Abstract
The development of the pharmaceutical industry, driven by progress in chemistry, biology, and technology, ranks as one of the most successful of human endeavors. However, serious health problems persist, among which are diseases caused by protozoan parasites, largely ignored in modern times. Advances in genomic sciences, molecular and structural biology, and computational and medicinal chemistry now set the scene for a renewed assault on such infections. A structure-centric approach to support discovery of antiparasitic compounds promises much. Current strategies and benefits of a structure-based approach to support early stage drug discovery will be described.
Molecular structure and chemical interactions define how cells and organisms live and die. Paul Ehrlich worked this out over a century ago and coined the term “pharmacophore” to define those properties of a compound responsible for a pharmacological response. Ehrlich addressed problems caused by the parasites Plasmodium sp. and Trypanosoma brucei, the organisms responsible for malaria and African sleeping sickness, respectively. His links to industry, in particular dyestuff manufacturers, were important because their chemistry expertise provided compounds. Investigations with dyes extended to arsenicals and ultimately produced Salvarsan, the first chemotherapeutic agent, for treatment of a bacterial infection, syphilis, and African sleeping sickness (1). The foundation of the pharmaceutical industry was therefore based in large part on discovery of drugs against protozoan parasites. Despite progress, diseases caused by protozoan parasites remain a serious health issue in much of the developing world. In particular, malaria (2), African sleeping sickness (3), Chagas disease (caused by Trypanosoma cruzi) (4), and cutaneous, mucocutaneous, and visceral leishmanises (Leishmania major and Leishmania donovani) (4, 5) profoundly influence the lives of millions of people.
Vaccination, an ideal method of dealing with pathogens, represents a holy grail in parasitology especially for malaria, but investment in antiprotozoan vaccine development has gained scant reward. Plasmodium, Leishmania, and T. cruzi infect and shelter from the immune system within host cells. Plasmodium species display variability and antigenic variation in their membrane-bound surface proteins (6). This prolongs parasite circulation in the blood, increases the likelihood of transmission, and helps evasion of the immune system. The African trypanosome T. brucei lives in the bloodstream fully exposed to the immune system. However, this organism possesses a repertoire of genes, only one of which is switched on at any given time, encoding a VSG.2 The abundant VSG cloaks the parasite and prevents access to potential antigens embedded in the surface membrane. Antigenic variation changes the VSG at regular intervals, allowing the pathogen to circumvent the immune response (7).
In some cases, e.g. malaria, a few acceptable drugs are available. However, for trypanosomatid infections, the current treatments are woefully inadequate due to toxicity, high cost, and poor efficacy (8). Increasing levels of drug resistance exacerbate the problem of parasitic diseases (9), and in addition, ∼10% of emerging infectious diseases are caused by protozoans (10). These factors, in conjunction with the lack of vaccines, emphasize the urgent need for new drugs. Recent significant advances in molecular parasitology and modern drug research provide opportunities to progress in this. I will outline how the industry evolved from Ehrlich's pioneering work, describe modern approaches (heavily reliant on access to structural data) to early stage drug discovery, and explain concepts and current ideas in the field. The emphasis will be on what modern approaches offer the area of neglected parasitic diseases.
Post-Ehrlich
Drug discovery is inexorably linked to society's requirements and scientific advances. At first, the need for anti-infectives dominated; the emphasis was on testing compounds, mainly natural products, against organisms. Advances in extraction and separation technologies, with analytical methods extending to structure determination, enhanced natural product and synthetic chemistry and the elucidation of SAR. A particularly successful approach was derivatization of natural products: metabolites and constituents of biomolecules. The legacy is that 75% of antibacterials in the clinic are natural products or their semisynthetic derivatives (11). The research had wide reaching benefits, and readers might care to reflect on what the Journal of Biological Chemistry would look like without the antibiotics that assist modern life sciences research.
The industry successfully developed drugs and vaccines and revolutionized health care. A downside of this success was the perception that microbial diseases were consigned to the past or less developed (less profitable) countries, and complacency set in. Commercial pressures forced the industry toward “modern” diseases such as cancer, diabetes, and cardiovascular conditions. Although the occasional war provoked activity against malaria research, antiparasite drug research was a limited activity, discarded when companies reassessed priorities or consolidated activities.
