Abstract
Leucascandrolide A and neopeltolide are structurally homologous marine natural products that elicit potent antiproliferative profiles in mammalian cells and yeast. The scarcity of naturally available material provided a significant barrier for their biochemical and pharmacological evaluation. We developed practical synthetic access to this class of natural products that enabled the determination of their mechanism of action. We demonstrated effective cellular growth inhibition in yeast, which was significantly enhanced by substituting glucose with galactose or glycerol. These results along with genetic analysis of determinants of drug sensitivity suggested that leucascandrolide A and neopeltolide may inhibit mitochondrial ATP synthesis. Evaluation of the activity of the four mitochondrial electron-transport chain complexes inyeast and mammalian cells revealed cytochrome bc1 complex as the principal cellular target. This result provided the molecular basis for the potent antiproliferative activity of this class of marine macrolides identifying them as new biochemical tools for investigation of eukaryotic energy metabolism.
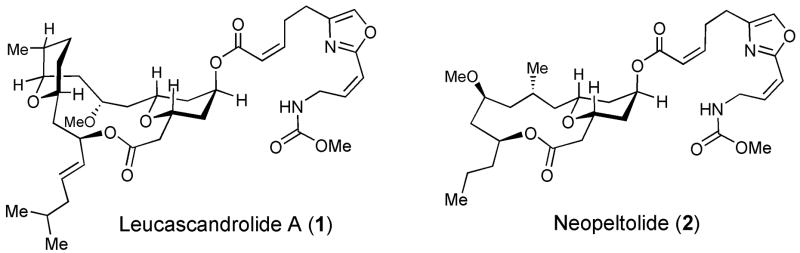
Recent progress in bioassay-guided isolation and structure elucidation of natural products has placed increasing demands on the efficiency of modern organic synthesis, which is required to provide practical access to rare and biosynthetically inaccessible compounds in order to enable their biological and pharmacological evaluation en route to the ultimate development of new therapeutic agents.[1] In 1996, the search for new antiproliferative marine natural products resulted in the discovery of leucascandrolide A (1, Fig. 1), which was isolated from Leucascandra caveolata sponge and proved highly effective at suppressing growth of KB and P388 tumor cells, as well as the pathogenic yeast Candida albicans.[2] Isolation of neopeltolide (2, Fig. 1) from Daedalopelta sponge and a similarly potent antiproliferative profile of this marine macrolide were reported in 2007.[3] Leucascandrolide A (1) and neopeltolide (2) share several notable structural features including the trisubstitited tetrahydropyran subunit and an identical, highly unsaturated, oxazole-containing side-chain, suggesting that the two natural products may inhibit cellular proliferation by a similar mechanism. The scarcity of the naturally available material, however, has provided a significant barrier to establishing the mode of action of either natural product. Indeed, efforts to reisolate leucascandrolide A from its natural source have failed, perhaps reflecting that this polyketide-based natural product is produced by a bacterial symbiote rather than the sponge itself.[2] Thus, a significant synthetic effort has been made in developing chemical approaches for preparation of these complex natural products.[4–18] Initial efforts confirmed the proposed structure of leucascandrolide A (1)[4] and revised the relative stereochemical assignment of neopeltolide (2),[15,16] but the difficulty of preparing sufficient synthetic material has slowed discovery of molecular target(s).
Figure 1.
Structures of antiproliferative marine natural products leucascandrolide A (1) and neopeltolide (2) isolated from Leucascandra caveolata and Daedalopelta sponges, respectively.
In this article, we describe elucidation of the mechanism of action of this class of marine natural products, which was enabled by practical chemical syntheses of a simplified congener of leucascandrolide A (3) and a recently revised structure of neopeltolide (2). Subsequent cell-based, genetics and biochemical studies of the two compounds in both mammalian cells and the brewers yeast Saccharomyces cerevisiae unambiguously established cytochrome bc1 complex as the principal cellular target ofthis class of highly potent antiproliferative marine macrolides.
RESULTS
Synthesis of a simplified congener of leucascandrolide A
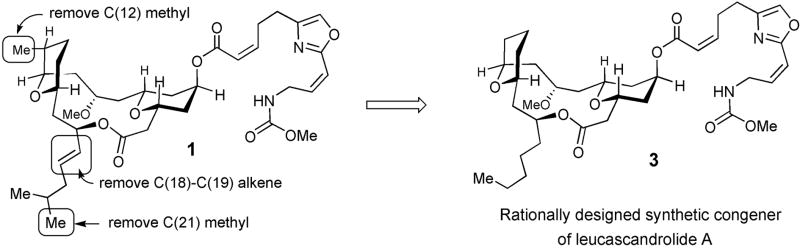
Evaluation of the opposite enantiomers of leucascandrolide A (1)[17] in a number of cancer cell lines and S. cerevisiae revealed that the naturally occurring (+)-leucascandrolide A proved to be only 2–3 fold more potent than the unnatural enantiomer (Supplementary Fig. 1). This observation, verified by additional structure-activity relationship studies,[19] indicated that the oxazole-containing side chain may be primarily responsible for the toxicity of leucascandrolide A and that the macrolide portion might be rationally simplified without affecting activity. Conformational analysis indicated that the axial C(12) methyl group (Fig. 2) could be eliminated without affecting the conformation of the tricyclic macrolide subunit. Furthermore, the C(17)–C(22) alkenyl moiety could be further simplified to a linear alkyl chain. However, removal of this alkyl group or inversion of the relative configuration at the C(17) resulted in complete loss of biological activity.[19] This analysis suggested a revised target 3 in which the structural simplification offered significant improvement of overall synthetic efficiency. Taking into account the similar activity of either enantiomer, synthesis of analog 3 was designed to deliver the final target in racemic form.
Figure 2.
Design of a simplified leucascandrolide A congener (3).
