Abstract
The lipopolysaccharide of Pseudomonas aeruginosa PAO1 contains an unusual sugar, 2,3-diacetamido-2,3-dideoxy-d-mannuronic acid (d-ManNAc3NAcA). wbpB, wbpE, and wbpD are thought to encode oxidase, transaminase, and N-acetyltransferase enzymes. To characterize their functions, recombinant proteins were overexpressed and purified from heterologous hosts. Activities of His6-WbpB and His6-WbpE were detected only when both proteins were combined in the same reaction. Using a direct MALDI-TOF mass spectrometry approach, we identified ions that corresponded to the predicted products of WbpB (UDP-3-keto-d-GlcNAcA) and WbpE (UDP-d-GlcNAc3NA) in the coupled enzyme-substrate reaction. Additionally, in reactions involving WbpB, WbpE, and WbpD, an ion consistent with the expected product of WbpD (UDP-d-GlcNAc3NAcA) was identified. Preparative quantities of UDP-d-GlcNAc3NA and UDP-d-GlcNAc3NAcA were enzymatically synthesized. These compounds were purified by high-performance liquid chromatography, and their structures were elucidated by NMR spectroscopy. This is the first report of the functional characterization of these proteins, and the enzymatic synthesis of UDP-d-GlcNAc3NA and UDP-d-GlcNAc3NAcA.
Gram-negative organisms such as Pseudomonas aeruginosa produce lipopolysaccharide (LPS)4 as an essential component of the outer leaflet of the outer membrane. LPS can be conceptually divided into three parts: lipid A, which anchors LPS into the membrane; core oligosaccharide, which contributes to membrane stability; and the O-antigen, which is a polysaccharide that extends away from the cell surface. In P. aeruginosa, two types of O-antigen are observed: A-band O-antigen, which is common to most strains, and B-band O-antigen, which is variable and therefore used as the basis of the International Antigenic Typing Scheme (1). P. aeruginosa serotypes O2, O5, O16, O18, and O20 collectively belong to serogroup O2, because they all share common backbone sugar structures in their O-antigen repeat units consisting of two di-N-acetylated uronic acids and one 2-acetamido-2,6-dideoxy-d-galactose (N-acetyl-d-fucosamine). The minor structural variations in the O-antigen repeat units that differentiate this serogroup into five serotypes are: the type of glycosidic linkage between O-units (alpha versus beta) that is formed by the O-antigen polymerase (Wzy), isomers present (d-mannuronic or l-guluronic acid), and acetyl group substituents (2–4). The B-band O-antigen of P. aeruginosa PAO1 (serotype O5) contains a repeating trisaccharide of 2-acetamido-3-acetamidino-2,3-dideoxy-d-mannuronic acid (d-ManNAc3NAmA), 2,3-diacetamido-2,3-dideoxy-d-mannuronic acid (d-ManNAc3NAcA), and 2-acetamido-2,6-dideoxy-d-galactose (3).
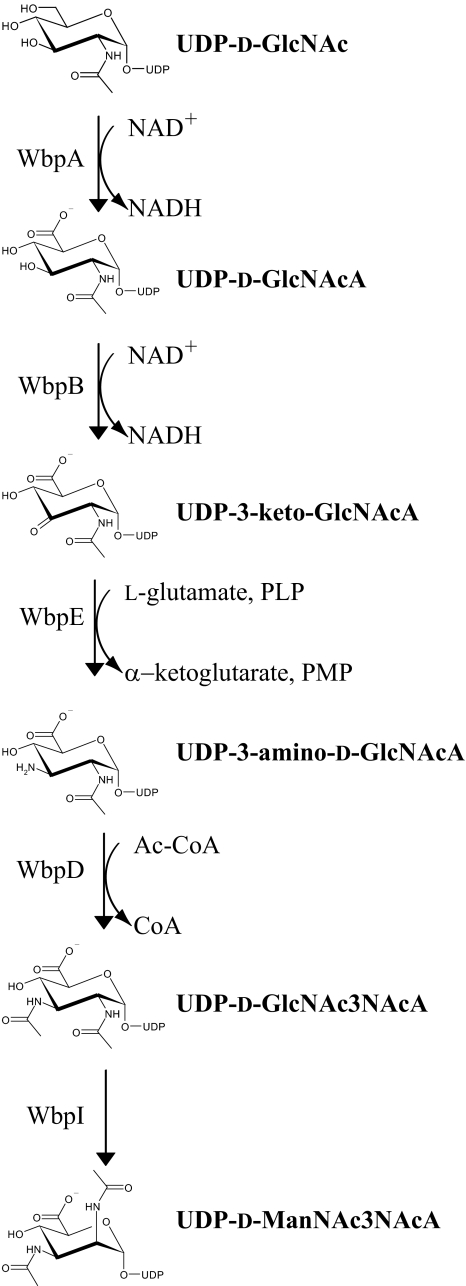
The biosynthesis of the two mannuronic acid derivatives has yet to be fully understood and has been the subject of investigation by our group. To produce UDP-d-ManNAc3NAcA, a five-step pathway has been proposed (Fig. 1) that requires the products of five genes localized to the B-band O-antigen biosynthesis cluster (5). The O-antigen biosynthesis cluster was shown to be identical for all serotypes within serogroup O2, which further underscores the high similarity between these serotypes (5). The five genes, including wbpA, wbpB, wbpE, wbpD, and wbpI, have been shown to be essential for B-band LPS biosynthesis, because knockout mutants of each of these genes are deficient in B-band O-antigen (6–8). Homologs of all five of the proteins required for the UDP-d-ManNAc3NAcA biosynthesis pathway are conserved in other bacterial pathogens, including Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica. Cross-complementation of P. aeruginosa knockout mutants lacking wbpA, wbpB, wbpE, wbpD, or wbpI with the homologues from B. pertussis could fully restore LPS production in the P. aeruginosa LPS mutants, suggesting that the genes from B. pertussis are functional homologs of the wbp genes (7). Homologs of these genes could be identified in diverse bacterial species, demonstrating the importance of UDP-d-ManNAc3NAcA biosynthesis beyond its role in P. aeruginosa (7).
FIGURE 1.
Proposed pathway for the biosynthesis of UDP-d-ManNAc3NAcA in P. aeruginosa PAO1. The full names of the sugars are as follows: GlcNAc, 2-acetamido-2-deoxy-d-glucose; GlcNAcA, 2-acetamido-2-deoxy-d-glucuronic acid; 3-keto-d-GlcNAcA, 2-acetamido-2-deoxy-d-ribo-hex-3-uluronic acid; GlcNAc3NA, 2-acetamido-3-amino-2,3-dideoxy-d-glucuronic acid; GlcNAc3NAcA, 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid; ManNAc3NAcA, 2,3-diacetamido-2,3-dideoxy-d-mannuronic acid. Adapted from Ref. 8.