Genomic science heralded insight into the blueprint of life and, together with advances in molecular biology, shaped a strategy where the emphasis changed to identification and exploitation of specific targets for therapeutic intervention. Combinatorial chemistry and automated HTS provided the technology to synthesize and test vast numbers of compounds, searching out the first new ligands or hits. This was seen as a panacea for the failure of existing programs to find new chemical entities directed against specific targets. However, the rate of new compounds entering clinical trials did not increase relative to the huge investments in compound libraries, the number of screens being run, or the targets addressed. Indeed, in the last 25 years, only a single de novo drug based on combinatorial chemistry has been reported (12).
A critical failing was the use of libraries that were large collections of similar molecules, assembled on the basis of ease of synthesis and cost rather than a likelihood of being biomedically relevant (13). This was recognized within the industry. Christopher Lipinski in particular identified the general features of oral and passively absorbed compounds most relevant to early stage drug discovery. They are relatively small (≤500 Da) and lipophilic, with five or fewer H-bond donors, 10 or fewer H-bond acceptors, and an octanol/water partition coefficient of <5 (14, 15). These guidelines are not applicable to natural products and where active transport occurs. The trend now is toward smaller, “smarter,” combinatorial, and focused libraries containing drug-like compounds (16). An increased emphasis is placed on synthetic feasibility and omission of moieties known to cause problems with respect to ADMET (17). To circumvent toxicity issues, the choice of compounds used for testing in general avoids functional groups likely to render molecules reactive, covalent inhibitors. This issue highlights a dilemma in drug discovery: the exceptions. Although experience dictates that covalent binding enzyme inhibitors are likely to cause ADMET problems, many drugs are just such compounds (18), e.g. aspirin!
There have been tremendous advances in macromolecular structure determination by x-ray diffraction and NMR methods. Automated crystallization facilities and larger magnets have brought more samples forward for structural studies. Modern diffraction data collection facilities (in-house or at synchrotron radiation sources), automated sample changers, and semiautomated computational pipelines allow structure determination in high-throughput fashion. This means that three-dimensional structures of key therapeutic targets and ligand complexes can be obtained quickly, producing data crucial for early stage drug discovery.
Protozoan-specific Requirements Dictate the Nature of the End Product
When targeting human proteins for drug discovery, the desired effect is to increase or decrease some biological activity, i.e. activation or inhibition. For protozoan parasites, we are solely concerned with a deleterious effect to the organism, and the goal is a stable compound, cheap, orally bioavailable, and potent. This molecule should have high affinity for one or more targets in the parasite, disrupt an essential aspect of metabolism, and kill the pathogen quickly. There should be no or little toxicity against humans (19).
Oral bioavailability is a priority. This provides a practical benefit of using tablets in developing countries and optimizes the chances of treating pathogens occupying an intracellular niche or infecting the central nervous system. The Lipinski guidelines (14) are therefore particularly relevant to antiparasite drug research. For protozoan parasites, drug research is also affected by the simple fact that selectivity for one eukaryotic cell over another (the host) has to be found, and in this respect, the search for such therapies is more akin to anticancer than antibacterial drug research.
Exploiting Structural Information for Ligand Discovery
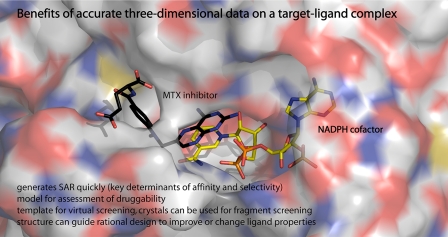
A small molecule binds and interacts with a target exploiting chemical interactions such as hydrophobic van der Waals associations, hydrogen bonds, ionic attractions, charge transfer complexes, and, sometimes, covalent bonds. The cumulative effect of these interactions determines affinity, specificity, and the level and mode of action. A structure of the complex is the most direct way to gain a detailed understanding of how a ligand binds a target (Fig. 1). This immediately informs on SAR and provides a template for screening and modeling to improve affinity. The growing maturity of structure-based ligand discovery methods (20, 21) offers the potential to reduce the number of compounds being synthesized and tested by revealing early and valuable insight into the interactions that influence selectivity and binding affinity. This can be a significant saving in resource.
