372 and Ser373 would sterically hinder binding of the Cx43 Ser373 peptide to 14-3-3, since the dually phosphorylated Cx43 peptide (phosphorylated at Ser372 and Ser373) could not be accommodated well in the 14-3-3 substrate binding pocket in our model of rat/mouse 14-3-3 θ (Park et al. 2006).
Previously we demonstrated that preventing the activation of Akt in cells pretreated with a PI3 kinase inhibitor did not prevent the disruption of Cx43 function that was induced by an exposure to LPA (lysophosphatidic acid) (Park et al. 2006). This suggested that an Akt-mediated interaction with 14-3-3 was not required for the growth factor-induced disruption of Cx43 function. The confocal images of Rat-1 cells co-stained for Cx43 and 14-3-3 θ or with Cx43 and Akt provided several clues to a possible functional significance of this interaction, since the proteins were co-localized at the periphery of gap junctional plaques (Fig. 4(b)). This is where newly synthesized Cx43 is reported to be incorporated into existing gap junctional plaques (Gaietta et al. 2002). Although we cannot rule out the possibility that steric hindrance prevented the interaction with Akt or 14-3-3 at the center of the plaques, we do not think this was the case. The Cx43 antibody had access to Cx43 throughout the plaque and the co-localization with 14-3-3 and Akt was only detected at the periphery. The data would suggest that these interactions with Cx43 are taking place either “concomitant with” or “prior to” the insertion of Cx43 hemichannels into the plasma membrane and their subsequent incorporation into gap junctional plaques. Recent studies by Shaw et al. (2007) support a targeted microtubule-mediated delivery of newly synthesized Cx43 to adherens junctions at regions of cell-cell contact, where gap junctional plaques are located.
Co-localization of Cx43 with 14-3-3 θ was detected in untreated Rat-1 cells as well as in EGF-treated cells, consistent with a more constitutive role for the interaction. This was demonstrated by the presence of the phospho-Ser 14-3-3-binding motif on Cx43 from both untreated and EGF-treated cells (Fig. 3(a)), the apparent binding of His-14-3-3 θ to Cx43 in whole cell lysates obtained from untreated and EGF-treated cells in an overlay assay (Fig. 5(a) and data not shown), and by the co-localization of the proteins in untreated and EGF-treated cells (Fig. 4 (b), lower panels). Importantly, the validity of the interaction is strongly supported by data demonstrating that endogenously phosphorylated Cx43 bound to 14-3-3 θ in the overlay assay and in GST pull-downs. Many studies of the binding interactions of 14-3-3 proteins have been carried out in cells that over-express 14-3-3 and the reputed binding partner. Over-expression of the proteins and/or expression in compartments where they might not normally be expressed could suggest interactions that may not occur at normal levels of protein expression. The interaction of Cx43 with 14-3-3 θ at the endogenous levels of protein expression and with protein phosphorylation that occurred in live cells provides more convincing evidence for an interaction in vivo.
Cx43 is widely expressed in different tissue types and has been shown to have a critical role in the development and physiology of the heart, in male fertility, and in the control of body size (Plum et al. 2000). The interaction with 14-3-3 proteins appears to be Cx43-specific and may be an example of a connexin regulatory step that provides added quality control during connexin processing that is not generalized to other connexins. A possible function for this interaction is in the assembly of Cx43 monomers into oligomers in a manner that may be monitored by a mechanism that allows for the escape of the Cx43 multimers from the endoplasmic reticulum (ER), following the binding of 14-3-3 to one or more phosphorylated sites on Cx43. Arginine-based sorting signals (R-X-R) can mediate ER-localization of a protein until they are masked by the proof reading of multimeric complexes by 14-3-3 proteins (Michelsen et al. 2005). This has been demonstrated for the major histocompatibility complex (MHC) invariant chain (Khalil et al. 2003; Kuwana et al. 1998) and for the Kir6.2 KATP channel protein (Nufer and Hauri 2003; O'Kelly et al. 2002; Yoo et al. 2005; Yuan et al. 2003). The R-P-R sequence at Arg374-Pro-Arg376 is adjacent to Ser373 of Cx43 and the phosphorylation of Ser373 and a subsequent interaction with 14-3-3 may mask an ER retention signal and allow for the forward trafficking of Cx43 and the escape of multimers from the ER.