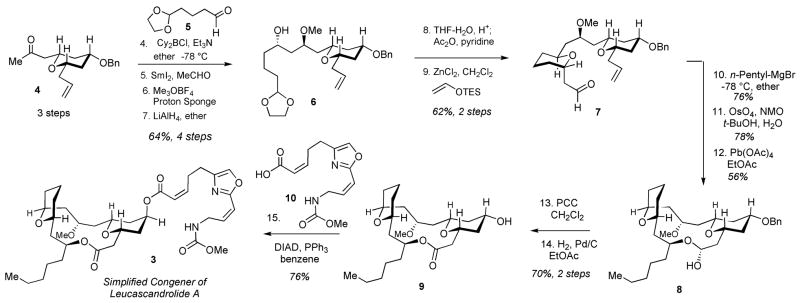
The synthesis of the engineered leucascandrolide A congener 3 is shown in Scheme 1. The approach followed our previously developed strategy[6,17] beginning with the Paterson-Evans 1,5-anti selective aldol condensation[20,21] of ketone 4 with aldehyde 5, SmI2-mediated reduction, methylation and reductive removal of the acetate to give alcohol 6. Deprotection of the dioxolane, acetylation of the resulting lactol, followed by C-glycosidation furnished aldehyde 7 with excellent overall efficiency and stereocontrol. Incorporation of the required alkyl fragment was achieved via the addition of n-pentyl magnesium bromide to aldehyde 7. Although both diastereomeric alcohols were obtained in equal amounts, the undesired diastereomer was readily separated by column chromatography and inverted via Mitsunobu reaction (not shown) to give the desired alcohol in 76% overall yield. Dihydroxylation, followed by Pb(OAc)4 cleavage of the vicinal diol afforded macrolactol 8 as a result of a spontaneous macrocyclization, which was originally observed during our synthesis of the parent natural product.[6] Subsequent oxidation, hydrogenolysis of the benzyl ether and final Mitsunobu esterification with oxazole-containing acid 10[6] afforded the simplified congener of leucascandrolide A (3) with the longest linear sequence of 15 steps and a total of 24 steps. As expected, due to significant simplification of the natural product’s structure, the synthetic efficiency for the assembly of 3 compared favorably to our original synthesis of leucascandrolide A (1), which required a total of 34 steps with 18 steps in the longest linear sequence.[6,17]
Scheme 1.
Synthesis of neopeltolide A employing a common strategy
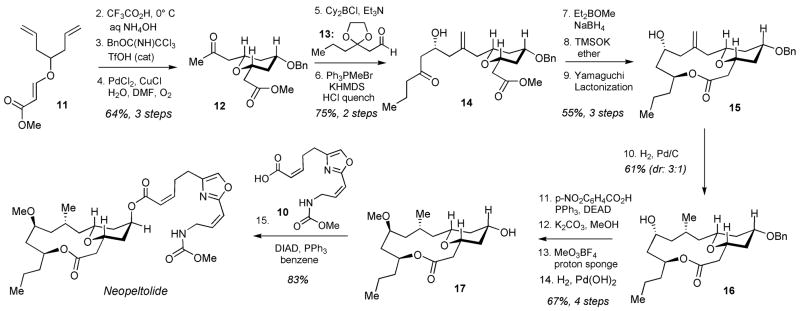
The synthesis of neopeltolide (2) is shown in Scheme 2. The assembly process began with the Prins desymmetrization[18] of diene 11, followed by benzyl protection and Wacker alkene oxidation to give ketone 12. Generation of boron enolate from ketone 12, followed by addition of aldehyde 13 (for preparation, see Supplementary Methods) gave the expected aldol product with excellent diastereoselectivity (>98:2),[20,21] which was subjected to Wittig methylenation, followed by acidic removal of the dioxolane during the work-up to give ketone 14 in 75% yield for the two operations. 1,3-Syn-selective reduction [Et2BOMe, NaBH4],[22] followed by saponification of ester with TMSOK gave the diol, which was subjected to the Yamaguchi macrolactonization to successfully assemble macrolide 15. As we expected, the macrocyclization proceeded with complete chemoselectivity to give the desired 14-membered macrolide in preference of the significantly more strained 12-membered lactone. Hydrogenation of alkene 15 using Pd/C catalyst proceeded with 3:1 diastereoselectivity to give the alcohol 16 as the major product isolated in 61% yield. Inversion of alcohol 16 was efficiently accomplished under the Mitsunobu conditions, followed by hydrolysis of the resulting p-nitrophenyl benzoate with K2CO3 in MeOH. Subsequent methylation of the alcohol with MeO3BF4 and hydrogenolysis of the benzyl ether afforded fully functionalized macrolide 17. The final step entailed coupling of two fragments 10[6] and 17, which was conducted under our standard Mitsunobu conditions[6] to deliver neopeltolide (2) with the longest linear sequence of 15 steps. The spectral data of this natural product were in excellent agreement to that described in the literature.[3,15,16]
Scheme 2.
Antiproliferative activity and cell cycle arrest
We examined the effect of a simplified analog of leucascandrolide A (3) on proliferation of mammalian cells and the yeast S. cerevisiae. The dose-dependent suppression of growth of human tumor cell lines A549 (lung), PC3 (prostate), and HCT116 (colon) was established using a standard assay monitoring total cellular ATP (18), and was further confirmed by counting cells in several representative experiments. Analog 3 elicited a highly potent antiproliferative profile and inhibited growth of cells by 50% at concentrations (GI50) of 0.3–0.8 nM (Supplementary Fig. 2a–c), equal to that of the parent natural product 1. To evaluate the effect of leucascandrolide A on yeast cell growth, we performed disc-permeation assays using the W303-1A laboratory strain of S. cerevisiae as wild-type. Analog 3 inhibited yeast growth in a zone with a diameter of 35 mm (11.2 μg of 3 per disc), comparable to the diameter of 33 mm inhibited by natural leucascandrolide A (11.2 μg of 1 per disc).