FIGURE 1.
Benefits of structural information. The complex of a potent inhibitor, methotrexate (MTX), in the active site of T. brucei pteridine reductase (PTR1), the NADPH complex (48), is shown. The protein surface is depicted as a semitransparent van der Waals surface colored according to atom type (carbon, white; nitrogen, blue; oxygen, red). The ligands are shown as stick models, with the methotrexate and NADPH cofactor positions in black and yellow, respectively.
FS methods are increasingly used to identify hits (22–24), where collections of small compounds, typically with a mass of 100–250 Da, are assayed for binding by NMR or differential scanning fluorometry, for example. Crystallography can be a particularly powerful method to apply here by employing co-crystallization, or if robust crystals are available, then fragments can be soaked in. Such molecules have weak binding affinities in the range 10 mm to 100 μm. Such low affinity for a target is far removed from that required (typically low nm) to elicit a potent biological effect. The critical fact is, however, that fragments have high ligand binding efficiency, i.e. a high average free energy of binding per heavy atom (25). The benefits of FS include ready availability and generally good solubility of a FS compound collection, together with simple and robust binding assays. The output can be the accurate placement of novel chemical matter in a target-binding site. This provides a template for ligand design from a tractable starting point and feeds an iterative process of design that guides synthetic and medicinal chemistry decisions concerning modifications and rounds of synthesis and testing as affinity for the target is improved.
Computational methods are increasingly important in ligand identification and complement empirical screening approaches. VS is the process whereby large in silico compound libraries are interrogated and filtered, based on a pharmacophore hypothesis, to identify potential ligands (26). Prior structural data are essential to define the pharmacophore and the template for screening. Selected compounds are docked into the binding site to provide models of target-ligand complexes. Advances in development of computing grids, large central processing unit clusters, and structure-based tools mean that VS is a cost-efficient reliable way to investigate diversity in chemical space and to find hits. Docking calculations can also be applied to hits found by HTS to quickly investigate aspects of molecular recognition. Molecular modeling and design strategies then generate ideas to improve affinity for a target.
Process of Structure-based Ligand Design
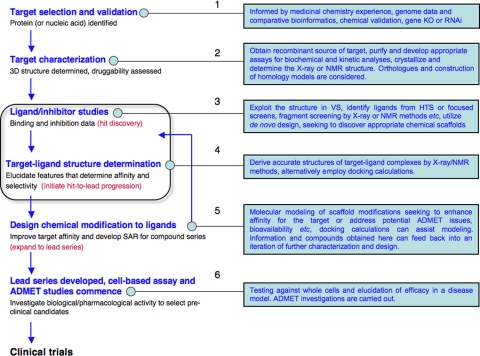
SBLD can be outlined in six stages (Fig. 2).
FIGURE 2.
Outline of a structure-based approach to the design and discovery of ligands that underpins early stage drug discovery. Compounds acquired in Stage 5 go into Stages 3 and 4 in an iterative process. KO, knock-out; RNAi, RNA interference.
Stage 1: Target Selection and Validation—In antimicrobial drug research, a target is sought that is proven essential for growth, survival, or infectious capability of the pathogen and that is either absent from humans or sufficiently distinct that species-specific inhibition is possible. Target selection was based previously on known medicinal chemistry and historical use of drugs. With genome sequencing and comparative bioinformatics, potentially important metabolic block points and targets can be predicted, and information about the importance of a potential pathogen target can be found by gene knock-out, RNA interference (gene knockdown), or chemical validation using an already known specific ligand. A catch-22 situation exists in that a target is not properly validated until a drug is found. The issue of species-specific inhibition requires detailed biochemical studies, the next stage of the process.
Stage 2: Target Characterization—A recombinant source of active material is sought for detailed biochemical and biophysical study, including structure determination by x-ray or NMR methods. The material would be used to develop appropriate activity or ligand binding assays. If the potential target has an ortholog in humans, then comparisons can assess if species-specific inhibition is likely.