It is also of interest to note that interactions with PDZ domain-containing proteins have been shown to mask an Arg-based signal at the C-terminus of the NR1-3 subunit of the N-methyl-D-aspartate receptor (Standley et al. 2000). Cx43 as well as Cx40, Cx45, Cx46, and Cx50 interact at their C-termini with the PDZ domain-containing protein, ZO-1 (Giepmans and Moolenaar 1998; Jin et al. 2004; Laing et al. 2001; Nielsen et al. 2003). Cx45 also has a dibasic motif near the C-terminal domain that could act as an ER retention signal and Cx45 also interacts with ZO-1. Thus, more than one mechanism for proofreading the formation of multimers may exist for Cx43 and for other connexins. In the case of Cx43, it seems likely that an interaction with 14-3-3 at the phosphorylated Ser373 site and an interaction with PDZ domain-containing proteins at flanking sequences would be mutually exclusive, due to steric considerations.
Some activated Akt was detected in untreated cells and the confocal images in Fig. 4(b) demonstrate the physical proximity of Cx43 and Akt in the untreated cells. As demonstrated by the appearance of the phosphorylated Akt-substrate motif on a protein that migrated as Cx43 (Fig. 3(a)), the interaction of Cx43 with Akt appeared to be enhanced in Rat-1 cells by a treatment with EGF and Akt activation. The motif antibody to “mode-1” phospho-Ser 14-3-3-binding sites recognized Cx43 from both untreated and EGF-treated Rat-1 cells (Fig. 3(a)) and Cx43 was also co-localized with 14-3-3 θ in untreated and EGF-treated cells (Fig. 4(b)). Although an extensive analysis of protein co-localization was not carried out as a part of this study, an exposure to EGF appeared to significantly enhance the co-localization of Cx43 with Akt, while the co-localization of Cx43 with 14-3-3 appeared to be more constitutive.
Interactions of Cx43 with 14-3-3 dimers could occur in an inter- or intra-molecular manner. 14-3-3 dimers could cross-link Cx43 monomers within a connexon by binding to two phospho-Ser373 sites on different molecules, or cross-link Cx43 molecules in adjacent connexons. If the phospho-Ser244 site, which does not support a binding interaction on its own, were able to bind to a second 14-3-3 monomer through an induced proximity following the binding of one monomer to phospho-Ser373, this could result in an intra-molecular interaction (Yaffe 2002). 14-3-3 could also serve as a molecular scaffold to bring another protein to Cx43. Both PKCγ and Erk5 have been shown to interact with 14-3-3 in a manner that regulates their function (Nguyen et al. 2004; Zheng et al. 2004) and these kinases interact with and phosphorylate Cx43 (Cameron et al. 2003; Nguyen et al. 2003).
A number of kinases that target Cx43 and their target sites have been identified (Lampe and Lau 2004; Warn-Cramer and Lau 2004) and in some cases, specific antibody reagents have been developed to allow assessment of the phosphorylation state of Cx43 in vivo. Earlier studies by Yogo et al. (2006) implicated the Ser265, Ser368, Ser369, and Ser373 sites of Cx43 as targets for PKA in rat granulosa cells stimulated with follicle-stimulating hormone (FSH). The studies reported here support a role for Akt as an additional Ser/Thr kinase that can target Cx43 in the C-terminal domain, at the Ser373 and Ser369 sites and provide evidence that Cx43 phosphorylated endogenously is recognized by an antibody to Akt-phosphorylated substrates. They also suggest that Cx43 interacts with 14-3-3 in vivo, an interaction that might be involved in the forward trafficking of Cx43 multimers by masking an Arg-based ER retention signal near Ser373. A recent study by Thomas et al. (2005) has demonstrated differences in the biogenesis of Cx43 and Cx26. These connexins have common secretory pathways and common intermediates, however they differ in their dependence on microtubules for transport to the plasma membrane and in their mobilities within the membrane. These authors suggested that some of these differences might be due to differences in the binding of these connexins to scaffold proteins.