The synthetic neopeltolide (2) inhibited cell growth with highly potent GI50 values of 0.3–0.5 nM, in agreement with previously reported data[3] and with a pattern nearly identical to that observed with leucascandrolide A analog 3 (Supplementary Fig. 2d–f). For example, HCT116 cell line proved to be the most sensitive to the two compounds, which inhibited the growth of these cells by 90% at 1.0 nM. In contrast, PC3 and A549 cell growth was inhibited only by 60% at the same concentration of 2 or 3.
Having established a highly potent cell-based antiproliferative profile of simplified leucascandrolide A congener 3, we evaluated the effect of this compound on cell cycle regulation. Exposure of HCT116 cells to analog 3 (100 nM) for 24 h resulted in a marked increase in the G0/G1 population to 61% from 45% in the control, and a decrease in the G2/M cell population to 6% from 12% (Supplementary Fig. 3a). A similar G1 cell cycle delay was observed with the parent natural product 1 (Supplementary Fig. 3b). Taking into account the reported G1 cell cycle arrest induced by neopeltolide,[3] the combination of similar cell-based activity profile and the cell-cycle inhibition by 2 and 3 strongly supported the hypothesis that the two natural product-based agents inhibit cell growth by the same mechanism.
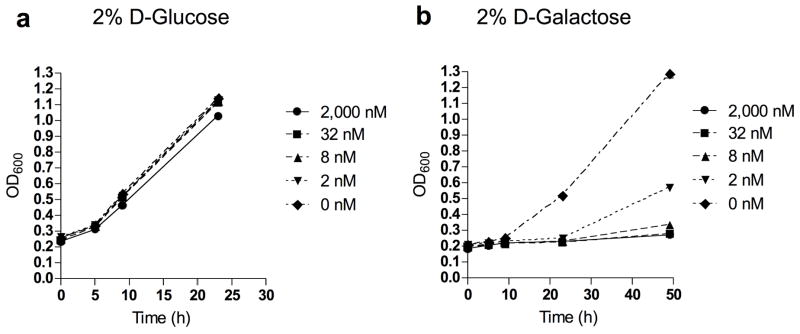
While the W303-1A wild-type strain of S. cerevisiae displayed marked sensitivity to leucascandrolide analog 3 and neopeltolide 2 on agar media containing 2% glucose (19), almost no sensitivity to 3 was observed in the 2% glucose liquid medium, YPD (Fig. 3a). Replacing glucose with ethanol (20) or glycerol (21) slightly increased sensitivity to analog 3 while galactose (22) increased sensitivity to 3 over 10,000-fold (Fig. 3b). In galactose media, analog 3 induced a similar G1-shifted cell cycle profile in the budding yeast cells (Supplementary Fig. 4). Interestingly, adding glucose to the galactose-containing media restored resistance to the drug and normal growth kinetics.
Figure 3.
Effect of analog 3 on yeast S. cerevisiae growth in D-glucose (a) and D-galactose (b) liquid media. Data represent one typical experiment.
Genome-wide yeast deletion screen
Previous work using yeast genetics to identify cellular targets of small molecules demonstrates the value of an organism with low complexity, powerful genetics and conserved cell cycle.[23,24] We evaluated the activity of leucascandrolide A analog in the library of 4900 yeast strains comprising the yeast haploid non-essential gene deletion collection. The screen was aimed at providing additional information about the mechanism of action en route to the ultimate discovery of the cellular target. We examined the effect of analog 3 on growth inhibition of each member of the yeast haploid deletion library grown in a 96-well format, aiming to identify the supersensitive organisms. Growth-inhibition of each mutant strain cultured in liquid media containing 2% galactose was measured by monitoring the optical density at 600 nm at 24 h intervals in the presence of three concentrations of analog 3 (0 nM, 5 nM, 25 nM). Quantitative analysis of growth rates revealed several supersensitive deletion mutants, which were subdivided into three categories, including (a) genes involved in pathways for drug inactivation via transport and/or secretion of metabolites: ATG11, ERG3, ERG6, KCS1, PPA1, RVS161, and VMA8; (b) key regulators of glucose metabolism, SNF4, SNF5, and SNF6; and (c) genes involved in amino acid biosynthesis, HOM3, SER1, and SER2. Among these mutants, the strain carrying a deletion of the SNF4 gene also demonstrated slow growth on galactose alone (Supplementary Fig. 5a), but was particularly sensitive to leucascandrolide A analog 3 (Supplementary Fig. 5b). SNF4 encodes a regulatory subunit of the yeast homolog of the heterotrimeric AMP-activated protein kinase, AMPK. Mammalian AMPK is stress responsive, in part via sensing the cellular AMP:ATP ratio and is thus activated by metabolic stresses that inhibit ATP production or stimulate consumption. Similarly, Snf4 along with its binding partner, the protein serine/threonine kinase Snf1, are required for adaptation to growth on low glucose, unfavorable sugars such as galactose and non-fermentable carbon sources such as glycerol and ethanol.[25,26] Though galactose is fermentable, yeast grown semi-anaerobically on galactose medium demonstrate increased respiration and elevated mitochondrial cytochromes.[27] The pattern of sensitivity to non-glucose carbon sources and the marked supersensitivity of the snf4 knockout strain raised the possibility that leucascandrolide A may antagonize a pathway required for ATP biosynthesis in the absence of glucose fermentation, such as mitochondrial oxidative phosphorylation.