Often the specific target from a pathogen does not provide material in a suitable form for characterization, and orthologs are used to circumvent this problem. The attrition rate in protein crystallization is particularly high, and it is routine for structural biologists to jump across species seeking crystals. The structure of an ortholog provides a template for homology modeling. An outcome of this stage is a structural template, obtained either directly or by modeling and understanding of the structure-function relationship of the target in a biological context. The model immediately informs on the issue of druggability. Caution is exercised when using homology models with experimental data sought to test the model. Although homology modeling might not be the optimal scenario, industry relies on it in particular for membrane-bound targets like G-protein-Coupled receptors (27).
Stage 3: Ligand/Inhibitor Studies—The first inhibitors or hits for the target are then sought. Hits, typically with IC50 values in the 1–30 μm range, can be identified by different means, and binding properties can be elucidated. HTS of compound libraries can be carried out if an appropriate assay is available. De novo design methods can be applied simply on the basis of possessing a template, but these methods have been replaced mainly by VS and docking calculations, which provide a more advanced starting point. Computational docking is estimated to fit ∼70–80% of ligands correctly (26), but bear in mind that a pre-selection of the type of active site being studied by this approach will have been made. SBLD can be applied directly if structural and biochemical information is available in particular on biologically relevant ligands that bind in an active site.
Stage 4: Target-Ligand Structure Determination—Based on results from Stage 3, structures of target-ligand complexes are sought to provide direct evidence of the molecular interactions used in binding. High-resolution crystal structures would be preferred. If suitable crystals are available, then FS can be carried out to directly identify hits, which can then be characterized as in Stage 3. If it is not possible to obtain an experimentally determined structure, then docking calculations can provide models for assessment and the next stage.
Stage 5: Chemical Design—Structures from Stage 4 are exploited to develop SAR around chemical scaffolds or ligand families. Taking into account medicinal chemistry experience and synthetic tractability, chemical modifications are designed, modeled, and put forward for synthesis. Computational methods and data base searches of commercially available compounds can identify ligands and fast track the development of SAR. Stages 4/5/6 are closely integrated, are highly dependent on accurate structural information, and form an iterative process that progresses from hits to leads, ligands with IC50 values <1 μm and well understood SAR.
Stage 6: Lead Series Development—The reliance on structural information is reduced, and the emphasis is on chemical modification of compound series based on the established SAR. A lead series (10–20 examples of a lead molecule with structural variations) is sought as an outcome here. The IC50 values of a useful lead series are likely to be <100 nm. The series should have the potential to supply candidates for preclinical assessment where aspects of ADMET are considered.
Druggability
An increasing awareness of what chemical starting point might be most appropriate for drug discovery has resulted in a better appreciation of what constitutes a useful target and the concept of druggability (28, 29). A druggable target is one able to bind drug-like compounds with lowμm affinity. The concept, which serves to reduce the number of probable drug targets, is dominated by a qualitative assessment of inhibition of protein families.
Quantitative measures of druggability would be extremely valuable. The steric fit in a binding site is straightforward to estimate, van der Waals contacts and H-bonds can be evaluated, and estimates of contributions to binding can be made. It becomes much more difficult to consider charge distributions, conformational flexibility, and solvation effects, and even determination of pKa values can prove troublesome. Despite uncertainties, developments in exploiting structural data together with physicochemical data on binding sites to predict druggability are encouraging (30).
Aspects of Protozoan Biology Where SBLD Might Prosper and What Might Help
Genomic science has transformed parasitology, with sequences of several protozoan parasites (Babesia, Eimeria, Giardia, Leishmania, Plasmodium, Toxoplasma, and Trichomonas/Trypanosoma species) available. Combined with comparative bioinformatics and biochemical and biological studies, these data have informed on basic aspects of eukaryote and pathogen biology and provide opportunities to identify and exploit novel and essential aspects of parasite metabolism in drug discovery (31–33). Targets for which there is a precedent of drug discovery are also sought. Particular areas where relevant targets might be found or old targets might be resurrected and exposed to modern methods include the biosynthetic routes to isoprenoid precursors (34) and downstream in farnesylation (35), glycolipid and fatty acids (36), protein kinases (37), folate metabolism (38), and the regulation of oxidative stress (39).