An interaction with 14-3-3 proteins may also alter Cx43's association with other interacting proteins, such as ZO-1 (which interacts at the C-terminus of Cx43, near Ser373 (Giepmans and Moolenaar 1998; Jin et al. 2004; Laing et al. 2001)), CIP85 (which interacts with a proline-rich region (Pro253-Pro256 (Lan et al. 2005)) near a second possible site for 14-3-3 interaction, Ser244), or tubulin or CIP150 (which interact with the juxtamembrane region of the CT domain of Cx43 (Akiyama et al. 2005; Giepmans et al. 2001b) near Ser244). Cx43 trafficking appears to be complex, and, as recent studies have shown, involves a number of proteins and the transport of Cx43 multimers along microtubules to adherens junctions, prior to an incorporation into existing gap junctional plaques (Shaw et al. 2007). The studies presented here contribute to an understanding of how Cx43 is regulated by phosphorylation and phosphorylation-dependent protein interactions and they highlight the need for additional studies of the potential role for Cx43's interaction with 14-3-3 in Cx43 trafficking.
Acknowledgments
Supported by P20-RR016453 (R. Shohet, PI; BJW-C, Project PI), a grant from the Hawaii Community Foundation (BJW-C, PI), an equipment grant from the HS-BRIN (RR16467, to BJW-C), a Predoctoral Fellowship from the American Heart Association, Hawaii Affiliate (to CJ), and CA052098 (AFL, PI). The authors thank Dr. Eileen Freed for the 14-3-3 θ cDNA; Carrie Guyette and Anne Hernandez for technical assistance; graduate student Kelsie Takasaki; and undergraduates Dawn Matsui, Erica Nolte, Caroline Lai, Absalon Galat, and Dennis Hidalgo for their assistance during this study. We also thank Mr. Frank Lei (Leica Microsystems) for expert assistance in obtaining the confocal images.
REFERENCES
- Ai Z, Fischer A, Spray DC, Brown AM, Fishman GI. Wnt-1 regulation of connexin43 in cardiac myocytes. J Clin Invest. 2000;105:161–171. doi: 10.1172/JCI7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama M, Ishida N, Ogawa T, Yogo K, Takeya T. Molecular cloning and functional analysis of a novel Cx43 partner protein CIP150. Biochem Biophys Res Commun. 2005;335:1264–1271. doi: 10.1016/j.bbrc.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Bartel PL, Chien C, Stenglanz R, Field S. Using the two-hybrid system to detect protein-protein interactions. In. In: Hartley DA, editor. Cellular interactions in development: A practical approach. Oxford University Press; New York: 1993. pp. 153–179. [Google Scholar]
- Bridges D, Moorhead GB. 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE. 20042004:re10. doi: 10.1126/stke.2422004re10. [DOI] [PubMed] [Google Scholar]
- Butkevich E, Hulsmann S, Wenzel D, Shirao T, Duden R, Majoul I. Drebrin is a novel connexin-43 binding partner that links gap junctions to the submembrane cytoskeleton. Curr Biol. 2004;14:650–658. doi: 10.1016/j.cub.2004.03.063. [DOI] [PubMed] [Google Scholar]
- Cameron SJ, Malik S, Akaike M, Lerner-Marmarosh N, Yan C, Lee JD, Abe J, Yang J. Regulation of epidermal growth factor-induced connexin 43 gap junction communication by big mitogen-activated protein kinase1/ERK5 but not ERK1/2 kinase activation. J Biol Chem. 2003;278:18682–18688. doi: 10.1074/jbc.M213283200. [DOI] [PubMed] [Google Scholar]
- Chaudhri M, Scarabel M, Aitken A. Mammalian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo. Biochem Biophys Res Commun. 2003;300:679–685. doi: 10.1016/s0006-291x(02)02902-9. [DOI] [PubMed] [Google Scholar]