The first enzyme of the UDP-d-ManNAc3NAcA biosynthesis pathway, WbpA, is a 6-dehydrogenase that converts UDP-2-acetamido-2-deoxy-d-glucose (N-acetyl-d-glucosamine; UDP-d-GlcNAc) to UDP-2-acetamido-2-deoxy-d-glucuronic acid (N-acetyl-d-glucosaminuronic acid, UDP-d-GlcNAcA) using NAD+ as a coenzyme (9) (Fig. 1). Following this, the second step in UDP-d-ManNAc3NAcA biosynthesis is proposed to be an oxidation reaction catalyzed by WbpB, forming UDP-2-acetamido-2-deoxy-d-ribo-hex-3-uluronic acid (3-keto-d-GlcNAcA), which in turn is used as the substrate for transamination by WbpE, creating UDP-2-acetamido-3-amino-2,3-dideoxy-d-glucuronic acid (d-GlcNAc3NA).
This residue is thought to be the substrate for WbpD, a putative N-acetyltransferase of the hexapeptide acyltransferase superfamily (10) that requires acetyl-CoA as a co-substrate (8). WbpD has been proposed to synthesize UDP-2,3-diacetamido-2,3-dideoxy-d-glucuronic acid (UDP-d-GlcNAc-3NAcA), which is utilized in the B-band O-antigen of P. aeruginosa serotype O1. In P. aeruginosa serogroup O2, the UDP-d-GlcNAc3NAcA is then epimerized by WbpI to create the UDP-d-ManNAc3NAcA required for incorporation into B-band LPS (11). A derivative of UDP-d-ManNAc3NAcA is also used in the synthesis of B-band O-antigen of P. aeruginosa serogroup O2. UDP-d-ManNAc3NAmA is thought to be produced through additional modification of UDP-d-ManNAc3NAcA via the action of WbpG, an amidotransferase, which has also been demonstrated to be essential for the production of B-band O-antigen (12, 13).
In the current study, our aim was to define the function of WbpB, WbpE, and WbpD, because only genetic evidence has previously been given for the involvement of wbpB and wbpE (7), and the reaction catalyzed by WbpD could not be demonstrated due to the unavailability of its presumed substrate, UDP-d-GlcNAc3NA (8). The functional characterization of these proteins is also important for understanding LPS biosynthesis in B. pertussis, because the genes in the LPS locus of this species, wlbA, wlbC, and wlbB, could cross-complement knockouts of wbpB, wbpE, and wbpD, respectively, when expressed in P. aeruginosa PAO1 (7). Furthermore, these three proteins form a cassette for the generation of C-3 N-acetylated hexoses and may be important for the biosynthesis of a variety of other sugars. Capillary electrophoresis and MALDI-TOF mass spectrometry were used to analyze reaction mixtures of WbpB and WbpE and showed that the expected products were produced only when both enzymes were present together. Achieving the enzymatic synthesis of the product of both enzymes, which was demonstrated to be UDP-d-GlcNAc3NA by 1H NMR spectroscopy, was a key breakthrough, because this rare sugar has never before been produced by any means. UDP-d-GlcNAc3NA was also essential for use as the substrate of WbpD, which not only allowed us to determine the enzymatic activity of this protein but also allowed the enzymatic synthesis of UDP-d-GlcNAc3NAcA to be achieved as well. Although this sugar had previously been produced through a 17-step chemical synthesis (11, 14), the 4-step concurrent enzymatic reaction demonstrates the advantage of linking chemistry with biology and represents a significant saving of both time and reagents as compared with chemical synthesis. Finally, our data also showed the success in reconstituting in vitro the 5-step pathway for the biosynthesis of UDP-d-ManNAc3NAcA in P. aeruginosa.
EXPERIMENTAL PROCEDURES
Overexpression and Purification of Wbp* Enzymes—All strains and plasmids used have been documented (Table 1). pET-His6-WbpB was expressed in Escherichia coli Tuner(DE3) induced with 0.01 mm isopropyl β-d-thiogalactopyranoside overnight at 15 °C. Frozen cell pellets were resuspended in buffer (50 mm sodium phosphate, pH 8.0, 300 mm NaCl, 5 mm β-mercaptoethanol, 5 mm imidazole, and 20% glycerol). After ultrasonication (Sonic Dismembrator model 500, Fisher Scientific), the soluble fraction was obtained by ultracentrifugation at 120,000 × g for 1 h (Beckman L8–55M ultracentrifuge) and then bound to 2 ml of nickel-nitrilotriacetic acid-agarose (Qiagen). The column was washed with buffer containing 50 mm imidazole, and then eluted with buffer containing 150 mm imidazole. A PD-10 column was used to exchange the protein into storage buffer (50 mm sodium phosphate, pH 8.0, 150 mm NaCl, 2 mm dithiothreitol, 5 mm MgCl2, and 25% glycerol).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strain | ||
| P. aeruginosa | ||
| PAO1 | Serotype O5, type strain | (27) |
| E. coli | ||
| BL21(DE3) | General expression strain F-ompT hsdSB(rB-mB-) gal dcm (DE3) | Novagen |
| Rosetta(DE3) | Expression strain with pRARE plasmid (CmR) encoding rare tRNAs | Novagen |
| Tuner(DE3) | Expression strain with lacY deletion for strict regulation of induction by IPTG | Novagen |
| Plasmid | ||
| pLysS | Expresses T7 lysozyme to suppress basal expression of T7 RNA polymerase prior to induction | Novagen |
| pET-28a | 5.4-kb T7 expression vector with N- or C-terminal His tags, KmR | Novagen |
| pET-23dr | Derivative of pET-23a T7 expression vector with short linker between N-terminal His tag and protein, AmpR | (18) |
| pCQW14 | PCR from PAO1 template inserted NdeI-XhoI into pET-28a to generate His6-WbpA expression plasmid, KanR | (9) |
| pET-His6-WbpB | PCR from PAO1 template inserted BamHI-EcoRI into pET-28a to generate His6-WbpB expression plasmid, KanR | (7) |
| pWMJL085 | PCR from PAO1 template DNA inserted NcoI-BamHI into pET-23dr vector to generate His6-WbpE expression plasmid, KanR | (7) |
| pCQW3 | PCR from PAO1 template DNA inserted NdeI-HindIII into pET-28a to generate His6-WbpD expression plasmid, KanR | (8) |
| pWMJL072 | pET-23dr-based His6-WbpI expression plasmid, AmpR | (11) |
His6-WbpE was expressed in E. coli Rosetta(DE3) cells using the scheme for His6-WbpB, except that cells were induced with 0.1 mm isopropyl β-d-thiogalactopyranoside, the resuspension buffer was 50 mm sodium phosphate pH 7.0, 150 mm NaCl, and 10 mm imidazole, and the column used was a 1-ml HiTrap Chelating HP column charged with NiSO4. Using FPLC, His6-WbpE was eluted with buffer containing 250 mm imidazole. The storage buffer was 50 mm sodium phosphate, pH 7.0, 150 mm NaCl, and 25% glycerol. Purified His6-WbpE was quantified using the Bradford assay (15).