Inhibition of oxidative phosphorylation
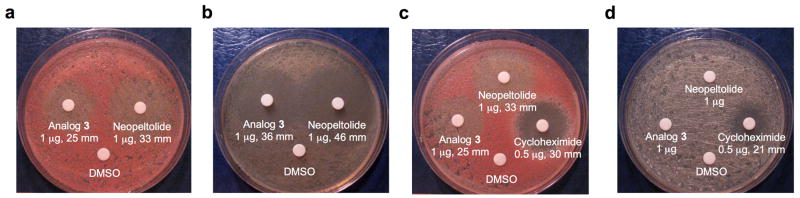
To further probe this hypothesis, we used a disc diffusion assay to examine yeast sensitivity to analog 3 when glucose was replaced with the non-fermentable carbon source glycerol (Fig. 4a,b). Glycerol metabolism requires a shift in ATP production to oxidative phosphorylation and cells lacking mitochondrial respiration (petites) are unable to grow on glycerol alone. Increased sensitivity to analog 3 in glycerol-containing medium was evident both in the larger diameter and complete lack of cell growth within the zone of inhibition. We next examined the effect of analog 3 on the growth of ρ0 petite yeast. Lacking mitochondrial DNA and thus genes encoding key components of the electron transport chain, ρ0 cells are incapable of producing ATP by oxidative phosphorylation and can grow only via glycolysis. Consistent with both drugs targeting oxidative phosphorylation, neither leucascandrolide A analog 3 nor neopeltolide 2 further inhibited growth of ρ0 petite yeast compared to their effects on the ρ+ control (Fig. 4c,d). As expected, cycloheximide (23) inhibited growth of ρ0 petite yeast.
Figure 4.
Effect of leucascandrolide A analog 3 and neopeltolide 2 on yeast S. cerevisiae on agar plates. (a) Growth inhibition of wild type strain by analog 3 and neopeltolide 2 on glucose agar plates. (b) Growth inhibition of wild type strain by analog 3 and neopeltolide 2 on glycerol agar plates. (c) Growth inhibition of wild type strain by analog 3, neopeltolide 2 and cycloheximide (23) on glucose agar plates. (d) Growth inhibition of petite strain by analog 3, neopeltolide 2 and cycloheximide on glucose agar plates.
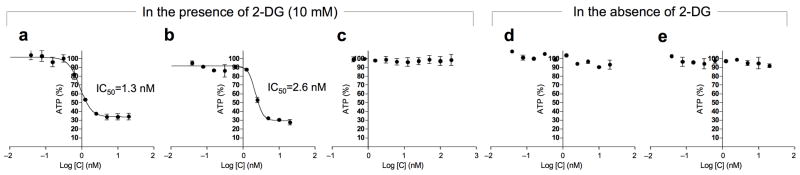
Because higher eukaryotes employ similar pathways for cellular ATP production, we next tested the effects of leucascandrolide A analog 3 and neopeltolide 2 on modulation of oxidative phosphorylation in mammalian cells. Our strategy was to employ 2-deoxy-glucose (24, 2-DG) to block glycolysis[28] and thereby reveal the effects of 2 and 3 on ATP production by oxidative phosphorylation. Treatment of A549 cells with either 2 or 3 for 30 min in the presence of 2-DG decreased ATP concentration with IC50 of 1.3 and 2.6 nM, respectively (Fig. 5a,b). As predicted, ATP levels were not affected by the spindle toxin taxol (25) in the presence of 2-DG (Fig. 5c). The effect appeared specific to oxidative phosphorylation as the level of ATP remained relatively constant during the first 30 min after incubation of cells with either 2 or 3 in the absence of 2-DG (Fig. 5d,e)
Figure 5.
Inhibition of ATP (18) production in A549 cells in response to treatment with neopeltolide 2 and leucascandrolide analog 3, in the presence or absence of 2-DG (24). Whole-cell ATP content in the presence and absence of 2-DG was 25×10−6 nmol/cell and 37 ×10−6 nmol/cell, respectively. All values are presented as % of vehicle treated sample. Each value is mean ± SEM of duplicate values from a representative experiment. (a) ATP depletion upon treatment with a combination of neopeltolide 2 and 2-DG (10 mM). (b) ATP depletion upon treatment with a combination of leucascandrolide analog 3 and 2-DG (10 mM). (c) Treatment of cells with a combination of taxol (25) and 2-DG does not effect ATP levels significantly within 30 min of incubation. (d) Constant level of ATP after treatment of cells with neopeltolide 2 in the absence of 2-DG. (e) Constant level of ATP after treatment of cells with analog 3 in the absence of 2-DG.
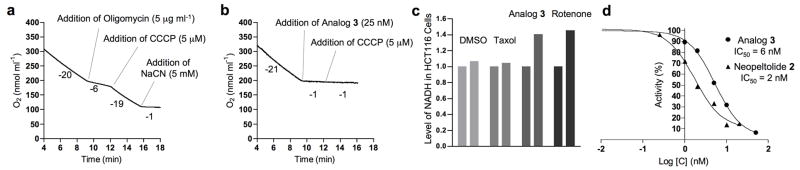
To confirm these results, we examined the effect of analog 3 cellular respiration by monitoring oxygen consumption in 143B cells. Oligomycin A (26), an inhibitor of the mitochondrial Fo/F1 ATP synthase,[29] decreased oxygen uptake, and treatment of these cells with carbonyl cyanide m-chlorophenyl hydrazone (27, CCCP)[30] to uncouple mitochondrial respiration from ATP synthesis restored respiration (Fig. 6a). As expected, treatment of cells with NaCN, which inhibits complex IV of the electron transport chain, blocked respiration irreversibly. Treatment of 143B cells with leucascandrolide A analog 3 inhibited respiration in a dose-dependent fashion. Unlike the oligomycin control, the respiration of the treated cells was not restored by CCCP, which indicated that analog 3 did not influence the activity of ATP synthase (Fig. 6b). Treatment of HCT116 cells with 3 resulted in a 40% increase in NADH (28) fluorescence of cells at 457 nm (Fig 6c), an indicator of inhibition of electron transport in mitochondria. Rotenone (29), which inhibits complex I, was employed as a positive control in these studies. In addition, treatment of isolated yeast sonicated mitochondria with leucascandrolide A analog 3 (10 nM) or neopeltolide 2 (10 nM) resulted in complete inhibition of NADH oxidation compared to DMSO control (Supplementary Fig. 6a). Given that NADH oxidation is dependent on the activity of complexes I, III and IV, our data indicated that leucascandrolide A and neopeltolide may elicit their potent antiproliferative activity by blocking one or more complexes in the mitochondrial electron-transport chain.