- Cline K, Mori H. Thylakoid DeltapH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J Cell Biol. 2001;154:719–729. doi: 10.1083/jcb.200105149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Saier MH., Jr. Ser/Thr/Tyr protein phosphorylation in bacteria - for long time neglected, now well established. J Mol Microbiol Biotechnol. 2005;9:125–131. doi: 10.1159/000089641. [DOI] [PubMed] [Google Scholar]
- Duffy HS, Delmar M, Spray DC. Formation of the gap junction nexus: binding partners for connexins. J Physiol Paris. 2002;96:243–249. doi: 10.1016/s0928-4257(02)00012-8. [DOI] [PubMed] [Google Scholar]
- Dummler B, Hemmings BA. Physiological roles of PKB/Akt isoforms in development and disease. Biochem Soc Trans. 2007;35:231–235. doi: 10.1042/BST0350231. [DOI] [PubMed] [Google Scholar]
- Fu CT, Bechberger JF, Ozog MA, Perbal B, Naus CC. CCN3 (NOV) interacts with connexin43 in C6 glioma cells: possible mechanism of connexin-mediated growth suppression. J Biol Chem. 2004;279:36943–36950. doi: 10.1074/jbc.M403952200. [DOI] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Gellhaus A, Dong X, Propson S, Maass K, Klein-Hitpass L, Kibschull M, Traub O, Willecke K, Peral B, Lye SJ, Winterhager E. Connexin43 interacts with NOV: a possible mechanism for negative regulation of cell growth in choriocarcinoma cells. J Biol Chem. 2004;279:36931–36942. doi: 10.1074/jbc.M404073200. [DOI] [PubMed] [Google Scholar]
- Giepmans BN. Gap junctions and connexin-interacting proteins. Cardiovasc Res. 2004;62:233–245. doi: 10.1016/j.cardiores.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Hengeveld T, Postma FR, Moolenaar WH. Interaction of c-Src with gap junction protein connexin-43. Role in the regulation of cell-cell communication. J Biol Chem. 2001a;276:8544–8549. doi: 10.1074/jbc.M005847200. [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Moolenaar WH. The gap junction protein connexin43 interacts with the second PDZ domain of the zona occludens-1 protein. Curr Biol. 1998;8:931–934. doi: 10.1016/s0960-9822(07)00375-2. [DOI] [PubMed] [Google Scholar]
- Giepmans BN, Verlaan I, Hengeveld T, Janssen H, Calafat J, Falk MM, Moolenaar WH. Gap junction protein connexin-43 interacts directly with microtubules. Curr Biol. 2001b;11:1364–1368. doi: 10.1016/s0960-9822(01)00424-9. [DOI] [PubMed] [Google Scholar]
- Hertig CM, Butz S, Koch S, Eppenberger-Eberhardt M, KemLer R, Eppenberger HM. N-cadherin in adult rat cardiomyocytes in culture. II. Spatio-temporal appearance of proteins involved in cell-cell contact and communication. Formation of two distinct N-cadherin/catenin complexes. J Cell Sci. 1996;109(Pt 1):11–20. doi: 10.1242/jcs.109.1.11. [DOI] [PubMed] [Google Scholar]
- Herve JC, Bourmeyster N, Sarrouilhe D. Diversity in protein-protein interactions of connexins: emerging roles. Biochim Biophys Acta. 2004;1662:22–41. doi: 10.1016/j.bbamem.2003.10.022. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Zhang P, Armstrong RN, Gilliland GL. The threedimensional structure of a glutathione S-transferase from the mu gene class. Structural analysis of the binary complex of isoenzyme 3-3 and glutathione at 2.2-A resolution. Biochemistry. 1992;31:10169–10184. doi: 10.1021/bi00157a004. [DOI] [PubMed] [Google Scholar]
- Jin C. Doctoral Dissertation. University of Hawaii; Manoa: 1998. Identification and characterization of Cx43-interacting proteins. [Google Scholar]
