Abstract
The homothallic ascomycete Aspergillus nidulans serves as model organism for filamentous fungi because of its ability to propagate with both asexual and sexual life cycles, and fatty acid-derived substances regulate the balance between both cycles. These so-called psi (precocious sexual inducer) factors are produced by psi factor-producing oxygenases (Ppo enzymes). Bioinformatic analysis predicted the presence of two different heme domains in Ppo proteins: in the N-terminal region, a fatty acid heme dioxygenase/peroxidase domain is predicted, whereas in the C-terminal region, a P450 heme thiolate domain is predicted. To analyze the reaction catalyzed by Ppo enzymes, PpoA was expressed in Escherichia coli as an active enzyme. The protein was purified by 62-fold and identified as a homotetrameric ferric heme protein that metabolizes mono- as well as polyunsaturated C16 and C18 fatty acids at pH ∼7.25. The presence of thiolate-ligated heme was confirmed on the basis of sequence alignments and the appearance of a characteristic 450 nm CO-binding spectrum. Studies on its reaction mechanism revealed that PpoA uses different heme domains to catalyze two separate reactions. Within the heme peroxidase domain, linoleic acid is oxidized to (8R)-hydroperoxyoctadecadienoic acid by abstracting a H-atom from C-8 of the fatty acid, yielding a carbon-centered radical that reacts with molecular dioxygen. In the second reaction step, 8-hydroperoxyoctadecadienoic acid is isomerized within the P450 heme thiolate domain to 5,8-dihydroxyoctadecadienoic acid. We identify PpoA as a bifunctional P450 fusion protein that uses a previously unknown reaction mechanism for forming psi factors.
The fungus Aspergillus nidulans (teleomorph Emericella nidulans) is a homothallic ascomycete that has a defined sexual and asexual developmental cycle. Therefore, it serves as a model system for the understanding of fungal development (1). Oxidized unsaturated fatty acids, so-called oxylipins, derived from endogenous fatty acids were found to influence the development of the asexual conidiophores and sexual cleistothecia (2–6). Moreover, they seem to regulate the secondary metabolism of the fungus (7). These substances were collectively named psi factors and are primarily a mixture of hydroxylated oleic (18:1Δ9Z; x:yΔz denotes a fatty acid with x carbons and y double bonds in position z counting from the carboxyl end), linoleic (18:2Δ9Z,12Z), and α-linolenic (18:3Δ9Z,12Z,15Z) acids. They are termed psiβ, psiα, and psiγ, respectively. Psi factors can be further classified by the number and positioning of hydroxy groups on the fatty acid backbone: psiB (OH at C-8, e.g. (8R)-HODE),2 psiA (OH at C-5 and C-8, e.g. (5S,8R)-DiHODE), and psiC (OH at C-8 and the δ-lactone ring) (8, 9).
The psi factor (8R)-HODE was first discovered in the fungus Laetisaria arvalis (10, 11); it was later also found in Gaeumannomyces graminis (12, 13), where the first enzyme, which is responsible for production of (8R)-HPODE, 7,8-LDS, was detected (13). This heme-containing enzyme is bifunctional because it oxidizes 18:2Δ9Z,12Z in a first reaction step to (8R)-HPODE and subsequently isomerizes this intermediate compound to (7S,8S)-DiHODE (13–15).
After the genome of A. nidulans was available, Keller and co-workers (6, 16, 17) found three genes that share a high homology with the sequence of 7,8-LDS, namely ppoA, ppoB, and ppoC. They showed that the deletion of these genes had a significant effect (i) on the developmental ratio between the asexual conidiospores and sexual ascospores; (ii) on the production of psi factors; and (iii) on the production of secondary metabolites, the mycotoxins (6, 7, 16, 17). Furthermore, the encoded proteins showed remarkable sequence homology to both mammalian PGHS isoforms, enzymes that are responsible for the synthesis of prostaglandins (18). Using the NCBI conserved domain search analysis tool, it turned out that ppoA amino acid residues 210–580 contain a domain similar to mammalian heme peroxidases, whereas residues 650–1050 contain a CYPX domain, similar to P450 heme thiolate enzymes (16). However, for 7,8-LDS from G. graminis, only the mammalian heme peroxidase domain is predicted. The identity of conserved catalytic domains between Ppo enzymes and mammalian PGHS ranges from 25 to 29% for PGHS-2 and from 25 to 26% for PGHS-1 (19). PpoA and 7,8-LDS show 42% amino acid identity.
Oliw and co-workers (20) observed that incubation of homogenates of mycelia of A. nidulans with 18:2Δ9Z,12Z converted the fatty acid to (8R)-HODE and (5S,8R)-DiHODE as the major products. (8R)-HPODE, (10R)-HODE, and (10R)-HPODE were detected as minor products. Incubation of mycelia of Aspergillus fumigatus with deuterium-labeled 18:2Δ9Z,12Z revealed that the synthesis of (8R)-HPODE is accomplished via pro-S-hydrogen abstraction at C-8 and antarafacial dioxygen insertion. (5S,8R)-DiHODE is generated via an additional pro-S-hydrogen abstraction at C-5 of the substrate (20, 21).
Additional studies with fungal knock-out strains led to the hypothesis that PpoA may be responsible for the synthesis of (8R)-hydroperoxides, which are partially reduced to (8R)-hydroxides (20). It was suggested that, analogous with 7,8-LDS, (8R)-hydroperoxides are then converted to 5,8-dihydroxides by PpoA. Furthermore, it was concluded that ppoC may code for linoleate (10R)-DOX (20). Analysis of Ppo enzymes from A. nidulans in studies published so far has been performed either by using knock-out mutants to demonstrate the absence of a subset of psi factors or by using crude mycelial extracts; both experimental setups have the disadvantage of observing multiple enzymatic reactions in parallel.
To characterize the biochemical properties of PpoA in more detail, we cloned and expressed recombinant PpoA in Escherichia coli. After purification of the enzyme by up to 62-fold, biochemical characterization was performed. The studies revealed mechanistic as well as structural similarities to and differences from 7,8-LDS from G. graminis. Both enzymes were found to be homotetrameric ferric heme proteins that catalyze the synthesis of (8R)-HPODE. Whereas G. graminis 7,8-LDS converts the intermediate formed to (7S,8S)-DiHODE, PpoA produces 5,8-DiHODE.
Using site-directed mutagenesis, we provide evidence that there are striking differences between both enzymes regarding the catalytic reaction cycle. Thus, we found that PpoA uses different domains to catalyze the two reaction steps. We suggest that the DOX reaction, yielding 8-HPODE, takes place in the N-terminal heme peroxidase domain. The isomerization of this intermediate product to the end product, 5,8-DiHODE, is accomplished, however, independently by the C-terminal P450 heme thiolate domain in an 8-hydroperoxide isomerase reaction.
In addition, we are able to provide evidence that, during the catalysis, PpoA generates a carbon-centered radical presumably at C-8, like G. graminis 7,8-LDS. Furthermore, we determined the kinetic parameters for the first reaction step.
EXPERIMENTAL PROCEDURES
Materials—Chemicals were obtained from Sigma and Carl Roth & Co. (Karlsruhe, Germany). Agarose was from Biozym Scientific GmbH (Hessisch Oldendorf, Germany). All fatty acids were from Sigma or Cayman Chemical (Ann Arbor, MI). Acetonitrile was from Fisher. Restriction enzymes were purchased from MBI Fermentas (St. Leon-Roth, Germany).
Cloning and Expression of Recombinant PpoA in E. coli—A. nidulans ppoA (GenBank™ accession number AY502073) was amplified from fungal cDNA of sexual and vegetative stages using gene-specific primers containing NheI and NotI recognition sites (forward primer, 5′-AGCTAGCATGGGTGAAGACAAAGAAAVAAATATC-3′; and reverse primer, 5′-AGCGGCCGCTTAAAAATCTTCCTTCAGTTGGGGAG-3′) via the Expand High Fidelity system (Roche Diagnostics) as described (22). PCR amplification was performed under the following conditions: 94 °C for 2 min, followed by 10 cycles at 94 °C for 30 s, 53 °C for 30 s, and 72 °C for 3 min. These cycles were followed by 15 cycles at 94 °C for 30 s, 53 °C for 30 s, and 72 °C for 3 min (5-s time increment) and terminated by 5 min at 72 °C.
The resulting fragment was cloned into pGEM-T (Promega), yielding the plasmid pGEM-T-PpoA. For expression in E. coli, ppoA was cloned into the pET24a expression vector (Novagen) by NheI and NotI, yielding the plasmid pET24-PpoA, and transformed into E. coli Bl21 Star cells (Invitrogen).
Cells were cultivated in 2×YT broth containing 25 μg/ml kanamycin and grown to A600 = 0.6–0.8. Expression of recombinant protein was then induced by 0.1 mm isopropyl β-d-thiogalactopyranoside, and cultures were cultivated by shaking at 28 °C for 18 h. Cells were harvested by centrifugation at 8000 × g for 10 min at 4 °C. The precipitate was shock-frozen in liquid nitrogen and stored at –20 °C.
Site-directed Mutagenesis—In vitro mutagenesis was carried out using the Phusion™ Hot Start High Fidelity DNA polymerase system (Finnzymes, Espoo, Finland). The following primers were used: H1004A, 5′-CACTTTGGCTTTGGGCCTGCCAAGTGTTTGGGCTTAG-3′ (sense) and 5′-CTAAGCCCAAACACTTGGCAGGCCCAAAGCCAAAGTG-3′ (antisense); and Y374F, 5′-CAGGTGTCAGCCGAATTCAATGTCGTGTTCCGGTGGCACGCTTGC-3′ (sense) and 5′-GCAAGCGTGCCACCGGAACACGACATTGAATTCGGCTGACACCTGG-3′ (antisense). Replacement of Cys1006 with Ala was achieved by overlap extension PCR as described (23). The above-mentioned gene-specific primers as well as the following mutagenic primers were used for PCR: C1006A, 5′-GGCTTTGGGCCCCACAAGGCCTTGGGCTTAGACCTATG-3′ (sense) and 5′-CATAGGTCTAAGCCCAAGGCCTTGTGGGGCCCAAAGCC-3′ (antisense). Mutation was confirmed by DNA sequencing. In the case of the C1006A mutant, an additional amino acid exchange according to the sequence of the wild-type enzyme was found: Tyr658 with Asp.