His6-WbpA, His6-WbpI, and His6-WbpD were purified as previously described (8, 9, 11), except that an FPLC strategy was used for His6-WbpD resulting in elution with 500 mm imidazole. All purified enzymes were stored at –20 °C, and quantified using spectrophotometric assay (A280 = εCl) using ε values calculated from ProtParam (16).
Analytical Enzyme Reactions—UDP-d-GlcNAcA was synthesized from commercial UDP-d-GlcNAc using purified His6-WbpA as previously described (17). His6-WbpB reactions contained 50 mm sodium phosphate, pH 7.0, 2.5 mm NAD+, 2.5 mm MgCl2, and 1 mm UDP-d-GlcNAcA. Reactions with His6-WbpE also contained 5 mm pyridoxal phosphate and 5 mm l-glutamate. Reactions with His6-WbpD contained additional 1 mm acetyl-CoA. All reactions used 0.05 mg/ml of each enzyme desired, were allowed to proceed overnight at 37 °C, and were frozen at –20 °C until capillary electrophoresis analysis. WbpI reactions were carried out as previously described (11).
Capillary Electrophoresis—Capillary electrophoresis was performed using a P/ACE MDQ glycoprotein system (Beckman Coulter) with UV detection at 254 nm. The capillary was bare silica 75 μm × 50 cm with a detector at 40 cm and was conditioned before each run by washing with 1 m sodium hydroxide for 2 min, and then running buffer (25 mm sodium tetraborate, pH 9.5) for 2 min. Samples were introduced by pressure injection for 16 s, and separation was performed at 22 kV.
Mass Spectrometry—Enzymes were removed from the reactions using Microcon YM-10 Centrifugal Filter Devices (Millipore), but no other purification was performed. Filtered samples (2 μl) were mixed directly with matrix solution (2 mg of 3,4-dihydroxybenzoic acid in 20% of ethanol) in an analyte: matrix 1:2 ratio (v/v), and 1 μl was spotted on the MALDI sample target and allowed to dry at room temperature. In some cases sample spots were covered by an additional 1-μl matrix solution. Analysis was performed using a Bruker Reflex III MALDI-TOF MS instrument (Bruker Daltonics, Germany) equipped with a 337 nm nitrogen laser. Samples were analyzed in reflectron and negative ion modes scanning from 0 to 1000 m/z and using ion suppression up to 150 m/z. For all experiments, the ion sources 1 and 2 were held at 20 kV and 16.35 kV, respectively, and guiding lens voltage at 9.75 kV. The nitrogen laser power was set to 66–90 μJ. Two-point external calibration was performed, using the [M – H]– (223.06 Da) and [2M – H]– (447.12 Da) peaks of sinapinic acid (Sigma-Aldrich) prepared in acetonitrile/water solution (1 mg of sinapinic acid in 100 μl of 1:1 H2O/CH3CN). The mass accuracy with external calibration using sinapinic acid is estimated to be better than 5 ppm.
Enzymatic Synthesis of UDP-GlcNAc3NA and UDP-GlcNAc3NAcA—For UDP-d-GlcNAc3NA, a 5-ml reaction contained 50 mm sodium phosphate, pH 7.0, 100 mm ammonium sulfate, 2.5 mm NAD+, 5 mm pyridoxal phosphate, 5 mm l-glutamate, 2.5 mm MgCl2, 1 mm UDP-d-GlcNAc, and 0.05 mg/ml each of His6-WbpA, His6-WbpB, and His6-WbpE. After incubation at 37 °C overnight, the reaction was filtered using VivaSpin 15R columns (Sartorius Stedim Biotech, Germany). The filtrate was diluted to 100 ml before anion-exchange chromatography using an Econo-Pac High-Q 5-ml cartridge and a gradient of 0–500 mm ammonium bicarbonate. Samples from peaks were lyophilized and screened by CE. Fractions containing putative UDP-d-GlcNAc3NA were pooled and lyophilized, and then stored at –20 °C until NMR analysis. For UDP-d-GlcNAc3NAcA, the same strategy was used except that 1 mm acetyl-CoA and 0.05 mg/ml of His6-WbpD were included in the 5-ml reaction mixture. The concentration for sugar pools was calculated by spectrophotometric assay (A262 = εCl) using ε = 10.1 mm–1cm–1. The chemical synthesis of UDP-d-GlcNAc3NAcA will be reported elsewhere.
NMR Spectroscopy—Lyophilized sugar samples were resuspended in 200 μl of D2O (Cambridge Isotopes Laboratories) and examined using a Varian Unity 500-MHz (1H) spectrometer (Palo Alto California) with a Varian Z-gradient 3-mm 15N, 31P probe. Standard homo- and heteronuclear correlated two-dimensional 1H NMR (COSY, TOCSY, and NOESY) and 13C HSQC (heteronuclear single quantum coherence) pulse sequences from Varian were used for general assignments. Selective one-dimensional TOCSY experiments with a Z-filter were used for complete residue assignments and measurement of proton coupling constants (JH,H) and nuclear Overhauser enhancements (NOEs). NMR experiments were performed with suppression of the HOD resonance at δH 4.78 ppm, and the methyl resonance of acetone was used as an internal reference (δH, 2.225 ppm; δC, 31.07 ppm).
RESULTS
In Silico Analysis of WbpB and WbpE—The Pseudomonas Genome Database V2 shows that WbpB contains PFAM domains common to the oxidoreductase family and predicts an NAD-binding Rossmann fold at the N terminus of the protein, while COG (Clusters of Orthologous Groups) predicted similarity to MviM, putative dehydrogenases, and related proteins. WbpE was predicted to have similarity to WecE, a putative pyridoxal phosphate-dependent enzyme apparently involved in regulation of cell wall biogenesis. TIGRFAM identified the same WecE domain. WecE is a TDP-4-keto-6-deoxy-d-glucose transaminase. The top-scoring PFAM was the DegT/DnrJ/EryC1/StrS aminotransferase family, but several other aminotransferase families were also identified as hits.