Figure 6.
Effect of leucascandrolide analog 3 and neopeltolide 2 on cellular respiration, NADH (28) consumption and activity of cytochrome bc1 complex. Data represent one typical experiment. (a) Effect of oligomycin A (26) on respiration of 143B cells. The initial inhibition of respiration was restored by the addition of proton uncoupler CCCP (27). (b) Inhibition of cellular respiration of 143B cells by leucascandrolide A analog 3. Subsequent treatment of cells with CCCP (27) did not restore respiration. (c) Effect of taxol (25, 2.5 μM), rotenone (29, 2.5 μM), and leucascandrolide A analog 3 (0.5 μM) on NADH level in HCT116 cells before and after treatment (bar graphs on the left and the right, respectively). (d) Dose-dependent inhibition of purified bovine cytochrome bc1 complex by analog 3 and neopeltolide 2.
Inhibition of cytochrome bc1 complex
To identify a specific electron-transport complex that is inhibited by leucascandrolide A and neopeltolide, we measured the enzymatic activity of each protein complex involved in oxidative phosporylation in sonicated yeast mitochondria. We found that treatment of isolated mitochondria with either 3 or 2 resulted in highly potent dose-dependent inhibition of the activity of complex III, also referred to as cytochrome bc1 complex or ubiquinol:cytochrome c oxidoreductase, with IC50 of 1.8 and 0.6 nM, respectively (Supplementary Fig. 6b,c). None of the other respiratory chain complexes were affected by the two natural products (Supplementary Fig. 7). To test the effect of leucascandrolide A analog (3) and neopeltolide (2) on the activity of purified bovine heart mitochondrial cytochrome bc1 complex, we monitored reduction of cytochrome c by the purified enzyme. Analog 3 inhibited the activity of this protein complex by 50% at a concentration of 6.0 nM (Fig. 6d). Neopeltolide (2) displayed a similar activity profile (IC50 2.0 nM, Fig 6d).
In order to gain an additional insight into the mechanism of inhibition of cytochrome bc1 complex, we carried out a series of spectroscopic studies that enable to monitor changes in the oxidation states of cytochromes. Treatment of sonicated yeast mitochondria with leucascandrolide-based analog 3 resulted in similar levels of reduced cytochrome b to that observed using typical P-type inhibitors (Supplementary Fig. 8).
DISCUSSION
The mitochondrial electron-transport chain consists of four major components, which are designated as complexes I, II, III and IV.[31] Ubiquinone (coenzyme Q10) and cytochrome c are responsible for the communication between the four trans-membrane complexes enabling the electron flow through the chain from NADH dehydrogenase (complex I) to cytochrome c oxidase (complex IV).[31] Cytochrome bc1 (complex III) is a transmembrane protein complex located in the inner mitochondrial membrane.[32–34] This multi-subunit enzyme is a central component of the mitochondrial respiratory electron transport chain.[35] The function of cytochrome bc1 is to reduce cytochrome c(Fe3+) into cytochrome c(Fe2+) using the membrane localized ubiquinol. The free energy of the proton-coupled electron transfer reaction, which is often referred as the protonmotive Q cycle,[36,37] contributes to the proton gradient across the mitochondrial membrane to enable subsequent ATP production by ATP synthase.
Our work demonstrated that practical chemical synthesis of leucascandrolide A (1) and neopeltolide (2), in the absence of any other means of accessing such compounds, enabled a comprehensive genetic and biochemical investigation of the molecular mechanism of action. This effort resulted in identification of cytochrome bc1 complex as the cellular target of the two marine natural products. Highly potent cell-based antiproliferative activity of leucascandrolide A and neopeltolide compares favorably to the most potent inhibitors of cytochrome bc1 complex known today, identifying such compounds as a new class of highly useful biochemical tools for investigation of eukaryotic energy metabolism.
It is noteworthy that leucascandrolide A (1) and neopeltolide (2) do not share any significant degree of structural homology with previously reported small-molecule inhibitors of cytochrome bc1 complex, including P-type inhibitors [i.e., myxothiazol (30) and stigmatellin (31)],[38–40] ubiquinone mimetics,[41] and N-type inhibitors [i.e., antimycin A (32)].[42,43] Our studies demonstrated that leucascandrolide-based analog 3 and neopeltolide (2) were exceedingly potent at inhibiting the activity of cytochrome bc1 complex. Such potent inhibition of isolated cytochrome bc1 complex by 2 and 3 was fully consistent with the ability of the two compounds to inhibit cell growth, ATP production, respiration and complex III activity in yeast mitochondria at similarly low concentrations. Our preliminary investigation revealed that the effect of compound 3 on reduction of cytochrome b in yeast mitochondria appeared to be similar to that observed for P-type inhibitors of complex III, i.e. myxothiazol and stigmatellin. The detailed biophysical and biochemical investigation of the mechanism of inhibition of cytochrome bc1 complex by leucascandrolide A and neopeltolide is currently in progress.
Oxidative phosphorylation and glycolysis are tightly coupled in eukaryotic cells. Due to the increased dependency of tumor cells on glycolysis, known as the Warburg effect,[44] and its likely role in promoting cell proliferation, survival and invasion,[45] disruption of energy metabolism in cancer cells is of significant current interest for the development of a new class of anticancer agents.[46] Several recent studies demonstrated that the induction of hypoxia-like conditions using inhibitors of oxidative phosphorylation hypersensitized tumor cells to treatment with glycolytic inhibitors.[47,48] Thus, a combination of suppressors of oxidative phosphorylation with inhibitors of glycolysis could represent a strategy for metabolic sensitization of tumor cells to cytotoxic and targeted therapies.