Protein Purification—Harvested cells from 100 ml of culture were resolved in 20 ml of 50 mm Tris-HCl (pH 7.6) containing 10% (v/v) glycerol and 5 mm EDTA. After lysozyme was added to a final concentration of 0.1 mg/ml and the protein inhibitor phenylmethylsulfonyl fluoride to a final concentration of 0.2 mm, cells were incubated for 30 min on ice. Subsequently, cell lysis was increased by subjecting the cells to three freeze/thaw cycles in liquid nitrogen and sonication. Cell debris was centrifuged at 45,000 × g for 20 min at 4 °C.
The supernatant from cell lysis was loaded onto Source 30Q resin (25 ml, XK 16/20 column; GE Healthcare) at a flow rate of 1 ml/min using anÄKTA FPLC system (GE Healthcare). The column was washed with 50 ml of 50 mm Tris-HCl (pH 7.6), and protein was eluted with linear gradient of 0–0.3 m NaCl in 50 mm Tris-HCl (pH 7.6) within 10 min at a flow rate of 3 ml/min. 2-ml fractions were collected. Fractions with the highest enzyme activity (as judged by Clark-type O2 electrode) and the highest purity (as judged by SDS-PAGE) were combined and concentrated to a final volume of 2 ml using Vivaspin 20 (molecular mass cutoff of 100,000 Da; Sartorius Stedim Biotech S.A., Göttingen, Germany).
For gel filtration, the enzyme concentrate was loaded onto a Superdex S200 26/60 prep grade column (GE Healthcare) equilibrated with 50 mm Tris-HCl (pH 7.6). Elution was performed at a flow rate of 1 ml/min. Fractions containing enzyme activity were combined and subsequently loaded onto a Mono Q 10/10 column (GE Healthcare). After loading and extensive washing with 50 mm Tris-HCl (pH 7.6), protein was eluted at 3 ml/min with a linear gradient to 50 mm Tris-HCl (pH 7.6) containing 300 mm NaCl within 10 min.
Spectroscopy—UV-visible spectroscopy was performed at room temperature using a dual-beam Uvikon 930 spectrophotometer (Kontron Instruments, Munich, Germany). CD spectroscopy was performed using Jasco J-810 chiro-optical spectrometer.
High spin states of the protein were induced by adding NaF to the enzyme solution. Analogous low spin states were induced by adding KCN (24). The iron of the protein was reduced by adding it to degassed buffer containing sodium dithionite. The reduced CO spectrum was obtained as described by Omura and Sato (25).
Activity Assay—250 μg of 18:2Δ9Z,12Z or 5.5 MBq of radiolabeled [1-14C]18:2Δ9Z,12Z were dissolved in 1 ml of 50 mm HEPES (pH 7.4). The reaction was started by the addition of 15 μg of enzyme to the solution and was allowed to proceed for 30 min at room temperature on a shaker. Incubations were stopped by the addition of 2 ml of diethyl ether, and fatty acids and their derivatives were extracted. A second extraction was performed with 2 ml of diethyl ether and 100 μl of glacial acid.
Incubations under an atmosphere of 18O2 were performed after degassing 1 ml of the reaction system (50 mm Na2HPO4/NaH2PO4 buffer (pH 7.2)). 18O2 (Campro Scientific, Berlin, Germany) was fed into the reaction system until it was saturated. 250 μg of 18:2Δ9Z,12Z were added, and the reaction was incubated for 30 min at room temperature. During the incubation, the reaction mixture was further enriched with 18O2 by continuous gas flow through the liquid. The reaction was terminated with diethyl ether and extracted as described above.
Determination of the pH Optimum—Using a Clark-type oxygen electrode (Rank Brothers, Bottisham, Cambridge, UK), the initial time-dependent O2 consumption at a given pH was determined with the sodium salt of 18:2Δ9Z,12Z as a substrate at a concentration of 100 μm. The following buffers were used for the different pH ranges: 0.2 m acetate buffer (pH 4.5–5.5), 0.2 m phosphate buffer (pH 5.5–8.0), and 0.2 m borate buffer (pH 8.0–10.5). It was verified that the different buffer systems had no influence on enzyme activity.
HPLC Analysis—Extracts from activity assays were dried under nitrogen stream and dissolved in 80 μl of acetonitrile/water/acetic acid (50/50/0.1, v/v/v). Separation of fatty acids and their derivatives was carried out via reversed phase (RP) HPLC using an EC250/2 Nucleosil 120-5 C18 column (2.1 × 250 mm, 5-μm particle size; Macherey-Nagel, Düren, Germany) on an 1100 HPLC system (Agilent, Waldbronn, Germany) coupled to a diode array detector. For detection of hydroxy fatty acids, the absorbance at 202 nm was recorded. For detection of 1-14C-radiolabeled derivatives, a Raytest radio detector was used.
The solvent systems were as follows: solvent system A, acetonitrile/water/acetic acid (50/50/0.1, v/v/v); and solvent system B, acetonitrile/acetic acid (100/0.1, v/v). The gradient elution profile was as follows: flow rate of 0.18 ml/min, 0–5 min, 100% A; 5–20 min from 100% A to 100% B; 20–22-min flow rate increase to 0.36 ml/min; 22–27 min, 100% B; 27–32 min from 100% B to 100% A; and 32–35 min, 100% A, flow rate decreased to 0.18 ml/min.
For steric analysis, the monohydroperoxide was reduced to its analogous hydroxy derivative with SnCl2 (300 μg/900 μl) by incubation for 10 min and further purified by straight phase HPLC. Straight phase HPLC was carried out on a Zorbax SIL column (4.6 × 250 mm, 5-μm particle size; Agilent) eluted with a solvent system of n-hexane/2-propanol/acetic acid (100/1/0.1, v/v/v) at a flow rate of 0.2 ml/min. Chiral phase HPLC of the hydroxy fatty acids was carried out on a Daicel CHIRALCEL OD-H column (2.1 × 150 mm, 5-μm particle size; VWR, Darmstadt, Germany) with a solvent system of n-hexane/2-propanol/acetic acid (100/5/0.1, v/v/v).
HPLC/MS Analysis—The HPLC assembly was a Surveyor HPLC system with an EC250/2 Nucleosil 100-5 C18 column (2.1 × 250 mm, 5-μm particle size; Macherey-Nagel). The solvent systems were as follows: solvent system A, acetonitrile/water/acetic acid (40/60/0.1, v/v/v); and solvent system B, acetonitrile/acetic acid (100/0.1, v/v). The gradient elution profile was as follows: flow rate of 0.2 ml/min, 0–10 min, 80% A and 20% B; 10–30 min from 80% A and 20% B to 100% B; 30–35-min flow rate increase to 0.3 ml/min; 35–40 min, 100% B; 40–44.5 min from 100% B to 80% A and 20% B; and 44.5–45 min, 80% A and 20% B, flow rate decreased to 0.2 ml/min.
The mass spectrometer used was a Thermo Finnigan LCQ ion trap mass spectrometer with electrospray ionization and monitoring of negative ions. The capillary temperature was 300 °C, and the capillary voltage was 27 kV. For tandem MS analysis, the collision energy was 1 V.
GC/MS Analysis—The identity of hydroxy fatty acids was verified by GC/MS. After separation via RP-HPLC, the products were extracted with 2 ml of diethyl ether and dried under nitrogen stream. For analysis by GC/MS, the hydroperoxy fatty acids were reduced with SnCl2 as described above and methylated by dissolving the samples in 400 μl of methanol containing 6.5 μl of 2 m (diazomethyl)trimethylsilane (in hexane). After incubation for 30 min on a shaker, the reaction was terminated by the addition of 0.2 μl of glacial acid. The fatty acid methyl esters were extracted with 2 ml of diethyl ether, dried under nitrogen stream, and subsequently dissolved in 3 μl of acetonitrile. All fatty acid methyl esters were trimethylsilylated with 1 μl of N,O-bi(trimethylsilyl)trifluoroacetamide. Analysis was carried out using an Agilent 5973 mass selective detector connected to an Agilent 6890 gas chromatograph equipped with a capillary DB-23 column (30 m × 0.25 mm, 0.25-μm coating thickness). Helium was used as carrier gas (1 ml/min). The temperature gradient was 150 °C for 1 min, 150–200 °C at 4 K min–1, 200–250 °C at 5 K min–1, and 250 °C for 6 min.
Determination of Kinetic Parameters—For analysis of the initial reaction rate data, the time-dependent O2 consumption was measured with a Clark-type O2 electrode at different substrate concentrations. The kinetic experiments were conducted at 24 °C in 50 mm phosphate buffer (pH 7.2). Reactions were initiated by the addition of 15 μg of purified enzyme to 1 ml of the rapidly stirring buffer containing a defined amount of substrate. The solution was pre-equilibrated against air at 1 atm. The following substrates were used: sodium salts of 16:1Δ9Z, 18:1Δ9Z, 18:2Δ9Z,12Z, and 18:3Δ9Z,12Z,15Z. Kinetic parameters were obtained by fitting five to seven data points to the Hanes-Woolf equation: v = (Vmax·[S])/(Km + [S]) ⇔ [S]/v = Km/Vmax + 1/Vmax·[S], where v is the initial reaction rate, Vmax is the maximum reaction rate, [S] is the concentration of substrate, and Km is the Michaelis constant.
Lipid-derived Radical Trapping by Nitroxyl Radicals—Lipid-derived radicals generated in the 18:2Δ9Z,12Z/PpoA system were trapped with nitroxyl radicals as follows. 250 μg of 18:2Δ9Z,12Z were dissolved in 1 ml of 50 mm HEPES buffer (pH 7.4) containing 2% EtOH and 5 mm EDTA. The solution was mixed with 500 μl of 5 mm CmΔP in HEPES buffer as described above. The reaction was started by the addition of 15 μg of PpoA from stock solution (10 μg/μl in 50 mm HEPES (pH 7.4)), and the reaction mixture was covered with argon. Products were extracted as described above and analyzed by HPLC/MS by monitoring of positive ions within a mass range of m/z 50–600 as described above.
Peroxidase Assay—To determine peroxidase activity, the method described by Kulmacz (26) was used. 100 μm TMPD and 100 μm hydroperoxide-free 18:2Δ9Z,12Z were dissolved in 1 ml 50 mm HEPES (pH 7.4) and incubated with 15 μg of PpoA at room temperature. The peroxidase activity was monitored by the increase in absorbance at 611 nm due to oxidized TMPD using the dual-beam Uvikon 930 spectrophotometer.