Antiparasite drug discovery will always have limited resources, and it would be beneficial if there were clarity with respect to validation (19) and prioritization of targets. More knock-out studies to identify essential gene products and access to specific inhibitors for chemical validation would underpin target assessment. More structural data on targets would allow assessments of druggability. In addition, improvements to in silico predictions of ADMET properties could reduce attrition levels and enhance chances of proceeding through the latter stages of the drug discovery process, where high attrition occurs (40).
With developments in HTS technologies and cell biology, it will become easier to carry out phenotypic screening using modern compound libraries. The hits, by virtue of being active, provide a head start but leave a requirement for further data if a target-dependent structure-based approach (to develop leads and lead series) is to be applied. The target(s) would have to be identified and then characterized. Without such data, traditional medicinal chemistry approaches would be required to develop SAR around the hits. With an increased appreciation about what types of structures affect parasites, then such a route has much to offer. In some cases, e.g. where target identification remains elusive or for multiprotein membrane-bound complexes, this may be the only way forward.
Nature's Bounty
Opinions are polarized about what natural products offer modern drug discovery. The complexity of such compounds makes chemists wary of having to develop demanding synthetic routes, and there are sourcing, ownership, and quality control issues. Nevertheless, such compounds continue to have an influence, and carefully considered natural product libraries have the potential to provide new scaffolds from which to work (41). New opportunities are likely to develop as automated relatively high-throughput phenotypic screening methods mature, so it would seem prudent to keep options open.
Polypharmacology
In recent times, even in this review, the emphasis has been on optimizing potency against an individual target, yet drugs often reach the clinic for an indication distinct from which they were first sought, and it would be rare for a drug to bind only one macromolecule in a cell. Experimental proof is beautifully encapsulated in a study that used similarities of the side effects observed for most marketed drugs as the phenotypic observation. This suggested that distinct molecular entities could elicit a biological effect by interaction with more than one target. Follow-up assays indeed show this to be the case (42). Given that some drugs act on multiple targets, then there may be significant benefits to adopting a polypharmacology approach that is the “promiscuous modulation of several molecular targets” (43). An important benefit of drugs that disrupt the activity of multiple targets could be increased potency and, in similar fashion to combination therapies, a decrease in drug resistance. The development of such an approach is at an early stage and more complex than dealing with a single target (44).
The Future
Hit finding is relatively straightforward with access to a compound library for HTS by interrogating a structure with VS or FS. However, ∼60% of small molecule drug discovery projects in industry have failed at the hit-to-lead stage (45). This might be due to a poor starting point and difficulties in synthesis but generally reflects the poor druggability or validity of the target. Poor targets need to be recognized and discarded as early as possible, allowing more effort and limited resource to be directed to the better targets. A difficulty arising from the success of genome sequencing is that the volume of data can complicate identification of the best targets. More genetic studies are required, and knowledge of potential target structures can also be informative. As our knowledge increases, the information garnered on drug metabolism and pharmacokinetics should inform on early decision-making and offers the potential to smooth the pathway through one of the major obstacles in drug discovery.
The pharmaceutical industry will not suddenly devote resources to antiparasite drug research and academic laboratories, although they possess a wealth of information on potential targets, and ligands are not professional drug finders. However, as in Ehrlich's time, alliances involving multinational government agencies and now charities are in place to broker deals and leverage input to support access to modern technologies such as HTS robotics, compound libraries, and expertise once confined to the pharmaceutical industry (46, 47). This provides opportunities for academics to contribute to early stage drug research and the expertise required to exploit and develop their discoveries. With the benefits that structure-based approaches offer, we can be cautiously optimistic about what we can do and therefore about what the future holds.
Supplementary Material
Author's Choice—Final version full access.