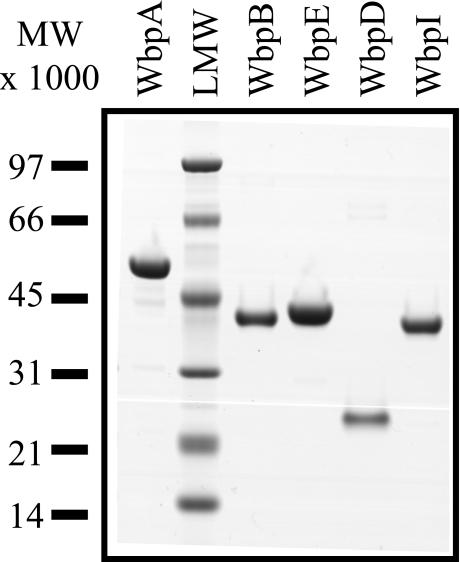
Purification of UDP-d-ManNAc3NAcA Pathway Enzymes—WbpA, WbpB, WbpE, WbpD, and WbpI were individually purified to near homogeneity and had apparent molecular masses consistent with their expected structures (Fig. 2). Initial trials with His6-WbpB indicated that the protein was expressed mainly as inclusion bodies. This problem was alleviated when the protein was expressed in Tuner(DE3) cells with very low concentration isopropyl β-d-thiogalactopyranoside induction and an adequate yield of soluble protein was obtained. His6-WbpB also frequently precipitates. To alleviate this problem, the purification was conducted in the presence of 20% glycerol and 12 mm β-mercaptoethanol at 4 °C. Typical yields of ∼0.7 mg of His6-WbpB were obtained from a 250-ml culture after exchange into storage buffer. Expression of His6-WbpE is extremely poor with most standard protein expression conditions used except with the use of Rosetta(DE3) cells and an overnight 15 °C induction to improve soluble yield. Under optimal conditions, ∼2.8 mg of His6-WbpE per 250-ml culture was obtained. An FPLC-based purification method used for His6-WbpD yielded ∼1.4 mg of protein from 250 ml of culture. When stored at –20 °C, the proteins were stable for at least 3 weeks, while precipitation occurred if the protein was left at ambient temperature or subjected to excessive freeze/thaw cycles. Purification of the previously characterized proteins His6-WbpA and His6-WbpI proceeded according to expectations (9, 11).
FIGURE 2.
SDS-PAGE analysis of purified enzymes of the UDP-d-ManNAc3NAcA biosynthesis pathway. Each protein was engineered with an N-terminal hexahistidine tag and overexpressed in E. coli. Predicted molecular masses were 50.3, 39.3, 40.2, 22.7, and 40.2 kDa for His6-WbpA, His6-WbpB, His6-WbpE, His6-WbpD, and His6-WbpI, respectively. A total of 2 μg of each protein was loaded, and the gel was stained with SimplyBlue™.
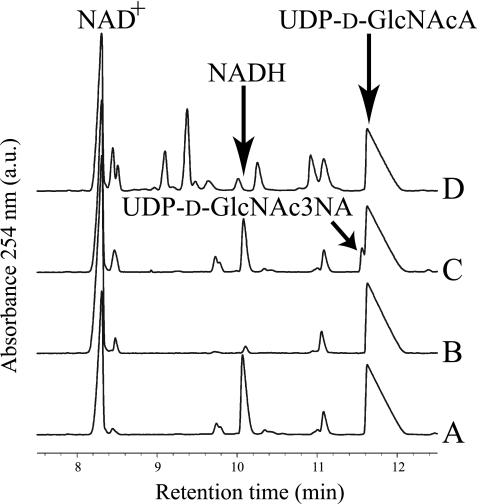
Capillary Electrophoresis Analysis of Enzyme Catalyzed Reactions—Many attempts to visualize a reaction product from His6-WbpB reactions under various conditions all failed, because no novel product peaks were observed when compared with no-enzyme controls. However, one new peak was observed, and it was identified as NADH based on spiking the reaction with additional NADH (Fig. 3). In the reaction between His6-WbpE and UDP-d-GlcNAcA, neither a new product peak nor cofactor turnover could be discerned (Fig. 3). However, when His6-WbpB and His6-WbpE were added together to a reaction mixture, two peaks were identified: NADH and a novel peak migrating just ahead of the UDP-d-GlcNAcA substrate (Fig. 3). This novel peak is thought to be UDP-d-GlcNAc3NA, which is the predicted substrate for WbpD. His6-WbpD reactions were attempted in conjunction with both His6-WbpB and His6-WbpE reactions to produce the putative substrate in situ. Reactions with His6-WbpB, His6-WbpE, and His6-WbpD also contained acetyl-CoA, because it is a required cofactor for His6-WbpD. When analyzed by CE, these reactions showed the presence of several new peaks and an absence of the putative UDP-d-GlcNAc3NA peak that was observed in the WbpB- and WbpE-coupled reactions (Fig. 3). Control reactions with UDP-d-GlcNAcA, acetyl-CoA, and His6-WbpD alone produced CE traces that were identical to those from the reactions with WbpB, WbpE, and WbpD (data not shown).
FIGURE 3.
Capillary electropherogram of WbpB, WbpE, and WbpD reactions. A, WbpB enzyme alone produced no product. B, WbpE enzyme alone produced no product. C, WbpB and WbpE incubated together yielded a novel peak that migrated just ahead of UDP-d-GlcNAcA, and this was identified as UDP-d-GlcNAc3NA. D, WbpB, WbpE, and WbpD incubated together do not show the presence of the novel peak, but the electropherogram shows several new peaks caused by the addition of UV-detectable acetyl-CoA (cofactor for WbpD). In all traces, the small peak at ∼11.1 min is a minor contaminant present in the NAD+.
Large scale reactions with His6-WbpA, His6-WbpB, and His6-WbpE, or His6-WbpA, His6-WbpB, His6-WbpE, and His6-WbpD, were screened by CE before NMR analysis. Analysis of the complete WbpA/WbpB/WbpE reaction mixture showed the same putative UDP-d-GlcNAc3NA peak migrating just before the UDP-d-GlcNAcA substrate peak. After separation by anion-exchange chromatography, selected aliquots were lyophilized, and small samples of these were analyzed by CE before and after the addition of UDP-d-GlcNAcA to the sample. The samples were shown to contain two peaks, one that co-migrated with UDP-d-GlcNAcA and one that migrated just ahead of the added UDP-d-GlcNAcA; these aliquots were pooled and subjected to NMR analysis. In the case of the unseparated WbpA/WbpB/WbpE/WbpD reaction, no putative product peak could be discerned due to poor signal-to-noise ratio created by the confounding acetyl-CoA peaks. After anion-exchange separation, selected aliquots were lyophilized, and samples of each were tested with and without the addition of chemically synthesized UDP-d-GlcNAc3NAcA. Aliquots represented by samples that co-migrated with the known UDP-d-GlcNAc3NAcA were pooled and further analyzed by NMR spectroscopy.
Reactions with His6-WbpI using the enzymatically synthesized UDP-d-GlcNAc3NAcA showed the production of a novel peak; this peak was absent in control reactions without enzymes. Comparison to a His6-WbpI reaction using chemically synthesized UDP-d-GlcNAc3NAcA as a standard indicated that the novel peak co-migrates with UDP-d-ManNAc3NAcA (data not shown).