RESULTS
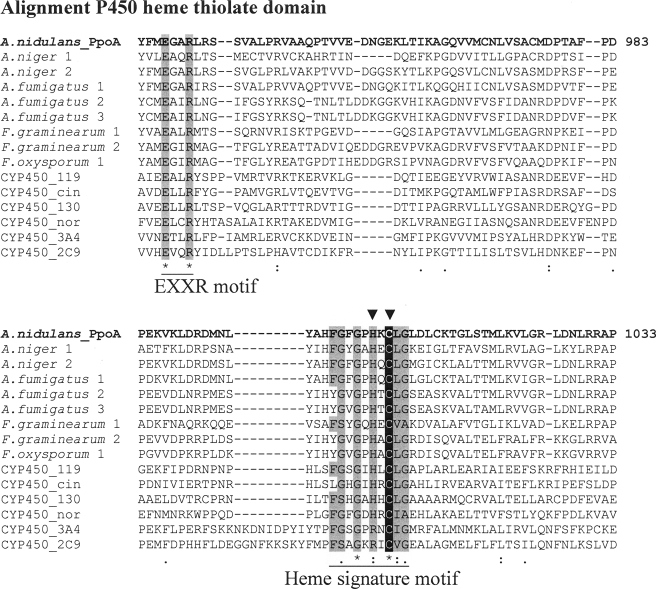
Domain Structure and Alignments—BLAST search analysis predicted for PpoA the presence of an N-terminal heme peroxidase as well as a C-terminal P450 heme thiolate domain. By aligning the sequence of the heme peroxidase domain of PpoA with those of 7,8-LDS (Swiss-Prot accession number Q9UUS2.1), sheep PGHS-1 (P05979.2), and sheep PGHS-2 (P79208.1), we found that most residues reported to be essential for the catalytic activity of 7,8-LDS and PGHS are conserved (supplemental Fig. 1A). Furthermore, alignments of the C-terminal P450 heme thiolate domain with sequences of known P450 heme thiolate enzymes from different organisms and with predicted P450 heme thiolate domains from different putative fungal Ppo-like enzymes also showed conserved residues (Fig. 1). Additionally, we compared the sequence of the P450 heme thiolate domain from PpoA with the known consensus sequence of the heme signature motif of P450 enzymes according to the PROSITE Database. All identified amino acid residues within the PpoA sequence are shaded gray in Fig. 1. Moreover, we also identified the highly conserved EXXR motif that is one of the most conserved residues in P450 heme thiolate enzymes. A full alignment of the P450 heme thiolate domain is shown in supplemental Fig. 1B. Therefore, PpoA was assigned as CYP6001A1 by Dr. D. R. Nelson of the P450 Nomenclature Committee (Department of Molecular Sciences, University of Tennessee).
FIGURE 1.
Partial alignment of the P450 domain from PpoA with sequences from different P450. A, partial alignment of the P450 domain from PpoA with known CYP450 enzymes (CYP450_119 (Protein Data Bank code 1F4T, B chain), CYP450_cin (code 1T2B, B chain), CYP450_130 (code 2UUQ, A chain), CYP450_nor (code 1JFB, A chain), CYP4503A4 (code 1TQN, A chain), and CYP4502C9 (code 1R9O, A chain)) and predicted CYP450 domains from different putative fungal Ppo-like enzymes (Aspergillus niger 1 (XP_001395220), A. niger 2 (XP_001401954) A. fumigatus 1 (EDP50447), A. fumigatus 2 (ABV21632.1), A. fumigatus 3 (EDP47075.1), Fusarium graminearum 1 (FGSG_02668; annotation code in accordance to the Fusarium Comparative Database), F. graminearum 2 (FGSG_10960)A, and F. oxysporum 1 (FOXG_12909). Amino acids matching the P450 heme signature consensus sequence according to the PROSITE Database as well as the known EXXR motif of P450 heme thiolate enzymes are shaded gray. Additionally, the cysteine responsible for the ligation of heme iron is shaded black. The mutated histidine and cysteine are labeled with arrowheads. The alignment was carried out using ClustalW.
Cloning and Functional Expression of PpoA—cDNA from vegetative and sexual stages of A. nidulans and gene-specific primers containing appropriate restriction sites were used to amplify via PCR a DNA fragment corresponding to the open reading frame of ppoA. After subcloning into pGEM-T, ppoA was finally cloned into the expression vector pET24, yielding plasmid pET24-PpoA. Sequencing confirmed the open reading frame with 3246 bp and 1081 amino acids, showing three amino acid residue exchanges according to the published genome sequence (1): Lys166 with Pro, Asp417 with Arg, and Ile859 with Val. Functional expression of PpoA was performed in E. coli Bl21 Star cells, and optimal cultivation conditions (28 °C, 18 h) were determined. Expression of PpoA was verified by SDS-PAGE, and for activity tests, oxygen consumption was measured using a Clark-type oxygen electrode. However, expression of tagged recombinant protein led to inactive protein.
Purification of PpoA—Extraction and solubilization of PpoA using crude extract from bacteria were done with 50 mm Tris-HCl (pH 7.6) containing 10% (v/v) glycerol and 5 mm EDTA. After each purification step, activity was determined using a Clark-type oxygen electrode. For the initial purification step, Source 30Q resin (25 ml, XK 16/20 column) was chosen to isolate and concentrate PpoA. Although >60% of PpoA was lost during this step (recovery rate of 33%), it also resulted in 4-fold purification (Table 1). The subsequent purification step using gel filtration was required to remove large and small aggregates of proteins. Most of the contaminants could be removed with this step, whereas the recovery of protein was very high at 28%. The last step using the Mono Q column was used to remove most of the remaining contaminants and resulted in 62-fold enrichment of PpoA (Table 1). This purification protocol yielded typically between 2 and 4 mg of protein/600 ml of culture.
TABLE 1.
Purification of recombinant A. nidulans PpoA
| Purification step | Total proteina | Specific activityb | Recovery | Purification |
|---|---|---|---|---|
| mg | μmol/min/mg | % | -fold | |
| Crude extract | 822 | 0.051 | 100 | 1 |
| Source 30Q | 64.8 | 0.215 | 33 | 4 |
| Superdex S200 26/60 | 8 | 1.47 | 28 | 28 |
| Mono Q 10/10 | 3.6 | 3.2 | 27 | 62 |
Protein concentration was determined using the method of Bradford (55) with bovine serum albumin as a standard.
Activity was determined using a Clark-type O2 electrode; oxygen consumption was monitored using sodium salt of linoleic acid (18:2Δ9Z,12Z) as a substrate at a concentration of 100 μm.
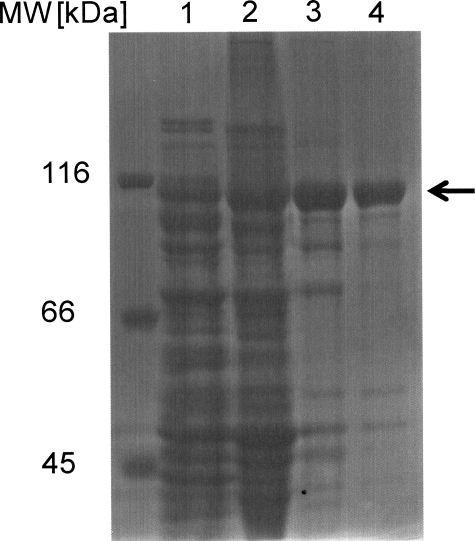
Molecular Mass and Oligomeric Structure—Protein obtained from the different stages of protein purification was subjected to SDS-PAGE (Fig. 2). After each purification step, a protein band at ∼110 kDa became more prominent, and the last purification step on Mono Q resulted in almost pure protein, where only traces of other proteins could be detected. The molecular mass for PpoA of 110 kDa as determined by SDS-PAGE appeared to be slightly smaller as calculated by its amino acid components (120 kDa).
FIGURE 2.
SDS-PAGE at different stages of purification. Aliquots after each purification step were analyzed by 8% SDS-PAGE. Lane 1, crude cell extract; lane 2, after Source 30Q; lane 3, after Superdex S200 (26/60); lane 4, after Mono Q (10/10).
Analysis and purification with the gel filtration column yielded a protein with a molecular mass of 440 kDa. This size was estimated to be 4-fold larger than that obtained upon SDS-PAGE, which led to the conclusion that the native protein consists of four subunits. When SDS-PAGE analysis was performed without treatment of reducing agent, the band of the corresponding molecular mass did not change (data not shown). This may indicate that the four subunits are not covalently bound by disulfide bridges.
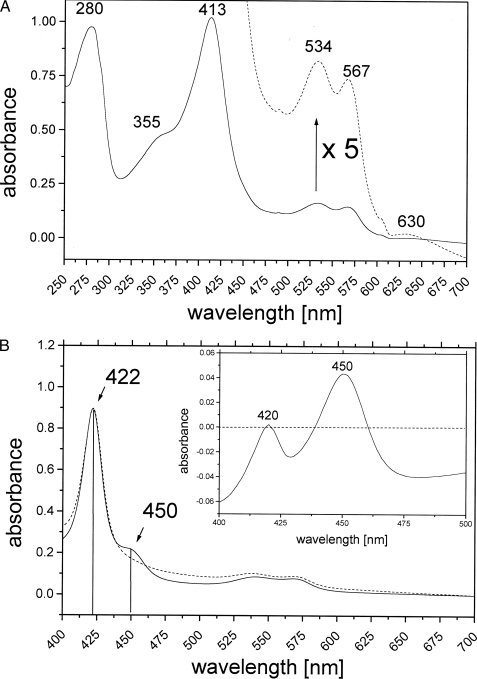
Spectral Analysis and Prosthetic Groups—The UV-visible absorption spectrum of purified PpoA is shown in Fig. 3A. The highest absorption maxima were at 280 and 413 nm (γ, Soret band). Additionally, four broader but weaker maxima could be detected at 534 (β), 567 (α), 355 (δ), and 630 nm, indicative of a cytochrome b group. The absorption maxima at 413 and 630 nm suggest that PpoA is a ferric heme protein in the high spin state (24). The ferric iron of heme proteins can be transformed by ions like CN– and F– into low and high spin states, respectively (24). CN– formed low spin derivatives of PpoA in which the γ-band was shifted to 418 nm, whereas F– had no effect on PpoA, confirming that PpoA contains ferric heme in the high spin state. Treatment with sodium dithionite shifted the Soret maximum to 422 nm.
FIGURE 3.