METHODS
General
Dichloromethane (HPLC grade), ethyl acetate (ACS grade), hexanes (ACS grade), and diethyl ether (ACS grade) were purchased from Fisher Scientific and used without further purification. Anhydrous tetrahydrofuran was purified by distillation from sodium-benzophenone. Commercially available reagents were used without further purification. Reactions were monitored by thin layer chromatography (TLC) using Whatman precoated glass silica gel plates. Flash column chromatography was performed over Silacycle silica gel (230–400 mesh). 1H NMR and 13C NMR spectra were recorded on Bruker DRX-400 or DMX-500 spectrometers using residual solvent peaks as an internal standard. Mass Spectra were recorded with a VG Instruments Model 7070EQ tandem mass spectrometer. All cell lines, media, serum and supplements were purchased from ATCC. CellTiter-Glo® luminescent cell viability assay was purchased from Promega. SensoLyte™ Culture dishes, 96-well plates and all other supplies were purchased from Fisher Scientific. Luminescence was measured on Perkin-Elmer Victor 3 plate reader. Oligomycin A (26), 2-deoxy-D-glucose (24), taxol (25), CCCP (27), rotenone (29), cyclohexamide (23) and cytochrome c from equine heart were purchased from Sigma-Aldrich
Cell lines and culture
A549, PC3, HCT116 cell lines were purchased from ATCC. A549 and PC3 cells were maintained in F12-K medium; HCT116 cells were maintained in McCoy’s 5A modified medium. Media were supplemented with 10% FBS and 1% Penicillin-Streptomycin-Glutamine solution. 143B cell line was maintained in MEM medium supplemented with 10% FBS, 1% Penicillin-Streptomycin solution, 0.1% BrdU and 2% sodium pyruvate.
Yeast strain and media
S. cerevisiae strain was W303-1A (MATa ade2–1 can1–100 ura3–1 leu2–3, 112 his3–11, 15 trp1–1). Yeast cells were grown in standard rich YPD (1% yeast extract, 2% peptone, 2% glucose), YPGal (1% yeast extract, 2% peptone, 2% galactose) and YPGly (1% yeast extract, 2% peptone, 3% v/v glycerol) media. Petite mutants of W303-1A strain were obtained by treatment with 25 μg of ethidium bromide per ml in 0.67% Difco yeast nitrogen base with 2% glucose, relevant nucleotides and amino acids for W303-1A. After culture was grown to saturation, a small inoculum was transferred to ethidium-bromide free medium and cells were grown to saturation and streaked on YPD. ρ0 petite colonies were identified by colony color (W303-1A wild type colonies are red on YPD media, petite mutants are white) and by assessing their lack of growth on YPGly agar plates.
Disc permeation assay
Yeast was grown to saturation in YPD media at RT. Cultures were diluted by the factor of 10, and 0.25 ml of cell suspension was spread onto YPD or YPGly plates. Small molecules and DMSO were dissolved in 20 μl of water and added to the paper discs, which were placed on top of agar. Plates were incubated for 2–3 days at 30°C.
Yeast growth inhibition assay in liquid media
All assays were performed using two replicate wells for each compound concentration test. Yeast suspension in logarithmic phase was distributed in 96 well-plate. Cells were treated with graded concentrations of the small molecule, and growth rates were measured by determination of OD600 as a function of time by using a PowerWave™ microplate spectrophotometer (BioTek).
ATP measurement
All assays were performed using two replicate wells for each small molecule concentration test. A549 cells were seeded in 96 well-plates at density of 2500 cells/well and incubated for 4h. Cells were then treated with graded concentrations of small molecules and incubated for 30 min. CellTiter-Glow® kit (Promega) was used to measure ATP levels. Luminescence was quantified as described before. Whole-cell ATP content (nmol/cell) was determined using ATP standard curve.
Measurement of cellular respiration
Cellular O2 consumption rates were measured in aliquots of 143 B cells (4 × 106 cells) in serum free medium and studied in a magnetically stirred, water-jacked (37°C) respirometer chamber by using an oxygen electrode (Hansatech Instruments, Norfolk, England). Oxygraph Plus software was used to determine oxygen consumption rate.
Measurement of cellular NADH level
Changes in cellular NADH concentrations were assessed from changes in the autofluorescence of cells by confocal microscopy at an excitation wavelength of 350 ± 25 nm and an emission wavelength of 457 ± 25 nm. Autofluorescence was measured by using DSU Spinning disk confocal microscope (Olympus). HCT116 cells were seeded to collagen-coated dishes (MatTek) at density of 200 000 cells/dish. Cells were allowed to proliferate for 41 h at 37 °C. Fresh medium was added, and cells were incubated for 30–60 min. Samples were placed into dish-holder maintained at 37 °C, and fluorescence was measured every 30 s. As soon as initial autofluorescence was obtained, small molecules were added and measurements were done for additional 4 min. Percent change in NADH content was calculated by dividing average initial autofluorescence by average autofluorescence after drug addition.
Isolation of cytochrome bc1 complex
Bovine mitochondria were isolated as previously described.[49] Mitochondria were stored at −80°C until used for purification. Mitochondrial cytochrome bc1 complex was purified as described before.[50] However, the starting NaCl concentration used here for the wash was 150mM, followed by a linear gradient of 150–400 mM NaCl. The DEAE Sepharose column was obtained from GE Healthcare (XK 16/40).
Inhibition of purified bovine cytochrome bc1 complex
Enzyme was diluted to 0.686 mg/ml in 50 mM phosphate buffer, pH 7.5, 300 mM NaCl, 0.1 g/l n-dodecyl β-maltoside. The enzymatic activity was assayed in 40 mM potassium phosphate buffer, pH 7.4, 0.5 mM EDTA. Complex III activity was measured while monitoring the reduction of 50 μM cytochrome c (λ 550 nm, ε 19.1 mM−1 cm−1) in the presence of 3 mM NaN3 to block its reoxidation, starting the reaction with 20 μM decylubiquinol (33). Complex III activity was 19.6 μmol min−1mg−1.