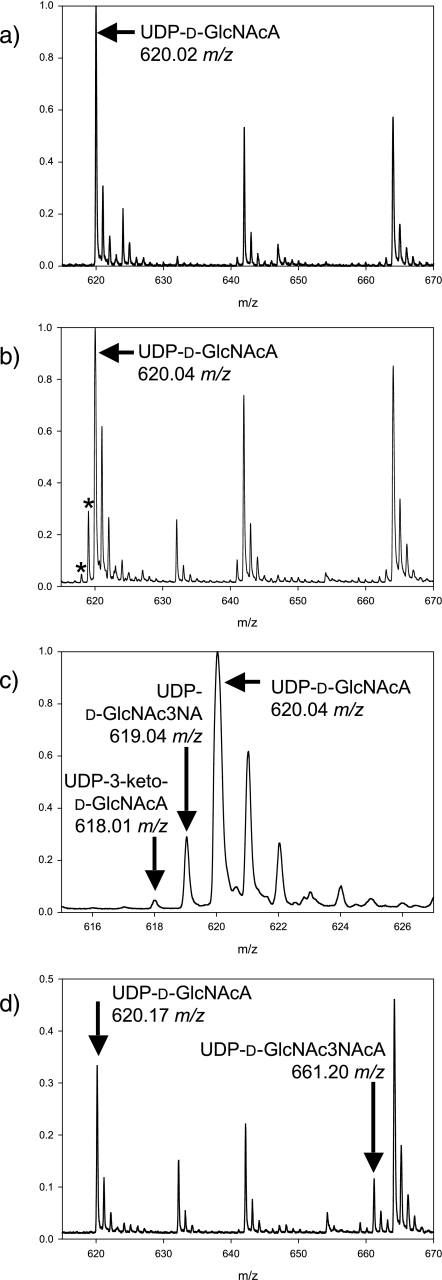
MS Analysis of WbpB, WbpE, and WbpD Reactions—Mass spectrometry was used to analyze the same reaction mixes screened by CE. Reactions containing only His6-WbpB or His6-WbpE, respectively, showed the presence of an ion at 620.01 m/z or 620.03 m/z (data not shown), like that of the no-enzyme control (620.02 m/z, Fig. 4a). In contrast, the reaction containing both His6-WbpB and His6-WbpE demonstrated the presence of three different ions: 620.09, 618.07, and 619.10 m/z (Fig. 4b). Reactions with His6-WbpB, His6-WbpE, and His6-WbpD contained ions 620.17 m/z and 619.24 m/z, as well as a new ion at 661.20 m/z (Fig. 4c). The ion at 661.20 m/z was absent in control reactions that lacked His6-WbpD, because only 620.13 m/z and 619.14 m/z were observed (data not shown). A positive control consisting of chemically synthesized UDP-d-GlcNAc3NAcA resuspended in reaction buffer without enzymes contained a peak at 661.1 m/z (Fig. 4).
FIGURE 4.
Mass spectra of reactions incubated with WbpB, WbpE, and WbpD. a, the substrate, UDP-d-GlcNAcA (621.34 g/mol), yields a mass spectrum in negative ion mode with observed peaks at 620.02 m/z and its natural abundant isotopes at ∼621, ∼622, and ∼623 m/z. b, after incubation with both WbpB and WbpE, two additional peaks appear at 618.01 and 619.04 m/z (marked by an asterisk), corresponding to UDP-3-keto-d-GlcNAcA (619.33 g/mol) and UDP-d-GlcNAc3NA (620.36 g/mol), respectively. Neither peak was observed when either WbpB or WbpE was reacted alone (data not shown). c, enlarged view of panel b, showing the peaks that correspond to UDP-3-keto-d-GlcNAcA and UDP-d-GlcNAc3NA. d, after incubation with WbpB, WbpE, and WbpD, a new peak not present in the no-enzyme control appears at 661.20 m/z corresponding to UDP-d-GlcNAc3NAcA (662.40 g/mol). The 661 m/z peak was not observed when WbpD was absent from the reaction mixture (data not shown).
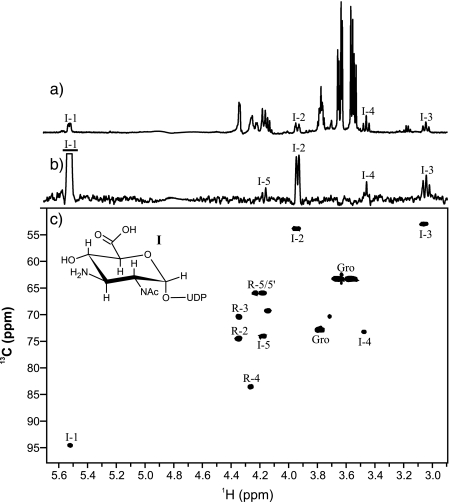
NMR Spectroscopy of Enzymatically Synthesized Putative UDP-α-d-GlcNAc3NA (I) and UDP-α-d-GlcNAc3NAcA (II)—The purified product of the coupled WbpB-WbpE reaction (I) was lyophilized, resuspended in 175 μl of D2O, and characterized by NMR spectroscopy (Fig. 5). The proton NMR spectrum revealed one major anomeric signal for I resonating at δH 5.52 ppm (Fig. 5a and Table 2). Selective one-dimensional TOCSY of H1 (90 ms) was used to identify resonances for H2, H3, H4, and H5, as well as to measure coupling constants (Fig. 5b). Large J2,3 (11.1 Hz), J3,4 (10.2 Hz), and J4,5 (10.0 Hz) couplings indicated a gluco ring configuration, which was corroborated by strong H2–H4 and H3–H5 NOEs observed using a two-dimensional NOESY experiment (800 ms, not shown). Using a 1H-13C HSQC experiment, C3 was determined to be N-substituted based on its upfield resonance at δC 53.1 ppm (Fig. 5c). In contrast, C3 in UDP-α-d-GlcNAcA was reported to resonate at δC 71.5 ppm (11). Based on the findings of mass spectrometric analyses as well as these NMR results, the product of the WbpB-WbpE reaction was concluded to be UDP-α-d-GlcNAc3NA (I). Based on the results of proton, COSY, TOCSY, NOESY, and 1H-13C HSQC experiments, the purified reaction product of the WbpD reaction (II) was determined to be UDP-α-d-GlcNAc3NAcA (Table 2). The identity of II was supported by data of proton and carbon chemical shifts as well as coupling constants that were nearly identical to those reported for UDP-α-d-GlcNAc3NAcA (11, 14).
FIGURE 5.
NMR spectroscopy of the purified WbpB/WbpE reaction product UDP-α-d-GlcNAc3NA (I). a, proton spectrum (128 scans). b, one-dimensional TOCSY (80 ms) of I H1. c, 1H-13C HSQC spectrum (128 scans, 128 increments, 1JC,H = 140 Hz, 20 h). For selective one-dimensional experiments, excited resonances are underlined. R is ribose and Gro is glycerol.
TABLE 2.
NMR data for the pyranose rings of the WbpE reaction product UDP-α-d-GlcNAc3NA (I), and WbpD reaction product UDP-α-d-GlcNAc3NAcA (II), respectively
NMR spectra were referenced to an internal acetone standard (dH, 2.225 ppm; δC, 31.07 ppm).
|
Compound
|
1H and 13C chemical shifts (δ ppm) and proton
coupling constants (JH,H Hz)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H4 | H5 | H6 | |||
| C1 | C2 | C3 | C4 | C5 | C6 | NAc-CH3 | NAc-CH3 | |
| J1,2 | J2,3 | J3,4 | J4,5 | |||||
| UDP-α-d-GlcNAc3NA (I) | ||||||||
| δH | 5.52 | 3.94 | 3.05 | 3.46 | 4.17 | 2.08 | ||
| δC | 94.6 | 54.0 | 53.1 | 73.3 | 74.3 | NDa | 22.9 | |
| JH,H | 3.2 | 11.1 | 10.2 | 10.0 | ||||
| JP,H | 7.5 | |||||||
| UDP-α-d-GlcNAc3NAcA (II) | ||||||||
| δH | 5.55 | 4.12 | 4.14 | 3.66 | 4.23 | 1.97 | 1.98 | |
| δC | 94.3 | 52.4 | 52.7 | 70.6 | 73.8 | ND | 22.5 | 22.5 |
| JH,H | 2.7 | 11.0 | 10.5 | 10.0 | ||||
| JP,H | 7.3 | |||||||
ND, not determined.