Spectral analysis of PpoA. A, UV-visible spectrum of the native enzyme in 50 mm HEPES (pH 7. 4). The UV-visible spectrum of the purified enzyme showed absorption maxima at 280 and 413 nm (γ, Soret) and smaller maxima at 355 nm (δ), 534 nm (β), 567 nm (α), and 630 nm. The dashed line represents a 5-fold enlargement of the spectrum. B, UV-visible spectrum of the dithionite-reduced heme-CO complex of PpoA. The spectrum was obtained as described by Omura and Sato (25). The spectrum of dithionite-reduced PpoA shows a Soret band at 422 nm (dashed line). Incubation of the reduced enzyme with CO resulted in the formation of an additional transient band at 450 nm (solid line) that is also illustrated by a corresponding maximum in the difference absorbance spectrum (inset). The heme-CO complex of PpoA appeared to be unstable and decayed to a species known as P420 with absorption maximum at 420 nm as also illustrated in the difference spectrum.
To confirm the postulated presence of a P450 heme thiolate domain as part of PpoA, the protein was reduced by the addition of dithionite and the subsequent reaction of the reduced enzyme with physically dissolved CO. The UV-visible absorption spectra of the reduced protein and the reduced heme-CO complex of the protein as well as a difference spectrum (inset) are shown in Fig. 3B. The spectrum of dithionite-reduced PpoA shows a bathochromic shift of the Soret band from 413 toward 422 nm. Incubation of the reduced enzyme with CO resulted in the formation of an additional transient band at 450 nm, which is also illustrated by a corresponding maximum in the difference absorbance spectrum. The absorbance of a reduced heme-CO complex at 450 nm is a generally known property of P450 heme thiolate enzymes (25, 27). In the case of PpoA, the CO complex appeared to be unstable and decayed after a few minutes, forming a species termed P420, as already described, for example, for P450bisd from Sphingomonas sp. (28) or prostacyclin synthase (29). This species is formed by weakening or distorting the heme thiolate bond and exhibits an absorption maximum of the reduced CO complex at 420 nm (25). The weak signal at 450 nm and at 420 nm is also indicative of an incomplete reaction, as has also been reported for other P450 heme thiolate enzymes such as the allene oxide synthases (30).
Identification of Products—Purified PpoA was incubated with [1-14C]18:2Δ9Z,12Z for 30 min on a shaker. After extraction of fatty acids and their derivatives, analysis was performed by RP-HPLC with a radio detector. Two new peaks were detected as shown in Fig. 4. The more polar one is labeled as product 2 and had a retention time of 10.8 min on the RP column. The less polar one eluted after 21.4 min from the column and is labeled as product 1. It is noteworthy that the amount of product 1 varied in different experiments from undetectable traces to high amounts, as is shown in Fig. 4.
FIGURE 4.
HPLC analysis of metabolites formed from [1-14C]18:2Δ9Z,12Z by PpoA. Shown are the results from RP-HPLC analysis after 60 of min incubation of 5.5 MBq of radiolabeled [1-14C]18:2Δ9Z,12Z with 15 μg of PpoA in 50 mm HEPES buffer (pH 7.4) and extractive isolation. Inset, chiral phase HPLC of product 1. LA, linoleic acid; ret., retention; rel., relative.
For further analysis, unlabeled 18:2Δ9Z,12Z was used. Product 1 was treated with SnCl2 and reanalyzed by RP-HPLC. The untreated compound showed a higher retention time on the column than the treated compound. The shift in the retention time between both compounds was ∼0.6 min.
The identities of the reduced products 1 and 2 could be solved by the mass spectra of their corresponding Me3Si ether methyl ester derivatives. The mass spectrum of product 2 is shown in supplemental Fig. 2A and displays signals at m/z 129 (CH(OSiCH3)3-C2H5), 203 (cleavage between C-5 and C-6), 239 (cleavage between C-7 and C-8), 269 (cleavage between C-10 and C-11), and 282 (cleavage between C-12 and C-13). The base peak was at m/z 73. These data indicate a dihydroxy metabolite with the OH groups at C-5 and C-8.
The mass spectrum of product 1 (after treatment with SnCl2) is shown in supplemental Fig. 2B and displays a fragmentation pattern analogous to the mass spectrum of product 2 with the exception of the disappearance of the m/z 203 signal. This result indicates a monohydroxylated metabolite with the OH group at C-8. Steric analysis of the hydroperoxy product (product 1) via chiral phase HPLC showed that it is generated in R-configuration with a specificity of 95%. The chiral phase HPLC chromatogram is shown in Fig. 4 (inset).
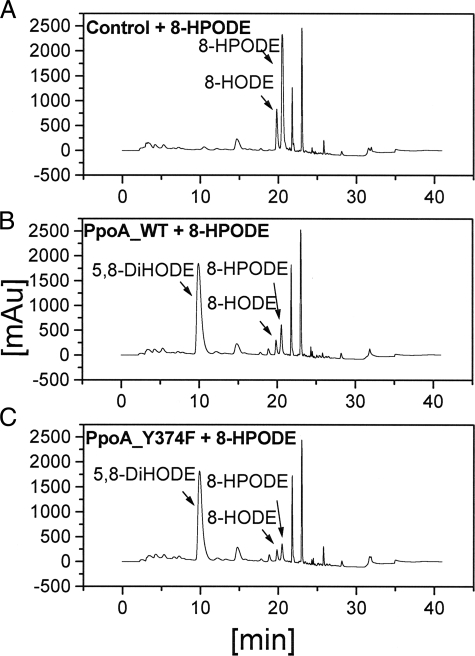
In most experiments, we were also able to detect small amounts of 8-HODE as a side product (see Fig. 7B). Additionally, when the formation of 5,8-DiHODE was high, we detected an additional product that eluted at ∼18 min. After methylation and derivatization, this product showed the same mass spectrum as 5,8-DiHODE. We concluded from this finding that 5,8-DiHODE can be nonenzymatically rearranged by an intramolecular esterification to 8-hydroxy-δ-lactone.
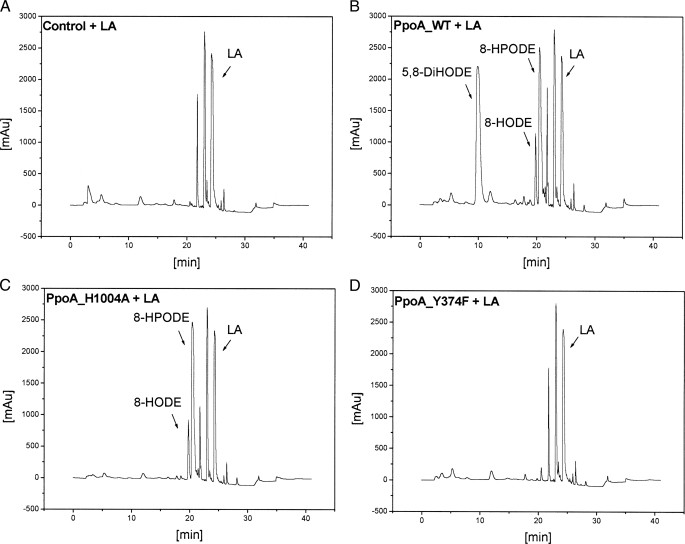
FIGURE 7.
HPLC analysis of products formed from 18:2Δ9Z,12Z by PpoA and mutants H1004A and Y374F. Shown are the results from RP-HPLC analysis after 60 min of incubation of 18:2Δ9Z,12Z with ∼15 μg of heat-inactivated PpoA as a control (A), wild-type (WT) PpoA (B), H1004A (C), and Y374F (D) in 50 mm HEPES buffer (pH 7.4) and extractive isolation. LA, linoleic acid; mAU, milliabsorbance units.
We next tested different fatty acids as substrates for the PpoA enzyme, especially fatty acids occurring endogenously in A. nidulans. In fatty acid profiles of the fungi, only fatty acids with a chain length of up to 18 carbon atoms could be detected (data not shown). As supposed, PpoA converted these additional fatty acids (16:1Δ9Z, 18:1Δ9Z, and 18:3Δ9Z,12Z,15Z) to their corresponding 8-monohydroperoxy and 5,8-dihydroxy derivatives (Table 2). In addition, we tested C20 fatty acids with at least one double bound at C-11, and we found that these compounds were converted to their corresponding 10-hydroperoxy and 7,10-dihydroxy derivatives (Table 2).
TABLE 2.
Oxygenation products of A. nidulans PpoA Products of C16 and C18 fatty acids were identified as Me3Si ether methyl ester derivatives by GC/MS analysis after separation by RP-HPLC.
HPHME, hydroperoxyhexadecenoic acid; HPOME, hydroperoxyoctadecenoic acid; HPOTrE, hydroperoxyoctadecatrienoic acid; HPEME, hydroperoxyeicosenoic acid; HPEDE, hydroperoxyeicosadienoic acid; HPETrE, hydroperoxyeicosatrienoic acid; DiHHME, dihydroxyhexadenoic acid; DiHOME, dihydroxyoctadecenoic acid; DiHOTrE, dihydroxyoctadecatrienoic acid; DiHEME, dihydroxyeicosenoic acid; DiHEDE, dihydroxyeicosadienoic acid; DiHETrE, dihydroxyeicosatrienoic acid.
| Substrate | Product 1 | Product 2 |
|---|---|---|
| 16:1Δ9Z | 8-HPHME | 5,8-DiHHME |
| 18:1Δ9Z | 8-HPOME | 5,8-DiHOME |
| 18:2Δ9Z,12Z | 8-HPODE | 5,8-DiHODE |
| 18:3Δ9Z,12Z,15Z | 8-HPOTrE | 5,8-DiHOTrE |
| 20:1Δ11Z | 10-HPEMEa | 7,10-DiHEMEb |
| 20:2Δ11Z,14Z | 10-HPEDEa | 7,10-DiHEDEb |
| 20:3Δ11Z,14Z,17Z | 10-HPETrEa | 7,10-DiHETrEb |
Hydroperoxide fatty acids were unequivocally identified; the position of hydroperoxide was deduced from the corresponding dihydroxides.
Dihydroxy products of C20 fatty acids were identified by HPLC/tandem MS analysis and by detection of characteristic fragments: cleavage between C-7 and C-8 with formation of a characteristic signal at m/z 143 (HOOC-(CH2)5COH-) and cleavage between C-10 and C-11 with a characteristic signal at m/z 201 (HOOC-(CH2)5-COH-(CH2)2-COH-).