This work was supported by the Biotechnology and Biological Sciences Research Council (Structural Proteomics of Rational Targets), Wellcome Trust Grants 082596 and 083481, and European Union FP7 Health-223461. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: VSG, variant surface glycoprotein; SAR, structure activity relationships; HTS, high-throughput screening; ADMET, adsorption, distribution, metabolism, excretion, and toxicity; FS, fragment screening; VS, virtual screening; SBLD, structure-based ligand design.
References
- 1.Travis, A. S. (1991) Biochemist 13 9–12 [Google Scholar]
- 2.Hay, S. I., Guerra, C. A., Tatem, A. J., Noor, A. M., and Snow, R. W. (2004) Lancet Infect. Dis. 4 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett, M. P., Boykin, D. W., Brun, R., and Tidwell, R. R. (2007) Br. J. Pharmacol. 152 1155–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stuart, K., Brun, R., Croft, S., Fairlamb, A., Gürtler, R. E., McKerrow, J., Reed, S., and Tarleton, R. (2008) J. Clin. Investig. 118 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reithinger, R., Dujardin, J. C., Louzir, H., Pirmez, C., Alexander, B., and Brooker, S. (2007) Lancet Infect. Dis. 7 581–596 [DOI] [PubMed] [Google Scholar]
- 6.Kyes, S. A., Kraemer, S. M., and Smith, J. D. (2007) Eukaryot. Cell 6 1511–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor, J. E., and Rudenko, G. (2006) Trends Genet. 22 614–620 [DOI] [PubMed] [Google Scholar]
- 8.Croft, S. L., Barrett, M. P., and Urbina, J. A. (2005) Trends Parasitol. 21 508–512 [DOI] [PubMed] [Google Scholar]
- 9.Croft, S. L., Sundar, S., and Fairlamb, A. H. (2006) Clin. Microbiol. Rev. 19 111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones, K. E., Patel, N. G., Levy, M. A., Storeygard, A., Balk, D., Gittleman, J. L., and Daszak, P. (2008) Nature 451 990–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler, M. S., and Newman, D. J. (2008) Prog. Drug Res. 65 3–44 [DOI] [PubMed] [Google Scholar]
- 12.Newman, D. J., and Cragg, G. M. (2007) J. Nat. Prod. (Lloydia) 70 461–477 [DOI] [PubMed] [Google Scholar]
- 13.Lipinski, C. A., and Hopkins, A. (2004) Nature 432 855–861 [DOI] [PubMed] [Google Scholar]
- 14.Lipinski, C. A., Lombardo, F., Dominy, B. W., and Feeney, P. J. (2001) Adv. Drug Delivery Rev. 46 3–26 [DOI] [PubMed] [Google Scholar]
- 15.Lipinski, C. A. (2004) Drug Discov. Today 1 337–341 [DOI] [PubMed] [Google Scholar]
- 16.Schnur, D. M. (2008) Curr. Opin. Drug Discovery Dev. 11 375–380 [PubMed] [Google Scholar]
- 17.Brenk, R., Schipani, A., James, D., Krasowski, A., Gilbert, I. H., Frearson, J., and Wyatt, P. G. (2008) Chem. Med. Chem. 3 435–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson, J. G. (2005) Biochemistry 44 5561–5571 [DOI] [PubMed] [Google Scholar]
- 19.Frearson, J. A., Wyatt, P. G., Gilbert, I. H., and Fairlamb, A. H. (2007) Trends Parasitol. 23 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Congreve, M., Murray, C. W., and Blundell, T. L. (2005) Drug Discov. Today 10 895–907 [DOI] [PubMed] [Google Scholar]
- 21.Muchmore, S. W., and Hajduk, P. J. (2003) Curr. Opin. Drug Discovery Dev. 6 544–549 [PubMed] [Google Scholar]