DISCUSSION
Each of the five enzymes required for the UDP-d-ManNAc3NAcA biosynthesis pathway in P. aeruginosa have now been overexpressed and purified. In this report, the purification of His6-WbpB and His6-WbpE was essential, because these upstream steps were required for the synthesis of the substrate for His6-WbpD, which had been previously overexpressed, but its biochemical function was not proven (8). Adequate yield and purity were obtained directly after affinity chromatography, but some material was lost during the buffer exchange process that was required to stabilize these proteins for storage. Use of an automated system such as FPLC streamlined the process of purifying four of the five proteins used in this study. Only His6-WbpB was unsuitable for FPLC purification, i.e. to prevent precipitation it required the presence of reducing agent and high glycerol concentrations that are incompatible with FPLC. His6-WbpE was also found to precipitate rapidly in elution buffer, but this issue was easy to overcome by swift exchange into the storage buffer. Similarly, prior work with His6-WbpD indicated precipitation issues, but in this study purification in the absence of glycerol was acceptable as long as the protein was immediately exchanged into storage buffer after elution. Addition of equimolar concentrations of acetyl-CoA to His6-WbpD has also been shown to enhance stability (8).
CE was used to determine whether the enzymatic reactions produced novel products. However, co-migration of UDP-d-GlcNAc3NAcA with acetyl-CoA in the WbpD reactions prevented the identification of clear signals that would be consistent with the emergence of new products. For this reason, we used a quick MS approach to screen this and other reactions for new products that might have co-migrating with other compounds on the CE trace. Typically, MS analysis of enzyme reaction mixtures has been performed using time-consuming liquid chromatography electrospray MS methods due to the complex nature of the components in these mixtures, including proteins, salts, buffers, and cofactors that require separation prior to ionization. Because MALDI-TOF analysis is more tolerant to the presence of salts and buffers, it provides a direct and rapid (10-s sampling rate) method for monitoring reactions that contained very small quantities of samples (5 μl). This method allowed the discrimination of a substrate and product with a single mass unit difference where CE showed co-migration.
Reactions with His6-WbpB and His6-WbpE together demonstrated the production of a novel peak by CE and an ion consistent with the mass of UDP-d-GlcNAc3NA was identified by MS. This provided initial evidence that His6-WbpB and His6-WbpE were producing the expected oxidoreductase and aminotransferase activity for the second and third step in the proposed UDP-d-ManNAc3NAcA biosynthesis pathway. Interestingly, an ion consistent with the mass of UDP-3-keto-d-GlcNAcA was detected by MS and only in the reaction containing both His6-WbpB and His6-WbpE. This gives direct evidence of the oxidation of UDP-d-GlcNAcA and suggests that WbpB and WbpE may have to interact to activate WbpB. Because ions consistent with the mass of UDP-3-keto-d-GlcNAcA and UDP-d-GlcNAc3NA were found only in reactions where His6-WbpB and His6-WbpE were both present, it is logical to assign the function of WbpB as the oxidoreductase and WbpE the aminotransferase. The lack of reaction when WbpE was incubated with UDP-d-GlcNAcA, screened by both CE and MS, demonstrated that WbpE could not utilize UDP-d-GlcNAcA as a substrate. This indicates that, in reactions with both enzymes, WbpE must be using a product made by WbpB to produce UDP-d-GlcNAc3NA. Finally, because in the co-reaction examined by MS, ions consistent with UDP-d-GlcNAcA, UDP-3-keto-d-GlcNAcA, and UDP-d-GlcNAc3NA were observed, our results indicated that WbpB is an oxidoreductase that acts first on UDP-d-GlcNAcA to catalyze the formation of UDP-3-keto-d-GlcNAcA, and WbpE is an aminotransferase that acts on UDP-3-keto-d-GlcNAcA to produce UDP-d-GlcNAc3NA. The amino acid sequences of WbpB and WbpE also support this assignment, because WbpB contains putative oxidoreductase motifs and a Rossman-fold region for binding NAD+/ NADP+, whereas the sequence of WbpE shared a high level of homology to proteins of aminotransferase families. Furthermore, the B. pertussis protein WlbC was shown to complement a wbpE knockout, indicating that they have the same function (7). WlbC has been shown to behave as a typical type-2 aminotransferase based on structural and spectroscopic data.5 WbpB is a novel enzyme that should be called UDP-2-acetamido-2-deoxy-d-glucuronic acid 3-dehydrogenase and given the IUPAC-IUBMB enzyme classification EC 1.1.1 (Oxidoreductases: Acting on the CH-OH Group of Donors: With NAD or NADP as Acceptor). WbpE also presents novel activity and should be called UDP-2-acetamido-2-dideoxy-d-ribo-hex-3-uluronic acid transaminase under EC 2.6.1 (Transferases: Transferring Nitrogenous Groups: Transaminases).
Reactions with His6-WbpD in addition to His6-WbpB and His6-WbpE screened by CE showed that the novel UDP-d-GlcNAc3NA peak was depleted, and the MS analysis showed that an ion consistent with UDP-d-GlcNAc3NAcA was produced. Evidence from NMR spectroscopy and successful utilization of this compound by His6-WbpI, which was previously shown to be stringently substrate-specific (11), further demonstrate that the product of His6-WbpD reaction is UDP-d-GlcNAc3NAcA. Together with previous evidence that WbpD belongs to the hexapeptide acyltransferase superfamily of proteins and uses acetyl-CoA as a cofactor (8), our results provide physical evidence of the enzymatic activity of WbpD for the first time. This unique enzyme should be given the name UDP-2-acetamido-3-amino-2,3-dideoxy-d-glucuronic acid N-acetyltransferase under EC 2.3.1 (Transferases: Acyltransferases: Transferring Groups Other Than Amino-acyl Groups).