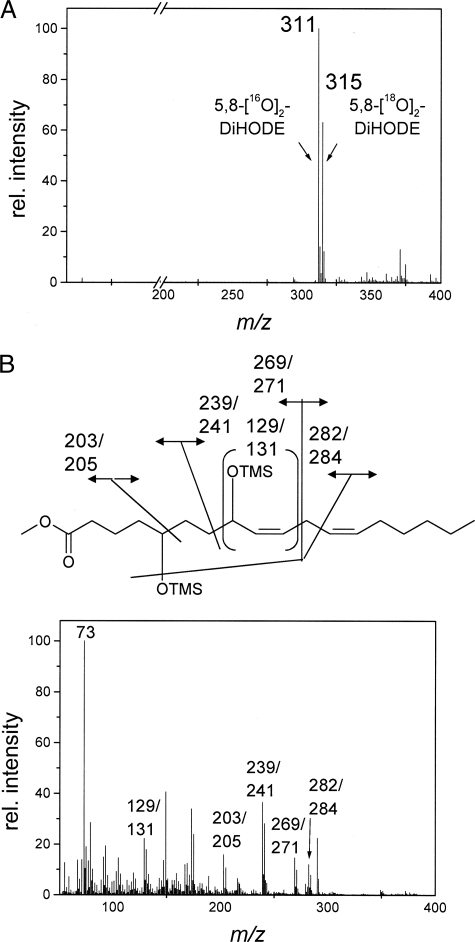
Analysis of Product Formation in the Presence of 18O2—To analyze whether both oxygen atoms of the 5,8-dihydroxy derivatives are derived from the same dioxygen molecule or from other co-substrates, 18:2Δ9Z,12Z was converted to 5,8-DiHODE under an atmosphere enriched in 18O2. The corresponding liquid chromatography/MS spectrum shows two major signals: m/z 311 (M– + 2 16O), corresponding to 5,8-[16O2]DiHODE, and 315 (M– + 2 18O), corresponding to 5,8-[18O2]DiHODE. No signal at m/z 313 was detected (Fig. 5A). This finding indicates that both hydroxy groups in the product contain either two atoms of 16O or two atoms of 18O, but nearly no species containing both isotopes could be detected. This further shows that oxygen atoms in the hydroxy groups of the product are derived from dioxygen. The GC/MS spectrum of labeled 5,8-DiHODE is shown in Fig. 5B. This spectrum of a mixture of the Me3Si ether methyl esters of 5,8-[18O2]DiHODE and 5,8-[16O2]DiHODE shows that the following fragments contain the labeled oxygen atom as judged by the m/z signal: 129/131 (CH(OSiCH3)3-C2H5), 203/205 (cleavage between C-5 and C-6), 239/241 (cleavage between C-7 and C-8), 269/271 (cleavage between C-10 and C-11), and 282/284 (cleavage between C-12 and C-13). The GC/MS data confirm the results derived from the HPLC/MS spectra.
FIGURE 5.
Mass spectrometric analysis of products formed by PpoA from 18:2Δ9Z,12Z under 18O2. A, HPLC/MS spectrum of 5,8-DiHODE obtained after incubation of 15 μg of PpoA with 250 μg of 18:2Δ9Z,12Z under 18O2 gas and extractive isolation. B, electron impact mass spectrum of a Me3Si ether methyl ester derivative of 5,8-DiHODE obtained after incubation of 15 μg of PpoA with 250 μg of 18:2Δ9Z,12Z under 18O2 gas and extractive isolation. rel., relative.
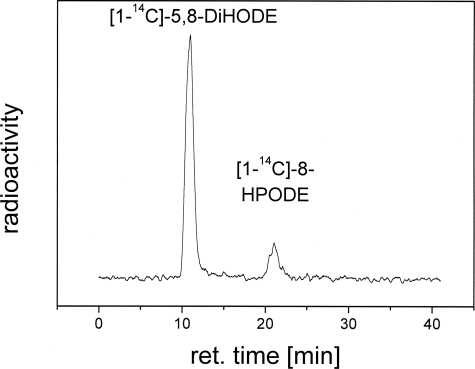
Conversion of 8-[1-14C]HPODE by PpoA—To elucidate further if the 8-hydroperoxide is a reaction intermediate, 8-[1-14C]HPODE was incubated with PpoA, which led to the generation of 5,8-[1-14C]DiHODE. The RP-HPLC chromatogram is shown in Fig. 6. Both the 18O2 experiment and the 8-HPODE experiment conversion indicate that 5,8-DiHODE is derived from the intermediate 8-HPODE. The observation that the amount of 8-HPODE differed in some experiments might therefore indicate that the catalytic 8-hydroperoxide isomerase activity is sensitive to perturbations of the protein folding.
FIGURE 6.
Conversion of 8-[1-14C]HPODE by PpoA to 5,8-[1-14C]DiHODE. Shown are the results from RP-HPLC analysis after 20 min of incubation of 8-[1-14C]HPODE with 15 μg of PpoA in 50 mm HEPES buffer (pH 7.4) and extractive isolation. ret., retention.
pH Optimum—The pH optimum was analyzed by measuring the initial rates of oxygen consumption for the reaction with 100 μm 18:2Δ9Z,12Z using a Clark-type O2 electrode. 15 μg of purified enzyme was used for this assay. Every measurement was performed in triplicates. Solutions of different pH values between 4.5 and 10.5 were used. The initial rate of O2 consumption increased rapidly from pH 5 to 6.0 and reached a maximum of 3 μmol/min/mg between pH 7.0 and 7.5. pH values >8.0 led to a rapid decrease in enzyme activity.
Kinetic Parameters—Kinetic constants were analyzed by measuring the initial rates of oxygen consumption for different substrate concentrations (five to seven concentrations in triplicates) in 50 mm phosphate buffer (pH 7.2). The substrates used were 16:1Δ9Z, 18:1Δ9Z, 18:2Δ9Z,12Z, and 18:3Δ9Z,12Z,15Z. The reactions were started by the addition of 15 μg of purified enzyme to the oxygen-saturated stirring solution. The Km and Vmax values were calculated by fitting the determined initial reaction rate to the Hanes-Woolf equation. The Michaelis-Menten plot and the corresponding Hanes-Woolf plot for the reaction with 18:2Δ9Z,12Z are shown in supplemental Fig. 3 (A and B, respectively). The different Km, Vmax, kcat, and kcat/Km values are summarized in Table 3. These results show that the substrate specificity of PpoA at least for the first step of the reaction decreases with the increasing number of double bonds of the fatty acid chain. Thus, 18:1Δ9Z seems to be the preferred substrate of PpoA.
TABLE 3.
Kinetic parameters for different substrates
Kinetic analysis was carried out using a Clark-type O2 electrode. Initial oxygen consumption was monitored for different substrate concentrations. The sodium salt of each fatty acid was used as a substrate.
| Substrate | Vmax | Km | kcat | kcat/Km |
|---|---|---|---|---|
| μmol/min/mg | μmol/liter | min-1 | min-1m-1 × 106 | |
| 16.1Δ9Z | 1.76 ± 0.102 | 5.00 ± 0.85 | 211 ± 12.3 | 42.2 ± 7.6 |
| 18.1Δ9Z | 2.48 ± 0.072 | 6.71 ± 0.74 | 297 ± 8.6 | 44.3 ± 5.05 |
| 18.2Δ9Z,12Z | 3.16 ± 0.18 | 18.30 ± 2.6 | 379 ± 21.6 | 20.3 ± 3.03 |
| 18.3Δ9Z,12Z,15Z | 2.97 ± 0.27 | 22.6 ± 4.3 | 356 ± 32 | 15.8 ± 3.3 |
Activation and Inactivation of PpoA—Measuring the conversion of 18:2Δ9Z,12Z by PpoA, we found a kinetic lag phase, which already has been described for other DOX-like PGHS enzymes (31). In the case of PpoA, this lag phase ranged from 3 to 5 s when the reaction was started with hydroperoxide-free 18:2Δ9Z,12Z. The lag phase was shortened or even abolished completely when less pure 18:2Δ9Z,12Z containing trace amounts of fatty acid hydroperoxides was used. This indicates that the native form of PpoA is inactive and needs to be activated by hydroperoxides. It is noteworthy that these hydroperoxides seem not to be necessarily 8-HPODE. Additionally, we found PpoA to be inactivated during the reaction and could not be reactivated by the addition of substrate. This finding is consistent with the results found for 7,8-LDS and PGHS (15, 32).
Peroxidase Activity—As mentioned above, we were able to detect 8-hydroxy derivatives in low amounts as a site product of the PpoA reaction. To determine whether the reduction of 8-hydroperoxide to the corresponding 8-hydroxide can be accomplished by PpoA, we used TMPD as a cosubstrate to analyze peroxidase activity. TMPD was used by Kulmacz (26) for determination of the peroxidase activity of PGHS. When 15 μg of PpoA were incubated with 100 μm 18:2Δ9Z,12Z and 100 μm TMPD in 50 mm HEPES (pH 7.4), we were able to follow the increase in the characteristic absorbance at 611 nm over 10 min. A typical reaction progress curve could be obtained and is shown in supplemental Fig. 4. After termination of the reaction, the products were extracted as described and analyzed by RP-HPLC. 8-HODE was the main product, whereas the amount of 5,8-DiHODE was significantly reduced. From this, we concluded that PpoA is able to reduce 8-HPODE to 8-HODE as well as to isomerase it into 5,8-DiHODE. In the presence of an appropriate electron donor like TMPD as a co-substrate, the peroxidase reaction seems to be the dominant reaction, whereas in the absence of a co-substrate, predominantly the 8-hydroperoxide isomerase reaction takes place.
Radical Spin Trapping Adducts—To investigate the formation of a carbon-centered radical at C-8 (15), we used the method of Koshiishi and co-workers (33–35) to scavenge the hypothetical C-8 radical with the radical scavenger CmΔP. In this case, a nitroxyl radical competes with the oxygen molecule for the lipid allyl radical on the enzyme, resulting in the formation of a lipid allyl radical-CmΔP adduct that can be detected by mass spectrometry.
As reported by Koshiishi et al. (36), a nitroxyl radical can trap the radical intermediate of the lipoxygenase reaction. It is known from the lipoxygenase/linoleate system that the product of this reaction consists of two regioisomers, namely 9- and 13-hydroperoxylinoleate (37). Thus, if CmΔP is used as a scavenger, which possesses no chiral center in its structure, two distinct peaks corresponding to both regioisomeric adducts can be detected on an HPLC system (33). In contrast to lipoxygenase, PpoA does not seem to produce regioisomers.