Supplementary Material
Acknowledgments
This work was supported by NCI R01 CA93457 to S.A.K. and NIH R01 GM60443 to S.J.K. S.A.K. thanks Alfred P. Sloan Foundation, Dreyfus Foundation, Amgen and GlaxoSmithKline for additional financial support. S.J.K. was a Leukemia & Lymphoma Society Scholar. J.J. acknowledges the support of Burroughs Wellcome Fund Interfaces #1001774.
Footnotes
COMPETING INTEREST STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Paterson I, Anderson EA. The Renaissance of natural products as drug candidates. Science. 2005;310:451–453. doi: 10.1126/science.1116364. [DOI] [PubMed] [Google Scholar]
- 2.D’Ambrosio M, Guerriero A, Debitus C, Pietra F. Leucascandrolide A, a new type of macrolide: The first powerfully bioactive metabolite of calcareous sponges Leucascandra caveolata, a new genus from the Coral Sea. Helv Chim Acta. 1996;79:51–60. [Google Scholar]
- 3.Wright AE, Botelho JC, Guzman E, Harmody D, Linley P, McCarthy PJ, Pitts TP, Pomponi SA, Reed JK. Neopeltolide, A macrolide from a Lithistid sponge of the family Neopeltidae. J Nat Prod. 2007;70:412–416. doi: 10.1021/np060597h. [DOI] [PubMed] [Google Scholar]
- 4.Hornberger KR, Hamblett CL, Leighton JL. Total synthesis of leucascandrolide A. J Am Chem Soc. 2000;122:12894–12895. [Google Scholar]
- 5.Kopecky DJ, Rychnovsky SD. Mukaiyama aldol-Prins cyclization cascade reaction: Formal total synthesis of leucascandrolide A. J Am Chem Soc. 2001;123:8420–8421. doi: 10.1021/ja011377n. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Janjic J, Kozmin SA. Synthesis of leucascandrolide A via a spontaneous macrolactolization. J Am Chem Soc. 2002;124:13670–13671. doi: 10.1021/ja028428g. [DOI] [PubMed] [Google Scholar]
- 7.Fettes A, Carreira EM. Total synthesis of leucascandrolide A. Angew Chem Int Ed Engl. 2002;41:4098–4101. doi: 10.1002/1521-3773(20021104)41:21<4098::AID-ANIE4098>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 8.Paterson I, Tudge M. Stereocontrolled total synthesis of (+)-leucascandrolide A. Angew Chem Int Ed Engl. 2003;42:343–347. doi: 10.1002/anie.200390112. [DOI] [PubMed] [Google Scholar]
- 9.Wipf P, Reeves JT. A Formal total synthesis of leucascandrolide A. Chem Commun. 2002;18:2066–2067. doi: 10.1039/b207383h. [DOI] [PubMed] [Google Scholar]
- 10.Williams DR, Plummer SV, Patnaik S. Formal synthesis of leucascandrolide A. Angew Chem Int Ed Engl. 2003;42:3934–3938. doi: 10.1002/anie.200351817. [DOI] [PubMed] [Google Scholar]
- 11.Crimmins MT, Siliphaivanh P. Enantioselective total synthesis of (+)-leucascandrolide A macrolactone. Org Lett. 2003;5:4641–4644. doi: 10.1021/ol035797o. [DOI] [PubMed] [Google Scholar]
- 12.Su Q, Panek JS. Total synthesis of (+)-leucascandrolide A. Angew Chem Int Ed Engl. 2005;44:1223–1225. doi: 10.1002/anie.200462408. [DOI] [PubMed] [Google Scholar]
- 13.Jung HH, Seiders JR, Floreancig PE. Oxidative cleavage in the construction of complex molecules: synthesis of the leucascandrolide A macrolactone. Angew Chem Int Ed Engl. 2007;46:8464–8467. doi: 10.1002/anie.200702999. [DOI] [PubMed] [Google Scholar]
- 14.Ferrie L, Reymond S, Capdevielle P, Cossy J. Formal chemoselective synthesis of leucascandrolide A. Org Lett. 2007;9:2461–2464. doi: 10.1021/ol070670a. [DOI] [PubMed] [Google Scholar]
- 15.Youngsaye W, Lowe JT, Pohlki F, Ralifo P, Panek JS. Total synthesis and stereochemical reassignment of (+)-neopeltolide. Angew Chem Int Ed Engl. 2007;46:9211–9214. doi: 10.1002/anie.200704122. [DOI] [PubMed] [Google Scholar]
- 16.Custar DW, Zabawa TP, Scheidt KA. Total synthesis and structural revision of the marine macrolide neopeltolide. J Am Chem Soc. 2008;130:804–805. doi: 10.1021/ja710080q. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Janjic J, Kozmin SA. Synthesis of leucascandrolide A. Pure Appl Chem. 2005;77:1161–1169. [Google Scholar]
- 18.Kozmin SA. Efficient Stereochemical relay en route to leucascandrolide A. Org Lett. 2001;3:755–758. doi: 10.1021/ol015514x. [DOI] [PubMed] [Google Scholar]
- 19.Janjic J, Leucascandrolide A. Ph D Dissertation. University of Chicago; 2006. Synthesis and studies toward identification of the biological target. [Google Scholar]
- 20.Paterson I, Gibson KR, Oballa RM. Remote, 1,5-Anti stereoinduction in the boron-mediated aldol reactions of β-oxygenated methyl ketones. Tetrahedron Lett. 1996;37:8585–8588. [Google Scholar]
- 21.Evans DA, Coleman PJ, Cote B. 1,5-Asymmetric induction in methyl ketone aldol addition reactions. J Org Chem. 1997;62:788–789. doi: 10.1021/ja027640h. [DOI] [PubMed] [Google Scholar]
- 22.Chen KM, Hardtmann GE, Prasad K, Repic O, Shapiro MJ. 1,3-diastereoselective reduction of β-hydroxyketones utilizing alkoxydialkylboranes. Tetrahedron Lett. 1987;28:155–158. [Google Scholar]