WbpB, WbpE, and WbpD work together to accomplish the formation of a 3-N-acetyl group on UDP-d-GlcNAcA, thus forming UDP-d-GlcNAc3NAcA. Other 3-amino or 3-acetamido sugars are known, such as UDP-2-acetamido-3-amino-2,3-dideoxy-d-glucose (UDP-d-GlcNAc3N), dTDP-3-acetamido-3,6-dideoxy-d-galactose (dTDP-d-Fuc3NAc), and dTDP-3-acetamido-3,6-dideoxy-d-glucose (dTDP-d-Qui3-NAc), and the biosynthesis of these sugars has been described (19–21). The formation of dTDP-d-Fuc3NAc requires fdtA, the isomerase; fdtB, the transaminase, and fdtC, the transacetylase genes (20). FdtB and FdtC are very similar to WbpE and WbpD, respectively, in terms of amino acid sequence, function, required cofactors, and conserved domains. FdtA, on the other hand, produces a 3-keto group via isomerization of dTDP-6-deoxy-d-xylo-4-hexulose (20) and shows no similarity to WbpB, the oxidoreductase required for the formation of UDP-d-ManNAc3NAcA. The biosynthesis of dTDP-d-Qui3NAc is similar to the pathway examined in this study, requiring qdtA, qdtB, and qdtC as the genes that encode isomerase, transaminase, and transacetylase, respectively (21). QdtB and QdtC are also similar to WbpE and WbpD, respectively, although QdtC shares only 52% similarity to WbpD while all the other pairs share at least 61% similarity. The formation of UDP-d-GlcNAc3N requires GnnA, an oxidoreductase, and GnnB, a transaminase (19). GnnB shows similarity to WbpE, but no significant similarity is detected between GnnA and WbpB.
In all these published reports, the 3-keto product could not be purified due to instability and degradation, and concurrent reactions were therefore used to yield the 3-amino product in a single reaction (19–21). Our approach was similar, as we studied concurrent reactions with the WbpB oxidoreductase and WbpE transaminase, but to our best knowledge, this is the first report of the detection of a 3-ketosugar nucleotide using the novel mass spectrometry approach described above.
Possible interaction between WbpB and WbpE was suggested by the fact that UDP-3-keto-d-GlcNAcA was not detected in WbpB-only reactions. It is possible that UDP-3-keto-d-GlcNAcA co-migrated with UDP-d-GlcNAcA during CE analysis, thus making it impossible to detect the product. This is probably not the case, because the MS analysis was also unable to detect an ion consistent with UDP-3-keto-d-GlcNAcA when only WbpB was used in a reaction. Hence it is clear that WbpE may activate WbpB through direct physical interaction, or the presence of WbpE may increase the rate of WbpB catalysis simply by rapidly utilizing the UDP-3-keto-d-GlcNAcA produced. Studies on QdtA isomerase reactions generating 3-keto products have been attempted using NMR, but a full assignment of the product's structure could not be achieved because of severe signal overlap with the remaining substrate and low signal intensity of the product (21). However, Pföstl et al. (21) concluded based on partial assignments that QdtA-mediated isomerization to 3-keto product peaked at ∼10–11% after 1 h. This suggested that the enzyme producing the 3-keto group was capable of working alone. Conversely, experiments with GnnA indicated that it could not function in the absence of GnnB (19). Experiments to determine whether physical interactions exist between WbpB and WbpE are currently underway.
A scheme similar to that described here for the creation of a 3-acetamido group was employed to create a 4-amino or 4-acetamido group in Shigella dysenteriae Type 7 and E. coli O7 (22). In this case, dTDP-d-glucose is converted to dTDP-6-deoxy-d-xylo-hex-4-ulose (dTDP-4-keto-d-Qui) by a 4,6-dehydratase, RmlB. VioA, an aminotransferase then catalyzes the conversion of dTDP-4-keto-d-Qui to dTDP-4-amino-4,6-dideoxy-d-glucose (dTDP-d-Qui4N), which is further modified by VioB, an acetyltransferase, to form dTDP-4-acetamido-4,6-dideoxy-d-glucose (dTDP-d-Qui4NAc) (22). WbpB, WbpE, and WbpD show only modest (<51%) similarity to these proteins, likely because both the substrate and site modified are different. These similar pathways establish the sequential roles of dehydration, transamination, and transacetylation as a three-part cassette required to construct an acetamido group at a given position.
The synthesis of UDP-d-GlcNAc3NA has never before been reported by either enzymatic or chemical means. This achievement required the use of three purified enzymes and a compromise of using a buffer that allows only moderate activity from each of the proteins. In such a reaction, it is difficult to use an optimal condition for all enzymes, because reagents that improve one of the reactions may be somewhat inhibitory to one or more of the other steps. In nature, depletion of UDP-d-ManNAc3NAcA as it is incorporated to produce LPS in P. aeruginosa may help pull along the overall reaction. Combined use of MS and NMR analyses of the purified material allowed us to determine the structure of UDP-d-GlcNAc3NA unambiguously. Similar methods were used to purify UDP-d-GlcNAc3NAcA from reactions using four enzymes (His6-WbpA, His6-WbpB, His6-WbpE, and His6-WbpD). Although UDP-d-GlcNAc3NAcA was previously produced by chemical synthesis, the protocol required 17 steps and gave an overall yield of ∼9% (14). The enzymatic process required four reactions that were accomplished simultaneously in a single overnight incubation. Following the incubation, anion-exchange purification was used to isolate UDP-d-GlcNAc3NAcA giving a yield of ∼40% (based on the molar quantity of starting substrate versus molar quantity of purified UDP-d-GlcNAc3NAcA). Data from both MS and NMR analysis unequivocally identified the structure of the product of WbpD reaction to be UDP-d-GlcNAc3NAcA, thereby demonstrating that this simplified enzymatic synthesis represents a significant improvement over the previous chemical synthesis method.
This is the first time that in vitro reconstitution of the UDP-d-ManNAc3NAcA biosynthesis pathway has been achieved, and these findings have importance beyond LPS biosynthesis in P. aeruginosa. As previously described, B. pertussis proteins WlbA, WlbC, and WlbB have been shown to have the same function as WbpB, WbpE, and WbpD, respectively (7). Thus, this evidence also supports the description of WlbA as a UDP-2-acetamido-2-deoxy-d-glucuronic acid 3-dehydrogenase, WlbC as a UDP-2-acetamido-2-dideoxy-d-ribo-hex-3-uluronic acid transaminase, and WlbB as a UDP-2-acetamido-3-amino-2,3-dideoxy-d-glucuronic acid N-acetyltransferase. The ability to enzymatically synthesize UDP-d-GlcNAc3NAcA and UDP-d-ManNAc3NAcA will enable additional studies. For example, B. pertussis is closely related to Bordetella bronchiseptica and Bordetella parapertussis. Both organisms also produce d-ManNAc3NAcA, and 2,3-diacetamido-2,3-dideoxy-l-galactosamine (l-GalNAc3NAcA), as constituents of the LPS (23, 24). Because UDP-l-GalNAc3NAcA is thought to be synthesized from UDP-d-ManNAc3NAcA by the enzymes of the wbm* gene cluster (25), further studies will benefit from enzymatic production of the predicted substrate. Purified UDP-d-GlcNAc3NA will also be made available to the community to support enzyme specificity tests or other applications, as reported for UDP-d-GlcNAc3NAcA (26).