We performed the trapping reaction with CmΔP. The linoleate allyl radical-CmΔP adducts at m/z 463 were identified by RP-HPLC/MS. Surprisingly, two distinct peaks could be detected: a major peak eluting at 28.8 min and a smaller one at 30.8 min. However, as expected, none of the adducts showed an absorbance maximum at 234 nm, and thus, none of them possessed a conjugated diene moiety in their structure. We suggest from this result that the major peak corresponds to the trapped C-8 radical adduct, whereas the weaker peak might correspond to the C-10 radical adduct. This compound could be generated by an intramolecular isomerization rearrangement of the double bond. Furthermore, we were able to detect trapping adducts at m/z 479. These adducts may correspond to nitroxyl radical-lipid epoxyallyl adducts similar to those of the lipoxygenase/linoleate system as reported (36).
It is noteworthy that the spectra of the trapping adducts of the PpoA/linoleate system showed a characteristic fragment at m/z 185. Such a fragment has already been reported for the lipoxygenase/linoleate system. The carbon-centered radial-CmΔP adducts fragmented into hydroxylamine (m/z 185); this arises from the reduced form of CmΔP through heterolysis of the C–O–N bond (36).
Site-directed Mutagenesis—Alignments of the N-terminal heme peroxidase domain and C-terminal P450 heme thiolate domain show conserved residues in both domains (Fig. 1 and supplemental Fig. 1). To determine which of these conserved residues might be essential for PpoA activity, site-directed mutagenesis was carried out. The most striking effects on the catalytic activity were observed with mutation Y374F, which is located in the heme peroxidase domain, and mutation H1004A, which is located in the P450 heme thiolate domain.
Alignments show that this tyrosine residue is conserved in 7,8-LDS (Tyr376), sheep PGHS-1 (Tyr385), and sheep PGHS-2 (Tyr370) (supplemental Fig. 1A), and this residue is known to be essential for the catalytic activity probably through the formation of a tyrosyl radical (38–40). Replacement of Tyr374 with Phe (Y374F) led to an enzyme that showed no detectable activity when incubated with 18:2Δ9Z,12Z as a substrate (Fig. 7D). This showed that the tyrosine residue may be important for the DOX activity, i.e. oxidation of 18:2Δ9Z,12Z to 8-HPODE.
In contrast, when the Y374F mutant enzyme was incubated with the intermediate product 8-HPODE, it was able to catalyze the formation of 5,8-DiHODE as shown in Fig. 8C. This indicates that the Tyr residue is not involved in the 8-hydroperoxide isomerase reaction.
FIGURE 8.
HPLC analysis of products formed from 8-HPODE by PpoA and mutant Y374F. Shown are the results from RP-HPLC analysis after 20 min of incubation of 8-HPODE with 15 μg of heat-inactivated PpoA as a control (A), wild-type (WT) PpoA (B), and Y374F (C) in 50 mm HEPES buffer (pH 7.4) and extractive isolation. mAU, milliabsorbance units.
On the contrary, replacement of His1004 with Ala (H1004A) led to an enzyme that catalyzed only the formation of 8-HPODE, whereas the 8-hydroperoxide isomerase reaction was absent as shown in Fig. 7C. His1004 is conserved in many P450 domains of putative Ppo-like proteins and also matches the P450 heme signature consensus sequence (Fig. 1 and supplemental Fig. 1B). Alignments show that this residue is in close proximity to the crucial cysteine residue (Cys1006), which is suggested to be the fifth heme iron ligand, as has been shown, for example, in CYP119 from Sulfolobus solfataricus (41). Upon replacement of Cys1006 with Ala, the same catalytic activity was observed as for the H1004A enzyme.
These results led us to the conclusion that the N-terminal heme peroxidase domain might be responsible for the DOX reaction as the first step of the PpoA reaction, i.e. oxidation of linoleic acid to 8-HPODE as an intermediate product. However, the C-terminal P450 domain catalyzes the second reaction step, the isomerization of 8-HPODE to 5,8-DiHODE, and was therefore termed the 8-hydroperoxide isomerase P450 domain.
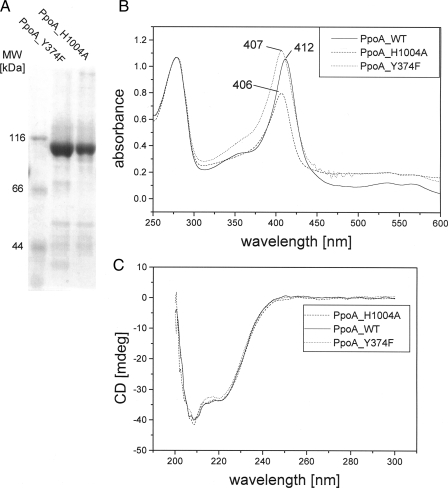
To ensure that the changed catalytic activity of H1004A and Y374F was not due to changed folding of the enzyme, we purified the mutant enzymes using the same purification protocol as for the wild-type enzyme. We found that both enzymes formed tetrameric complexes upon gel filtration like the wild-type enzyme. Judging by SDS-PAGE, both mutant enzymes showed the same degree of purity as the wild-type enzyme (Fig. 9A).
FIGURE 9.
SDS-PAGE of purified PpoA mutants H1004A and Y374F and spectroscopic analysis. Both mutants H1004A (dashed line) and Y374F (dotted line) were purified as described for the wild-type (WT) enzyme (solid line) and analyzed by 8% SDS-PAGE (A), UV-visible spectroscopy (B), and CD spectroscopy (C). mdeg, millidegrees.
In Fig. 9B, the UV-visible absorption spectra of both mutant enzymes in comparison with the wild-type enzyme are shown. Whereas Y374F showed a spectrum similar to that of the wild-type enzyme (with a shift in the Soret band to 407 nm and maxima at 534 nm (β), 567 nm (α), 355 nm (δ)), that of H1004A differed in some respects: the Soret band shifted to 406 nm, whereas the α-, β-, and δ-bands gave no distinct signals. The most striking difference is the changed ratio of ASoret to A280 nm. Enzyme preparations of the wild-type form showed a ASoret/A280 nm ratio between 1.0 and 1.1. In contrast, those of H1004A ranged from 0.5 to 0.7. We concluded from this finding that this mutant contains ∼50–70% of heme cofactor compared with the wild-type enzyme.
Moreover, we recorded a CD spectrum of the wild-type and mutant enzymes in the far-UV and near-UV regions. As shown in Fig. 9C, the wild-type enzyme (solid line) and the mutant enzymes (dashed and dotted lines) have very similar secondary and tertiary structures. Therefore, it seems unlikely that changes in the catalytic activity were due to an incorrectly folded enzyme structure.
The activity of the DOX reaction of the H1004A mutant was determined in a similar way as for the wild-type enzyme using the oxygen electrode as described above. We were able to determine kinetic constants for the reaction of H1004A incubated with 18:2Δ9Z,12Z (supplemental Fig. 5): Vmax = 1.16 ± 0.05 μmol/min/mg, Km = 4.90 ± 0.62 μm, kcat = 139 min–1, and kcat/Km = (28.3 ± 3.81) × 106 min–1 m–1. Vmax, kcat, and Km were reduced by 3–4-fold in comparison with the wild-type enzyme, giving similar kcat/Km values for the mutant (28.3 × 106 min–1 m–1) and wild-type enzyme (20.3 × 106 min–1 m–1). This finding may indicate that the first reaction step, the DOX reaction, is the rate-limiting step for the reaction. This goes along with the finding that, in some experiments, we detected only traces of 8-HPODE, whereas 5,8-DiHODE was the predominant product.
We also tested both mutant enzymes for peroxidase activity using TMPD as the electron donor. The Y374F enzyme showed no detectable peroxidase activity when incubated with 18:2Δ9Z,12Z because the production of hydroperoxides was abolished. In contrast, the H1004A mutant produced the characteristic increase at A611 nm over time. This shows that the H1004A mutation does not affect the peroxidase activity, whereas the 8-hydroperoxide isomerase activity is abolished.
DISCUSSION
This study aimed to characterize the biochemical properties of PpoA, a fungal DOX that is involved in the regulation of the life cycle of A. nidulans (43). Therefore, PpoA was expressed in E. coli. After purification of the enzyme by up to 62-fold, biochemical characterization was performed. We showed that PpoA is a ferric tetrameric heme protein that catalyzes the dioxygenation of linoleic acid to its 5,8-diol derivative via the formation of 8-HPODE. We also found 8-HODE to be formed in low amounts as a side product. Similar results were obtained by Oliw and co-workers (20), who concluded from incubation of 18:2Δ9Z,12Z with mycelia of A. nidulans lacking the ppoA allele that the ppoA gene might code for an 5,8-LDS that is similar to the 7,8-LDS from G. graminis.
Beside these principal similarities, we now provide evidence that there exist striking differences between both enzymes and their reaction mechanisms. Our study is based on the finding that PpoA appears to consist of two functional domains as predicted, namely an N-terminal heme peroxidase and a C-terminal P450 heme thiolate domain.
In 7,8-LDS, only the heme peroxidase domain can be found. Additional enzymes are known that use only a heme peroxidase domain for catalyzing similar reactions: plant α-DOX and mammalian PGHS. Whereas 7,8-LDS and PGHS have been found to be bifunctional enzymes because they catalyze either a dioxygenase and isomerase or a cyclooxygenase and peroxidase reaction, plant α-DOX is monofunctional because it catalyzes only a dioxygenation reaction, yielding a 2-hydroperoxy fatty acid (45). PpoA is another enzyme that harbors this domain, and therefore, we will first discuss similarities of these enzymes to PpoA and then focus on differences in the reaction mechanism due to the unique property of PpoA with an additional P450 heme thiolate domain.
Like 7,8-LDS, α-DOX, and PGHS, the native form of PpoA is inactive and needs to be activated by hydroperoxides. Whereas 7,8-LDS seems to be preferentially activated by its own product 8-HPODE (15), PGHS can be activated by many different hydroperoxide species (46). PpoA also seems to be relatively indifferent toward the activating hydroperoxide because we observed that not only the 8-hydroperoxides but also autoxidation products of fatty acids, e.g. racemic 13-hydroperoxides, activate the enzyme.
Furthermore, it is known that both 7,8-LDS and PGHS are inactivated during their reaction (15, 18). We observed that PpoA is subject to a similar kind of suicide inactivation and that the DOX activity fell to zero within a few minutes under our assay conditions, although substrates were available in sufficient amounts.