- 23.Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MJ, Arkin AP, Davis RW. Chemogenomic profiling: Identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baetz K, McHardy L, Gable K, Tarling T, Reberioux D, Bryan J, Andersen RJ, Dunn T, Hieter P, Roberge M. Yeast genome-wide drug-induced haploinsufficiency screen to determine drug mode of action. Proc Natl Acad Sci USA. 2004;101:4525–4530. doi: 10.1073/pnas.0307122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Celenza JL, Eng FJ, Carlson M. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol Cell Biol. 1989;9:5045–5054. doi: 10.1128/mcb.9.11.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Ann Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 27.Nagata I, Furuya E, Yoshida Y, Kanaseki T, Tagawa K. Development of mitochondrial membranes in anaerobically grown yeast cells. J Biochem. 1975;78:1353–1364. doi: 10.1093/oxfordjournals.jbchem.a131033. [DOI] [PubMed] [Google Scholar]
- 28.Brown J. Effect of 2-deoxyglucose on carbohydrate metabolism: review of the literature and studies in rats. Metabolism. 1962;11:1098–1112. [PubMed] [Google Scholar]
- 29.Linnet PE, Beechey RB. Inhibitors of the ATP synthase system. Methods Enzymol. 1979;55:472–518. doi: 10.1016/0076-6879(79)55061-7. [DOI] [PubMed] [Google Scholar]
- 30.Haytler PG, Prichard WW. A new class of uncoupling agents carbonyl cyanide phenylhydrazones. Biochim Biophys Res Commun. 1962;2:272–275. doi: 10.1016/0006-291x(62)90189-4. [DOI] [PubMed] [Google Scholar]
- 31.Wallace KB, Starkov AA. Mitochondrial targets of drug toxicity. Annu Rev Pharmacol Toxicol. 2000;40:353–388. doi: 10.1146/annurev.pharmtox.40.1.353. [DOI] [PubMed] [Google Scholar]
- 32.Xia D, Yu C, Kim H, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 35.Trumpower BL. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchel P. Protonmotive redox mechanism of the cytochrome bc-1 complex of the respiratory chain: protonmotive ubiquinone cycle. FEBS Lett. 1975;56:1–6. doi: 10.1016/0014-5793(75)80098-6. [DOI] [PubMed] [Google Scholar]
- 37.Trumpower BL. The Protonmotive Q cycle. J Biol Chem. 1990;265:11409–11412. [PubMed] [Google Scholar]
- 38.Von Jagow G, Ljungdahl PO, Graf P, Ohnishi T, Trumpower BL. An inhibitor of mitochondrial respiration which binds to cytochrome b and displaces quinone from the iron-sulfur protein of the cytochrome bc1 complex. J Biol Chem. 1984;259:6318–6326. [PubMed] [Google Scholar]
- 39.Thierbach G, Reichenbach H. Myxothiazol, a new inhibitor of the cytochrome bc1 segment of the respiratory chain. Biochim Biophys Acta. 1981;638:282–289. doi: 10.1016/0005-2728(81)90238-3. [DOI] [PubMed] [Google Scholar]
- 40.Thierbach G, Kunze B, Reichenbach H, Hofle G. The mode of action of stigmatellin, a new inhibitor of the cytochrome b–c1 segment of the respiratory chain. Biochim Biophys Acta. 1984;765:227–235. [Google Scholar]
- 41.Bower JR, Edwards CA, Ohnishi T, Trumpower BL. An analogue of ubiquinone which inhibits respiration by binding to the iron-sulfur protein of the cytochrome bc1 segment of mitochondrial respiratory chain. J Biol Chem. 1982;257:8321–8330. [PubMed] [Google Scholar]
- 42.Ohnishi T, Trumpower BL. Differential effects of antimycin on ubisemiquinone bound in different environments in isolated succinate-cytochrome c reductase complex. J Biol Chem. 1980;255:3278–3824. [PubMed] [Google Scholar]
- 43.Gutierrez-Cirlos EB, Merbitz-Zahradnik T, Trumpower BL. Inhibition of the yeast cytochrome bc1 complex by ilicicolin H, a novel inhibitor that acts at the Qn site of the bc1 complex. J Biol Chem. 2004;279:8708–8714. doi: 10.1074/jbc.M311805200. [DOI] [PubMed] [Google Scholar]
- 44.Warburg O. On the origin on cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 45.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Pelicano H, Martin DS, Xu RH, Hunag P. Glycolysis Inhibition for Anticancer Treatment. Oncogene. 2006;25:4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, Hu YP, Savaraj N, Priebe W, Lampidis TJ. Hypersensitization of tumor cells to glycolytic inhibitors. Biochemistry. 2001;40:5542–5547. doi: 10.1021/bi002426w. [DOI] [PubMed] [Google Scholar]
- 48.Maher JC, Krishan A, Lampidis TJ. Greater cell cycle inhibition and cytotoxicity induced by 2-deoxy-d-glucose in tumor cells treated under hypoxic vs aerobic conditions. Cancer Chemother Pharmacol. 2004;53:116–122. doi: 10.1007/s00280-003-0724-7. [DOI] [PubMed] [Google Scholar]
- 49.Smith AL. Preparation, properties, and conditions for assay of mitochondria: slaughterhouse material, small scale. Methods Enzymol. 1967;10:81–86. [Google Scholar]
- 50.Berry EA, Huang LS, DeRose VJ. Ubiquinol-cytochrome c oxidoreductase of higher plants. Isolation and characterization of the bc1 complex from potato tuber mitochondria. Biol Chem. 1991;266:9064–9077. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.