- 22.Hajduk, P. J., and Greer, J. (2007) Nat. Rev. Drug Discov. 6 211–219 [DOI] [PubMed] [Google Scholar]
- 23.Erlanson, D. A., McDowell, R. S., and O'Brien, T. (2004) J. Med. Chem. 47 3463–3482 [DOI] [PubMed] [Google Scholar]
- 24.Rees, D. C., Congreve, M., Murray, C. W., and Carr, R. (2004) Nat. Rev. Drug Discov. 3 660–672 [DOI] [PubMed] [Google Scholar]
- 25.Hopkins, A. L., Groom, C. R., and Alex, A. (2004) Drug Discov. Today 9 430–431 [DOI] [PubMed] [Google Scholar]
- 26.Shoichet, B. K. (2004) Nature 432 862–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuccinardi, T., and Martinelli, A. (2007) Curr. Med. Chem. 14 3105–3121 [DOI] [PubMed] [Google Scholar]
- 28.Hopkins, A. L., and Groom, C. R. (2002) Nat. Rev. Drug Discov. 1 727–730 [DOI] [PubMed] [Google Scholar]
- 29.Keller, T. H., Pichota, A., and Yin, Z. (2006) Curr. Opin. Chem. Biol. 10 357–361 [DOI] [PubMed] [Google Scholar]
- 30.Cheng, A. C., Coleman, R. G., Smyth, K. T., Cao, Q., Soulard, P., Caffrey, D. R., Salzberg, A. C., and Huang, E. S. (2007) Nat. Biotechnol. 25 71–75 [DOI] [PubMed] [Google Scholar]
- 31.Chaudhary, K., and Roos, D. S. (2005) Nat. Biotechnol. 23 1089–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeh, I., and Altman, R. B. (2006) Mini-Rev. Med. Chem. 6 177–202 [DOI] [PubMed] [Google Scholar]
- 33.Myler, P. J. (2008) Adv. Exp. Med. Biol. 625 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunter, W. N. (2007) J. Biol. Chem. 282 21573–21577 [DOI] [PubMed] [Google Scholar]
- 35.Gelb, M. H., Brunsveld, L., Hrycyna, C. A., Michaelis, S., Tamanoi, F., Van Voorhis, W. C., and Waldmann, H. (2006) Nat. Chem. Biol. 2 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodman, C. D., and McFadden, G. I. (2008) Curr. Pharm. Des. 14 901–916 [DOI] [PubMed] [Google Scholar]
- 37.Naula, C., Parsons, M., and Mottram, J. C. (2005) Biochim. Biophys. Acta 1754 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahu, N. K., Sahu, S., and Kohli, D. V. (2008) Chem. Biol. Drug Des. 71 287–297 [DOI] [PubMed] [Google Scholar]
- 39.Krauth-Siegel, R. L., Meiering, S. K., and Schmidt, H. (2003) Biol. Chem. 384 539–549 [DOI] [PubMed] [Google Scholar]
- 40.Dearden, J. C. (2007) Expert Opin. Drug Metab. Toxicol. 3 635–639 [DOI] [PubMed] [Google Scholar]
- 41.Newman, D. J. (2008) J. Med. Chem. 51 2589–2599 [DOI] [PubMed] [Google Scholar]
- 42.Campillos, M., Kuhn, M., Gavin, A. C., Jensen, L. J., and Bork, P. (2008) Science 321 263–266 [DOI] [PubMed] [Google Scholar]
- 43.Hopkins, A. L., Mason, J. S., and Overington, J. P. (2006) Curr. Opin. Struct. Biol. 16 127–136 [DOI] [PubMed] [Google Scholar]
- 44.Hopkins, A. L. (2008) Nat. Chem. Biol. 4 682–690 [DOI] [PubMed] [Google Scholar]
- 45.Brown, D., and Superti-Furga, G. (2003) Drug Discov. Today 8 1067–1077 [DOI] [PubMed] [Google Scholar]
- 46.Waka, S., and Hudson, A. (2006) Nat. Rev. Drug Discov. 5 941–955 [DOI] [PubMed] [Google Scholar]
- 47.Renslo, A. R., and McKerrow, J. H. (2006) Nat. Chem. Biol. 2 701–710 [DOI] [PubMed] [Google Scholar]
- 48.Dawson, A., Gibellini, F., Sienkiewicz, N., Tulloch, L. B., Fyfe, P. K., McLuskey, K., Fairlamb, A. H., and Hunter, W. N. (2006) Mol. Microbiol. 6 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.