Spectroscopic analysis showed that, like 7,8-LDS (15), PpoA contains a high spin ferriheme (Fig. 3A). This finding was confirmed by the Soret absorption of CN– and F– derivatives.
The most obvious similarity of PpoA to 7,8-LDS is the reaction process in which the formation of a diol product is accomplished via the intermediate formation of a hydroperoxide derivative. It was suggested that both enzymes abstract the pro-S-hydrogen atom, generating a carbon-centered radical at C-8 (20, 47), but the formation of protein radicals has been investigated so far only via EPR for 7,8-LDS (15). We were able to trap two carbon-centered radical species, presumably the C-8 and C-10 radical derivatives, providing additional evidence that supports the model of a radical as a reaction intermediate for both enzymes. The latter might be easily formed by an isomerization of the C-9 double bond. Interestingly, we could not observe the formation of any 10-hydroperoxide products in vitro by the wild-type enzyme.
To elucidate further the reaction mechanism, we mutated conserved amino acid residues in the heme peroxidase domain as well as in the P450 heme thiolate domain. Tyr374 in PpoA is conserved in 7,8-LDS (Tyr376), α-DOX (Tyr380), and PGHS (Tyr385 in sheep PGHS-1 and Tyr370 in sheep PGHS-2), and it is well known that DOX reactions are abolished when it is mutated in α-DOX, 7,8-LDS, or PGHS (39, 40, 42). Here, we have provided evidence that mutation of this Tyr to Phe abolished DOX activity, whereas 8-hydroperoxide isomerase activity was unaffected (Figs. 7D and 8C). This finding led us to conclude that both reactions (DOX and 8-hydroperoxide isomerase) occur at distinct sites of the protein, as has also been shown for the cyclooxygenase and peroxidase reaction of PGHS (18). Treatment of 7,8-LDS with tetranitromethane, a reagent that is known to convert tyrosine residues of proteins to 3-nitrotyrosine, results in loss of 7,8-LDS activity (15). However, the same group recently expressed recombinant 7,8-LDS in Pichia pastoris and observed not only that the expressed enzyme showed a different isomerase activity from the native enzyme but also that the mutation of Tyr376 to Phe in the recombinant enzyme abolished DOX activity, whereas isomerase activity was retained (48). A similar result was obtained when the same mutant was expressed in insect cells (39), suggesting a comparable function of the Tyr residue in 7,8-LDS and in PpoA.
One obvious difference between 7,8-LDS and PpoA is the presence of an additional P450 heme thiolate domain in PpoA. Therefore, we mutated conserved amino acid residues in the heme signature motif (Fig. 1) of this domain, which might be involved in binding of heme. Alignments show that His1004 is in close proximity to the crucial cysteine residue (Cys1006), which has been suggested to be the fifth heme iron ligand, as has been shown, for example, in CYP119 from S. solfataricus (41). Thus, substitution of His1004 with Ala might distort heme binding in this particular domain.
Replacement of His1004 with Ala led to an enzyme with abolished 8-hydroperoxide isomerase activity, whereas DOX activity was still present (Fig. 7C). Furthermore, the heme content was reduced to 50–70% as judged by the ASoret/A280 nm ratio in the UV-visible spectra (Fig. 9B). From that we concluded that PpoA may harbor two heme groups and that His1004 is involved in binding of the heme group that is localized in the P450 heme thiolate domain. Notably, this heme group seems to be essential for the 8-hydroperoxide isomerase reaction. Further proof for the 8-hydroperoxide isomerase activity of the P450 heme thiolate was provided by substitution of the crucial cysteine residue (Cys1006) with Ala. This enzyme also showed an abolished 8-hydroperoxide isomerase activity, whereas the DOX activity was retained.
Interestingly, the 8-hydroperoxide isomerase P450 domain appears to show functional as well as structural similarities to other P450 heme thiolate enzymes like the prostacyclin synthase and allene oxide synthase: P450 heme thiolate enzymes typically activate molecular oxygen by the use of external electron donors to catalyze monooxygenation reactions, e.g. hydroxylation, peroxidation, or epoxidation (49). In contrast to these enzymes, prostacyclin synthase and allene oxide synthase carry out isomerization of endoperoxides and hydroperoxides, respectively, and do not need either molecular oxygen or external electron donors. In analogy, the 8-hydroperoxide isomerase within the P450 domain of PpoA also catalyzes an isomerization, i.e. the conversion of 8-HPODE to 5,8-DiHODE. This reaction is also independent of molecular oxygen and external electron donors. Additionally, the thiolate-ligated heme group of PpoA gave a weak and transient UV-visible signal when incubated with dithionite and CO, similar to allene oxide synthase (44) and prostacyclin synthase, respectively. In the case of allene oxide synthase, it is believed that the weak binding of CO is a consequence of a structural conformation that hinders the axial coordination of the heme iron by ligands (30, 50). For prostacyclin synthase, it has been also been demonstrated that the transient formation of the ferrous CO complex (29) might be due to a small active-site volume (51).
As alignments of PpoA with 7,8-LDS show no homology in the region of the P450 domain and as no indications for an additional domain beside the heme peroxidase domain could be detected for 7,8-LDS, it is unlikely that the peroxidase and isomerase activities of this enzyme are allocated to distinct protein regions. It is known from PGHS that the peroxidase activity can operate independently of the cyclooxygenase activity (18), whereas in contrast, the cyclooxygenase reaction needs an initial oxidation of the heme group (31). Because we found that, in PpoA, both reactions, dioxygenation and 8-hydroperoxide isomerization, can work independently of each other (Figs. 7 and 8), further proof of the existence of an additional heme group was provided. This was also supported by the fact that the H1004A mutant lacking the 8-hydroperoxide isomerase activity showed DOX as well as peroxidase activity, indicating that heme oxidation was retained. Moreover, the UV-visible spectrum of the mutant enzyme still showed characteristic signals of a heme protein even though it was different from that of the wild-type enzyme. We suggest that the heme group of the N-terminal heme peroxidase domain might be needed for the activation of the enzyme by hydroperoxides, which are concomitantly reduced to the corresponding hydroxides, as is known for PGHS. Interestingly, in the presence of TMPD, we found peroxidase activity to be the dominant activity, whereas 8-hydroperoxide isomerase activity was decreased as judged by product analysis with HPLC. This finding may coincide with the fact that, in PGHS, compound I and intermediate II can be reduced by an exogenous electron donor, yielding compound II (52), and by further reduction back to heme, yielding the resting enzyme, from which a new peroxidase reaction can begin.
Taken together, our findings indicate that PpoA and the related fatty acid dioxygenase 7,8-LDS seem to have different reaction mechanisms. Whereas 7,8-LDS uses apparently just one domain for catalyzing its two reaction steps, PpoA seems to use two independent domains to carry out similar reaction steps. The DOX reaction appears to be accomplished within the N-terminal heme peroxidase domain, whereas the 8-hydroperoxide isomerase reaction takes place in the C-terminal P450 heme thiolate domain. From the data presented here, we suggest a catalytic mechanism that is different from that for 7,8-LDS, but is analogous to the widely accepted mechanisms of the PGHS and P450 enzymes as shown for linoleic acid in Fig. 10 (18, 53).
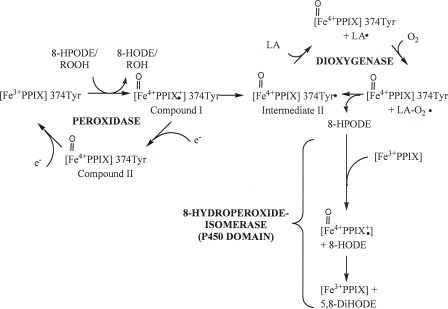
FIGURE 10.
Hypothetical catalytic mechanism of PpoA with linoleic acid as a substrate. The initiating step of catalysis is the reduction of hydroperoxides to the corresponding hydroxides with the formation of compound I. This species is either converted by an intermolecular reduction to intermediate II with the formation of the Tyr374 radical or alternatively reduced by an exogenous electron donor (e.g. TMPD) to compound II and further reduction back to heme, yielding the resting enzyme. The Tyr374 radical is then used for the generation of the linoleoyl radical, and subsequently, molecular oxygen is inserted, resulting in peroxylinoleate. The latter is converted to 8-HPODE by abstracting a hydrogen from Tyr374. Either 8-HPODE serves as the substrate for the 8-hydroperoxide isomerase reaction within the P450 heme thiolate domain in which it is isomerized via the intermediate formation of compound I to 5,8-DiHODE, or alternatively, it serves as a substrate for the peroxidase reaction, yielding 8-HODE. LA, linoleic acid; PPIX, ferric iron protoporphyrin IX.
The reduction of a relatively wide group of hydroperoxides to the corresponding hydroxides displays the initiating step of catalysis presumably with the formation of compound I. As is known for PGHS compound I, this species might then either be converted by an intramolecular reduction to intermediate II with the formation of the Tyr radical or alternatively be reduced by an exogenous electron donor (e.g. TMPD) to compound II. The Tyr radical is presumably used for the abstraction of the hydrogen at C-8, and subsequently, molecular oxygen is inserted, resulting in peroxylinoleate. The latter is converted to 8-HPODE by again abstracting a hydrogen from the Tyr residue. 8-HPODE then serves as the substrate for the 8-hydroperoxide isomerase reaction, in which it is isomerized to 5,8-DiHODE via the intermediate formation of compound I. Alternatively, 8-HPODE serves as a substrate for the peroxidase reaction (Fig. 10). EPR as well as stop flow studies are in progress and will be used for the exact determination of the catalytic mechanism.
Supplementary Material
Acknowledgments
We thank Sabine Freitag and Theres Riemekasten for expert technical assistance, André Nadler for help with CD spectroscopy, and Nambirajan Govindarajan for initial cloning experiments. The assignment of PpoA and comments on the PpoA P450 sequence by Dr. D. R. Nelson are also gratefully acknowledged.
This work was supported by the German Research Foundation (IRTG 1422).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: HODE, hydroxyoctadecadienoic acid; DiHODE, dihydroxyoctadecadienoic acid; LDS, linoleate diol synthase; HPODE, hydroperoxyoctadecadienoic acid; PGHS, prostaglandin H synthase; DOX, dioxygenase; HPLC, high pressure liquid chromatography; RP, reversed phase; GC/MS, gas chromatography/mass spectrometry; CmΔP, 3-carbamoyl-2,2,5,5-tetramethyl-3-pyrroline-N-oxyl; TMPD, N,N,N′,N′-tetramethyl-p-phenylenediamine.