- Jin C, Lau AF, Martyn KD. Identification of connexin-interacting proteins: application of the yeast two-hybrid screen. Methods. 2000;20:219–231. doi: 10.1006/meth.1999.0939. [DOI] [PubMed] [Google Scholar]
- Jin C, Martyn KD, Kurata WE, Warn-Cramer BJ, Lau AF. Connexin43 PDZ2 binding domain mutants create functional gap junctions and exhibit altered phosphorylation. Cell Commun Adhes. 2004;11:67–87. doi: 10.1080/15419060490951781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DH, Ley S, Aitken A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 1995;368:55–58. doi: 10.1016/0014-5793(95)00598-4. [DOI] [PubMed] [Google Scholar]
- Kanemitsu MY, Loo LW, Simon S, Lau AF, Eckhart W. Tyrosine phosphorylation of connexin 43 by v-Src is mediated by SH2 and SH3 domain interactions. J Biol Chem. 1997;272:22824–22831. doi: 10.1074/jbc.272.36.22824. [DOI] [PubMed] [Google Scholar]
- Khalil H, Brunet A, Saba I, Terra R, Sekaly RP, Thibodeau J. The MHC class II beta chain cytoplasmic tail overcomes the invariant chain p35-encoded endoplasmic reticulum retention signal. Int Immunol. 2003;15:1249–1263. doi: 10.1093/intimm/dxg124. [DOI] [PubMed] [Google Scholar]
- Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, Roth RA. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem. 2003;278:10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Peterson PA, Karlsson L. Exit of major histocompatibility complex class II-invariant chain p35 complexes from the endoplasmic reticulum is modulated by phosphorylation. Proc Natl Acad Sci USA. 1998;95:1056–1061. doi: 10.1073/pnas.95.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing JG, Manley-Markowski RN, Koval M, Civitelli R, Steinberg TH. Connexin45 interacts with zonula occludens-1 and connexin43 in osteoblastic cells. J Biol Chem. 2001;276:23051–23055. doi: 10.1074/jbc.M100303200. [DOI] [PubMed] [Google Scholar]
- Lampe PD, Lau AF. The effects of connexin phosphorylation on gap junctional communication. Int J Biochem Cell Biol. 2004;36:1171–1186. doi: 10.1016/S1357-2725(03)00264-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Z, Kurata WE, Martyn KD, Jin C, Lau AF. Novel rab GAP-like protein, CIP85, interacts with connexin43 and induces its degradation. Biochemistry. 2005;44:2385–2396. doi: 10.1021/bi048306w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kurata WE, Jin C, Lau AF. A novel connexin43-interacting protein, CIP75, that belongs to the Ubl-UBA protein family and regulates the turnover of connexin. 43 doi: 10.1074/jbc.M709288200. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Warn-Cramer BJ, Kurata WE, Lau AF. v-Src phosphorylation of connexin 43 on Tyr247 and Tyr265 disrupts gap junctional communication. J Cell Biol. 2001;154:815–827. doi: 10.1083/jcb.200102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo LW, Berestecky JM, Kanemitsu MY, Lau AF. pp60srcmediated phosphorylation of connexin 43, a gap junction protein. J Biol Chem. 1995;270:12751–12761. doi: 10.1074/jbc.270.21.12751. [DOI] [PubMed] [Google Scholar]
- Macek B, Mijakovic I, Olsen JV, Gnad F, Kumar C, Jensen PR, Mann M. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol Cell Proteomics. 2007;6:697–707. doi: 10.1074/mcp.M600464-MCP200. [DOI] [PubMed] [Google Scholar]