In conclusion, we have achieved the overexpression and purification of His6-WbpB, and His6-WbpE, which has never previously been established. Furthermore, biochemical and chemical evidence derived from CE, MS, and NMR analyses strongly support the assignment of the novel enzymes WbpB, WbpE, and WbpD as a UDP-2-acetamido-2-deoxy-d-glucuronic acid 3-dehydrogenase, UDP-2-acetamido-2-dideoxy-d-ribo-hex-3-uluronic acid transaminase, and UDP-2-acetamido-3-amino-2,3-dideoxy-d-glucuronic acid N-acetyltransferase, respectively. Use of these three enzymes in addition to His6-WbpA allowed for the single-reaction synthesis and subsequent purification of UDP-d-GlcNAc3NA or UDP-d-GlcNAc3NAcA, which were structurally validated by NMR. Reactions containing purified UDP-d-GlcNAc3NAcA and His6-WbpI, the known 2-epimerase, yielded UDP-d-ManNAc3NAcA. This represents the completion of in vitro reconstitution of the UDP-d-ManNAc3NAcA biosynthesis pathway from P. aeruginosa.
Acknowledgments
We thank Dr. Martin Rejzek for providing a standard of synthetic UDP-d-GlcNAc3NAcA.
This work was supported in part by the Canadian Institutes of Health Research (CIHR) (Grant MOP 14687 to J. S. L.). Studies in the Field laboratory are supported by the Biotechnology and Biological Sciences and Research Council.
Footnotes
The abbreviations used are: LPS, lipopolysaccharide; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; FPLC, fast-protein liquid chromatography; MS, mass spectrometry; TOCSY, total correlation spectroscopy; NOE, nuclear Overhauser effect; NOESY, nuclear Overhauser effect spectroscopy; CE, capillary electrophoresis.
R. A. Field, C. Wing, V. Sri Kannathasan, A. Preston, D. Maskell, and J. H. Naismith, unpublished observations.
References
- 1.Liu, P. V., and Wang, S. (1990) J. Clin. Microbiol. 28 922–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knirel, Y. A., Vinogradov, E. V., Kocharova, N. A., Paramonov, N. A., Kochetkov, N. K., Dmitriev, B. A., Stanislavsky, E. S., and Lanyi, B. (1988) Acta Microbiol. Hung. 35 3–24 [PubMed] [Google Scholar]
- 3.Knirel, Y. A., and Kochetkov, N. K. (1994) Biochemistry 59 1325–1382 [Google Scholar]
- 4.Newton, G. J., Daniels, C., Burrows, L. L., Kropinski, A. M., Clarke, A. J., and Lam, J. S. (2001) Mol. Microbiol. 39 1237–1247 [DOI] [PubMed] [Google Scholar]
- 5.Raymond, C. K., Sims, E. H., Kas, A., Spencer, D. H., Kutyavin, T. V., Ivey, R. G., Zhou, Y., Kaul, R., Clendenning, J. B., and Olson, M. V. (2002) J. Bacteriol. 184 3614–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrows, L. L., Pigeon, K. E., and Lam, J. S. (2000) FEMS Microbiol. Lett. 189 135–141 [DOI] [PubMed] [Google Scholar]
- 7.Westman, E. L., Preston, A., Field, R. A., and Lam, J. S. (2008) J. Bacteriol. 190 6060–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wenzel, C. Q., Daniels, C., Keates, R. A., Brewer, D., and Lam, J. S. (2005) Mol. Microbiol. 57 1288–1303 [DOI] [PubMed] [Google Scholar]
- 9.Miller, W. L., Wenzel, C. Q., Daniels, C., Larocque, S., Brisson, J. R., and Lam, J. S. (2004) J. Biol. Chem. 279 37551–37558 [DOI] [PubMed] [Google Scholar]
- 10.Vuorio, R., Hirvas, L., and Vaara, M. (1991) FEBS Lett. 292 90–94 [DOI] [PubMed] [Google Scholar]
- 11.Westman, E. L., McNally, D. J., Rejzek, M., Miller, W. L., Kannathasan, V. S., Preston, A., Maskell, D. J., Field, R. A., Brisson, J. R., and Lam, J. S. (2007) Biochem. J. 405 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burrows, L. L., Charter, D. F., and Lam, J. S. (1996) Mol. Microbiol. 22 481–495 [DOI] [PubMed] [Google Scholar]
- 13.Rocchetta, H. L., Burrows, L. L., and Lam, J. S. (1999) Microbiol. Mol. Biol. Rev. 63 523–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rejzek, M., Kannathasan, V. S., Wing, C., Preston, A., Westman, E. L., Lam, J. S., Naismith, J. H., Maskell, D. J., and Field, R. A. (2009) Org. Biomol. Chem. 7 1203–1210 [DOI] [PubMed] [Google Scholar]
- 15.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
- 16.Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., and Bairoch, A. (2005) in The Proteomics Protocols Handbook (Walker, J. M. ed) pp. 571–607, Humana Press, Totowa, NJ
- 17.Miller, W. L., Matewish, M. J., McNally, D. J., Ishiyama, N., Anderson, E. M., Brewer, D., Brisson, J. R., Berghuis, A. M., and Lam, J. S. (2008) J. Biol. Chem. 283 3507–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton, D. T., and Mangroo, D. (1999) Biochem. J. 339 63–69 [PMC free article] [PubMed] [Google Scholar]
- 19.Sweet, C. R., Ribeiro, A. A., and Raetz, C. R. (2004) J. Biol. Chem. 279 25400–25410 [DOI] [PubMed] [Google Scholar]
- 20.Pföstl, A., Hofinger, A., Kosma, P., and Messner, P. (2003) J. Biol. Chem. 278 26410–26417 [DOI] [PubMed] [Google Scholar]
- 21.Pföstl, A., Zayni, S., Hofinger, A., Kosma, P., Schaffer, C., and Messner, P. (2008) Biochem. J. 410 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, Y., Xu, Y., Perepelov, A. V., Qi, Y., Knirel, Y. A., Wang, L., and Feng, L. (2007) J. Bacteriol. 189 8626–8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preston, A., Allen, A. G., Cadisch, J., Thomas, R., Stevens, K., Churcher, C. M., Badcock, K. L., Parkhill, J., Barrell, B., and Maskell, D. J. (1999) Infect. Immun. 67 3763–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Fabio, J. L., Caroff, M., Karibian, D., Richards, J. C., and Perry, M. B. (1992) FEMS Microbiol. Lett. 76 275–281 [DOI] [PubMed] [Google Scholar]
- 25.King, J. D., Harmer, N. J., Preston, A., Palmer, C. M., Rejzek, M., Field, R. A., Blundell, T. L., and Maskell, D. J. (2007) J. Mol. Biol. 374 749–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sismey-Ragatz, A. E., Green, D. E., Otto, N. J., Rejzek, M., Field, R. A., and DeAngelis, P. L. (2007) J. Biol. Chem. 282 28321–28327 [DOI] [PubMed] [Google Scholar]
- 27.Hancock, R. E., and Carey, A. M. (1979) J. Bacteriol. 140 902–910 [DOI] [PMC free article] [PubMed] [Google Scholar]