References
- 1.Galagan, J. E., Calvo, S. E., Cuomo, C., Ma, L. J., Wortman, J. R., Batzoglou, S., Lee, S. I., Basturkmen, M., Spevak, C. C., Clutterbuck, J., Kapitonov, V., Jurka, J., Scazzocchio, C., Farman, M., Butler, J., Purcell, S., Harris, S., Braus, G. H., Draht, O., Busch, S., D'Enfert, C., Bouchier, C., Goldman, G. H., Bell-Pedersen, D., Griffiths-Jones, S., Doonan, J. H., Yu, J., Vienken, K., Pain, A., Freitag, M., Selker, E. U., Archer, D. B., Penalva, M. A., Oakley, B. R., Momany, M., Tanaka, T., Kumagai, T., Asai, K., Machida, M., Nierman, W. C., Denning, D. W., Caddick, M., Hynes, M., Paoletti, M., Fischer, R., Miller, B., Dyer, P., Sachs, M. S., Osmani, S. A., and Birren, B. W. (2005) Nature 438 1105–111516372000 [Google Scholar]
- 2.Champe, S. P., and el-Zayat, A. A. (1989) J. Bacteriol. 171 3982–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvo, A. M., Hinze, L. L., Gardner, H. W., and Keller, N. P. (1999) Appl. Environ. Microbiol. 65 3668–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo, A. M., Gardner, H. W., and Keller, N. P. (2001) J. Biol. Chem. 276 25766–25774 [DOI] [PubMed] [Google Scholar]
- 5.Calvo, A. M., Wilson, R. A., Bok, J. W., and Keller, N. P. (2002) Microbiol. Mol. Biol. Rev. 66 447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsitsigiannis, D. I., Kowieski, T. M., Zarnowski, R., and Keller, N. P. (2004) Eukaryot. Cell 3 1398–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsitsigiannis, D. I., and Keller, N. P. (2006) Mol. Microbiol. 59 882–892 [DOI] [PubMed] [Google Scholar]
- 8.Mazur, P., Meyers, H. V., Nakanishi, K., el-Zayat, A. A. E., and Champe, S. P. (1990) Tetrahedron Lett. 31 3837–3840 [Google Scholar]
- 9.Mazur, P., Nakanishi, K., el-Zayat, A. A. E., and Champe, S. P. (1991) J. Chem. Soc. Chem. Commun. 20 1486–1487 [Google Scholar]
- 10.Bowers, W. S., Hoch, H. C., Evans, P. H., and Katayama, M. (1986) Science 232 105–106 [DOI] [PubMed] [Google Scholar]
- 11.Brodowsky, I. D., and Oliw, E. H. (1993) Biochim. Biophys. Acta 1168 68–72 [DOI] [PubMed] [Google Scholar]
- 12.Brodowsky, I. D., and Oliw, E. H. (1992) Biochim. Biophys. Acta 1124 59–65 [DOI] [PubMed] [Google Scholar]
- 13.Brodowsky, I. D., Hamberg, M., and Oliw, E. H. (1992) J. Biol. Chem. 267 14738–14745 [PubMed] [Google Scholar]
- 14.Su, C., and Oliw, E. H. (1996) J. Biol. Chem. 271 14112–14118 [DOI] [PubMed] [Google Scholar]
- 15.Su, C., Sahlin, M., and Oliw, E. H. (1998) J. Biol. Chem. 273 20744–20751 [DOI] [PubMed] [Google Scholar]
- 16.Tsitsigiannis, D. I., Zarnowski, R., and Keller, N. P. (2004) J. Biol. Chem. 279 11344–11353 [DOI] [PubMed] [Google Scholar]
- 17.Tsitsigiannis, D. I., Kowieski, T. M., Zarnowski, R., and Keller, N. P. (2005) Microbiology (Read.) 151 1809–1821 [DOI] [PubMed] [Google Scholar]
- 18.Smith, W. L., DeWitt, D. L., and Garavito, R. M. (2000) Annu. Rev. Biochem. 69 145–182 [DOI] [PubMed] [Google Scholar]
- 19.Tsitsigiannis, D. I., Bok, J. W., Andes, D., Nielsen, K. F., Frisvad, J. C., and Keller, N. P. (2005) Infect. Immun. 73 4548–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garscha, U., Jerneren, F., Chung, D., Keller, N. P., Hamberg, M., and Oliw, E. H. (2007) J. Biol. Chem. 282 34707–34718 [DOI] [PubMed] [Google Scholar]
- 21.Garscha, U., and Oliw, E. H. (2007) Anal. Biochem. 367 238–246 [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann, M., Hornung, E., Busch, S., Kassner, N., Ternes, P., Braus, G. H., and Feussner, I. (2007) J. Biol. Chem. 282 26666–26674 [DOI] [PubMed] [Google Scholar]
- 23.Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K., and Pease, L. R. (1989) Gene (Amst.) 77 61–68 [DOI] [PubMed] [Google Scholar]
- 24.Antonini, E., and Brunori, M. (1971) Hemoglobin and Myoglobin and Their Reaction with Ligands, North Holland Publishing Co., Amsterdam
- 25.Omura, T., and Sato, R. (1964) J. Biol. Chem. 239 2379–2385 [PubMed] [Google Scholar]
- 26.Kulmacz, R. J. (1987) Prostaglandins 34 225–240 [DOI] [PubMed] [Google Scholar]
- 27.Omura, T., and Sato, R. (1964) J. Biol. Chem. 239 2370–2378 [PubMed] [Google Scholar]
- 28.Sasaki, M., Akahira, A., Oshiman, K., Tsuchido, T., and Matsumura, Y. (2005) Appl. Environ. Microbiol. 71 8024–8030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeh, H. C., Hsu, P. Y., Wang, J. S., Tsai, A. L., and Wang, L. H. (2005) Biochim. Biophys. Acta 1738 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau, S.-M. C., Harder, P. A., and O'Keefe, D. P. (1993) Biochemistry 32 1945–1950 [DOI] [PubMed] [Google Scholar]
- 31.Landino, L. M., Crews, B. C., Gierse, J. K., Hauser, S. D., and Marnett, L. J. (1997) J. Biol. Chem. 272 21565–21574 [DOI] [PubMed] [Google Scholar]
- 32.Kulmacz, R. J., Pendleton, R. B., and Lands, W. E. (1994) J. Biol. Chem. 269 5527–5536 [PubMed] [Google Scholar]
- 33.Koshiishi, I., Tsuchida, K., Takajo, T., and Komatsu, M. (2006) Biochem. J. 395 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takajo, T., Tsuchida, K., Ueno, K., and Koshiishi, I. (2007) J. Lipid Res. 48 1371–1377 [DOI] [PubMed] [Google Scholar]
- 35.Takajo, T., Tsuchida, K., Murahashi, T., Ueno, K., and Koshiishi, I. (2007) J. Lipid Res. 48 855–862 [DOI] [PubMed] [Google Scholar]
- 36.Koshiishi, I., Tsuchida, K., Takajo, T., and Komatsu, M. (2005) J. Lipid Res. 46 2506–2513 [DOI] [PubMed] [Google Scholar]
- 37.Liavonchanka, A., and Feussner, I. (2006) J. Plant Physiol. 163 348–357 [DOI] [PubMed] [Google Scholar]
- 38.Shimokawa, T., Kulmacz, R. J., DeWitt, D. L., and Smith, W. L. (1990) J. Biol. Chem. 265 20073–20076 [PubMed] [Google Scholar]
- 39.Garscha, U., and Oliw, E. H. (2008) FEBS Lett. 582 3547–3551 [DOI] [PubMed] [Google Scholar]
- 40.Koeduka, T., Matsui, K., Akakabe, Y., and Kajiwara, T. (2002) J. Biol. Chem. 277 22648–22655 [DOI] [PubMed] [Google Scholar]
- 41.Yano, J. K., Koo, L. S., Schuller, D. J., Li, H., Ortiz de Montellano, P. R., and Poulos, T. L. (2000) J. Biol. Chem. 275 31086–31092 [DOI] [PubMed] [Google Scholar]
- 42.Thuresson, E. D., Lakkides, K. M., Rieke, C. J., Sun, Y., Wingerd, B. A., Micielli, R., Mulichak, A. M., Malkowski, M. G., Garavito, R. M., and Smith, W. L. (2001) J. Biol. Chem. 276 10347–10357 [DOI] [PubMed] [Google Scholar]
- 43.Tsitsigiannis, D. I., and Keller, N. P. (2007) Trends Microbiol. 15 109–118 [DOI] [PubMed] [Google Scholar]
- 44.Song, W.-C., and Brash, A. R. (1991) Science 253 781–784 [DOI] [PubMed] [Google Scholar]
- 45.Sanz, A., Moreno, J. I., and Castresana, C. (1998) Plant Cell 10 1523–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohki, S., Ogino, N., Yamamoto, S., and Hayaishi, O. (1979) J. Biol. Chem. 254 829–836 [PubMed] [Google Scholar]
- 47.Hamberg, M., Zhang, L. Y., Brodowsky, I. D., and Oliw, E. H. (1994) Arch. Biochem. Biophys. 309 77–80 [DOI] [PubMed] [Google Scholar]
- 48.Garscha, U., and Oliw, E. H. (2008) Biochem. Biophys. Res. Commun. 373 579–583 [DOI] [PubMed] [Google Scholar]
- 49.Coon, M. J. (2005) Annu. Rev. Pharmacol. Toxicol. 45 1–25 [DOI] [PubMed] [Google Scholar]
- 50.Lee, D.-S., Nioche, P., Hamberg, M., and Raman, C. S. (2008) Nature 455 363–368 [DOI] [PubMed] [Google Scholar]
- 51.Chiang, C. W., Yeh, H. C., Wang, L. H., and Chan, N. L. (2006) J. Mol. Biol. 364 266–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu, G. Q., Tsai, A. L., Van Wart, H. E., and Kulmacz, R. J. (1999) J. Biol. Chem. 274 16162–16167 [DOI] [PubMed] [Google Scholar]
- 53.Kupfer, R., Liu, S. Y., Allentoff, A. J., and Thompson, J. A. (2001) Biochemistry 40 11490–11501 [DOI] [PubMed] [Google Scholar]
- 54.Picot, D., Loll, P. J., and Garavito, R. M. (1994) Nature 367 243–249 [DOI] [PubMed] [Google Scholar]
- 55.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.