- Michelsen K, Yuan H, Schwappach B. Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Re.p. 2005;6:717–722. doi: 10.1038/sj.embor.7400480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller DJ, Hand GM, Engel A, Sosinsky GE. Conformational changes in surface structures of isolated connexin 26 gap junctions. Embo J. 2002;21:3598–3607. doi: 10.1093/emboj/cdf365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musil LS, Goodenough DA. Biochemical analysis of connexin43 intracellular transport, phosphorylation, and assembly into gap junctional plaques. J Cell Biol. 1991;115:1357–1374. doi: 10.1083/jcb.115.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- Nagasawa K, Chiba H, Fujita H, Kojima T, Saito T, Endo T, Sawada N. Possible involvement of gap junctions in the barrier function of tight junctions of brain and lung endothelial cells. J Cell Physiol. 2006;208:123–132. doi: 10.1002/jcp.20647. [DOI] [PubMed] [Google Scholar]
- Nguyen TA, Boyle DL, Wagner LM, Shinohara T, Takemoto DJ. LEDGF activation of PKC gamma and gap junction disassembly in lens epithelial cells. Exp Eye Res. 2003;76:565–572. doi: 10.1016/s0014-4835(03)00049-6. [DOI] [PubMed] [Google Scholar]
- Nguyen TA, Takemoto LJ, Takemoto DJ. Inhibition of gap junction activity through the release of the C1B domain of protein kinase Cgamma (PKCgamma) from 14-3-3: identification of PKCgamma-binding sites. J Biol Chem. 2004;279:52714–52725. doi: 10.1074/jbc.M403040200. [DOI] [PubMed] [Google Scholar]
- Nielsen PA, Baruch A, Shestopalov VI, Giepmans BN, Dunia I, Benedetti EL, Kumar NM. Lens connexins alpha3Cx46 and alpha8Cx50 interact with zonula occludens protein-1 (ZO-1) Mol Biol Cell. 2003;14:2470–2481. doi: 10.1091/mbc.E02-10-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nufer O, Hauri HP. ER export: call 14-3-3. Curr Biol. 2003;13:R391–393. doi: 10.1016/s0960-9822(03)00318-x. [DOI] [PubMed] [Google Scholar]
- O'Kelly I, Butler MH, Zilberberg N, Goldstein SA. Forward transport. 14-3-3 binding overcomes retention in endoplasmic reticulum by dibasic signals. Cell. 2002;111:577–588. doi: 10.1016/s0092-8674(02)01040-1. [DOI] [PubMed] [Google Scholar]
- Obata T, Yaffe MB, Leparc GG, Piro ET, Maegawa H, Kashiwagi A, Kikkawa R, Cantley LC. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J Biol Chem. 2000;275:36108–36115. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteomewide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottmann C, Marco S, Jaspert N, Marcon C, Schauer N, Weyand M, Vandermeeren C, Duby G, Boutry M, Wittinghofer A, et al. Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol Cell. 2007;25:427–440. doi: 10.1016/j.molcel.2006.12.017. [DOI] [PubMed] [Google Scholar]
- Park DJ, Freitas TA, Wallick CJ, Guyette CV, Warn-Cramer BJ. Molecular dynamics and in vitro analysis of Connexin43: A new 14-3-3 mode-1 interacting protein. Protein Sci. 2006;15:2344–2355. doi: 10.1110/ps.062172506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MW, Lo Bello M, Federici G. Crystallization of glutathione S-transferase from human placenta. J Mol Biol. 1990;213:221–222. doi: 10.1016/s0022-2836(05)80183-4. [DOI] [PubMed] [Google Scholar]
- Plum A, Hallas G, Magin T, Dombrowski F, Hagendorff A, Schumaker B, Wolpert C, Kim J, Lamers WH, Evert M, et al. Unique and shared functions of different connexins in mice. Curr Biol. 2000;10:1083–1091. doi: 10.1016/s0960-9822(00)00690-4. [DOI] [PubMed] [Google Scholar]
- Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez JC, Nairn AC, Czernik AJ, Fishman GI, Spray DC, Hertzberg EL. Phosphorylation of connexin43 and the regulation of neonatal rat cardiac myocyte gap junctions. J Mol Cell Cardiol. 1997;29:2131–2145. doi: 10.1006/jmcc.1997.0447. [DOI] [PubMed] [Google Scholar]
- Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22:2954–2963. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- Schagger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Shaw RM, Fay AJ, Puthenveedu MA, von Zastrow M, Jan YN, Jan LY. Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell. 2007;128:547–560. doi: 10.1016/j.cell.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YH, Godlewski J, Bronisz A, Zhu J, Comb MJ, Avruch J, Tzivion G. Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Mol Biol Cell. 2003;14:4721–4733. doi: 10.1091/mbc.E02-12-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky GE, Nicholson BJ. Structural organization of gap junction channels. Biochim Biophys Acta. 2005;1711:99–125. doi: 10.1016/j.bbamem.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Standley S, Roche KW, McCallum J, Sans N, Wenthold RJ. PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron. 2000;28:887–898. doi: 10.1016/s0896-6273(00)00161-6. [DOI] [PubMed] [Google Scholar]
- Thomas T, Jordan K, Simek J, Shao Q, Jedeszko C, Walton P, Laird DW. Mechanisms of Cx43 and Cx26 transport to the plasma membrane and gap junction regeneration. J Cell Sci. 2005;118:4451–4462. doi: 10.1242/jcs.02569. [DOI] [PubMed] [Google Scholar]
- Toyofuku T, Akamatsu Y, Zhang H, Kuzuya T, Tada M, Hori M. c-Src regulates the interaction between connexin-43 and ZO-1 in cardiac myocytes. J Biol Chem. 2001;276:1780–1788. doi: 10.1074/jbc.M005826200. [DOI] [PubMed] [Google Scholar]
- Toyofuku T, Yabuki M, Otsu K, Kuzuya T, Hori M, Tada M. Direct association of the gap junction protein connexin-43 with ZO-1 in cardiac myocytes. J Biol Chem. 1998;273:12725–12731. doi: 10.1074/jbc.273.21.12725. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang L, Liddington R, Fu H. Mutations in the hydrophobic surface of an amphipathic groove of 14-3-3zeta disrupt its interaction with Raf-1 kinase. J Biol Chem. 1998;273:16297–16304. doi: 10.1074/jbc.273.26.16297. [DOI] [PubMed] [Google Scholar]
- Warn-Cramer BJ, Lau AF. Regulation of gap junctions by tyrosine protein kinases. Biochim Biophys Acta. 2004;1662:81–95. doi: 10.1016/j.bbamem.2003.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker E, Yaffe MB. 14-3-3 Proteins-a focus on cancer and human disease. J Mol Cell Cardiol. 2004;37:633–642. doi: 10.1016/j.yjmcc.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Yaffe MB. How do 14-3-3 proteins work?- Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Yogo K, Ogawa T, Akiyama M, Ishida-Kitagawa N, Sasada H, Sato E, Takeya T. PKA implicated in the phosphorylation of Cx43 induced by stimulation with FSH in rat granulosa cells. J Reprod Dev. 2006;52:321–328. doi: 10.1262/jrd.17107. [DOI] [PubMed] [Google Scholar]
- Yoo D, Fang L, Mason A, Kim BY, Welling PA. A phosphorylation-dependent export structure in ROMK (Kir 1.1) channel overrides an endoplasmic reticulum localization signal. J Biol Chem. 2005;280:35281–35289. doi: 10.1074/jbc.M504836200. [DOI] [PubMed] [Google Scholar]
- Yuan H, Michelsen K, Schwappach B. 14-3-3 dimers probe the assembly status of multimeric membrane proteins. Curr Biol. 2003;13:638–646. doi: 10.1016/s0960-9822(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Yin G, Yan C, Cavet M, Berk BC. 14-3-3beta binds to big mitogen-activated protein kinase 1 (BMK1/ERK5) and regulates BMK1 function. J Biol Chem. 2004;279:8787–8791. doi: 10.1074/jbc.M310212200. [DOI] [PubMed] [Google Scholar]


