Abstract
Recently we reported that N-glycans on the β-propeller domain of the integrin α5 subunit (S-3,4,5) are essential for α5β1 heterodimerization, expression, and cell adhesion. Herein to further investigate which N-glycosylation site is the most important for the biological function and regulation, we characterized the S-3,4,5 mutants in detail. We found that site-4 is a key site that can be specifically modified by N-acetylglucosaminyltransferase III (GnT-III). The introduction of bisecting GlcNAc into the S-3,4,5 mutant catalyzed by GnT-III decreased cell adhesion and migration on fibronectin, whereas overexpression of N-acetylglucosaminyltransferase V (GnT-V) promoted cell migration. The phenomenon is similar to previous observations that the functions of the wild-type α5 subunit were positively and negatively regulated by GnT-V and GnT-III, respectively, suggesting that the α5 subunit could be duplicated by the S-3,4,5 mutant. Interestingly GnT-III specifically modified the S-4,5 mutant but not the S-3,5 mutant. This result was confirmed by erythroagglutinating phytohemagglutinin lectin blot analysis. The reduction in cell adhesion was consistently observed in the S-4,5 mutant but not in the S-3,5 mutant cells. Furthermore mutation of site-4 alone resulted in a substantial decrease in erythroagglutinating phytohemagglutinin lectin staining and suppression of cell spread induced by GnT-III compared with that of either the site-3 single mutant or wild-type α5. These results, taken together, strongly suggest that N-glycosylation of site-4 on the α5 subunit is the most important site for its biological functions. To our knowledge, this is the first demonstration that site-specific modification of N-glycans by a glycosyltransferase results in functional regulation.
Glycosylation is a crucial post-translational modification of most secreted and cell surface proteins (1). Glycosylation is involved in a variety of physiological and pathological events, including cell growth, migration, differentiation, and tumor invasion. It is well known that glycans play important roles in cell-cell communication, intracellular signal transduction, protein folding, and stability (2, 3).
Integrins comprise a family of receptors that are important for cell adhesion. The major function of integrins is to connect cells to the extracellular matrix, activate intracellular signaling pathways, and regulate cytoskeletal formation (4). Integrin α5β1 is well known as a fibronectin (FN)3 receptor. The interaction between integrin α5 and FN is essential for cell migration, cell survival, and development (5–8). In addition, integrins are N-glycan carrier proteins. For example, α5β1 integrin contains 14 and 12 putative N-glycosylation sites on the α5 and β1 subunits, respectively. Several studies suggest that N-glycosylation is essential for functional integrin α5β1. When human fibroblasts were cultured in the presence of 1-deoxymannojirimycin, which prevents N-linked oligosaccharide processing, immature α5β1 integrin appeared on the cell surface, and FN-dependent adhesion was greatly reduced (9). Treatment of purified integrin α5β1 with N-glycosidase F, which cleaves between the innermost N-acetylglucosamine (GlcNAc) and asparagine N-glycan residues of N-linked glycoproteins, prevented the inherent association between subunits and blocked α5β1 binding to FN (10).
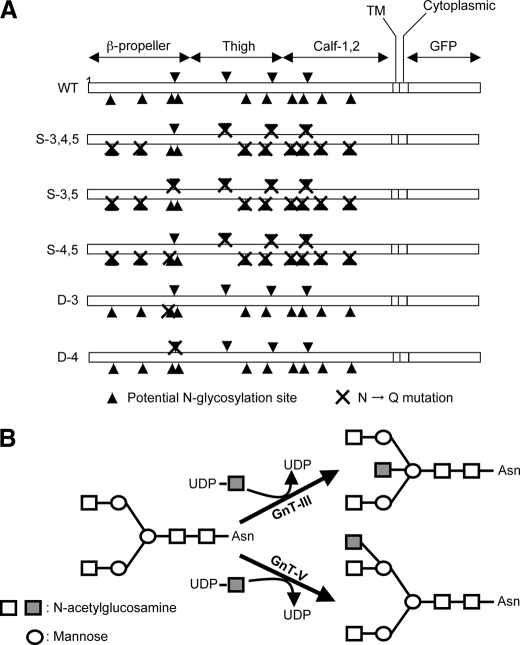
A growing body of evidence indicates that the presence of the appropriate oligosaccharide can modulate integrin activation. N-Acetylglucosaminyltransferase III (GnT-III) catalyzes the addition of GlcNAc to mannose that is β1,4-linked to an underlying N-acetylglucosamine, producing what is known as a “bisecting” GlcNAc linkage as shown in Fig. 1B. GnT-III is generally regarded as a key glycosyltransferase in N-glycan biosynthetic pathways and contributes to inhibition of metastasis. The introduction of a bisecting GlcNAc catalyzed by GnT-III suppresses additional processing and elongation of N-glycans. These reactions, which are catalyzed in vitro by other glycosyltransferases, such as N-acetylglucosaminyltransferase V (GnT-V), which catalyzes the formation of β1,6 GlcNAc branching structures (Fig. 1B) and plays important roles in tumor metastasis, do not proceed because the enzymes cannot utilize the bisected N-glycans as a substrate. Introduction of the bisecting GlcNAc to integrin α5 by overexpression of GnT-III resulted in decreased in ligand binding and down-regulation of cell adhesion and migration (11–13). Contrary to the functions of GnT-III, overexpression of GnT-V promoted integrin α5β1-mediated cell migration on FN (14). These observations clearly demonstrate that the alteration of N-glycan structure affected the biological functions of integrin α5β1. Similarly characterization of the carbohydrate moieties in integrin α3β1 from non-metastatic and metastatic human melanoma cell lines showed that expression of β1,6 GlcNAc branched structures was higher in metastatic cells compared with non-metastatic cells, confirming the notion that the β1,6 GlcNAc branched structure confers invasive and metastatic properties to cancer cells. In fact, Partridge et al. (15) reported that GnT-V-modified N-glycans containing poly-N-acetyllactosamine, the preferred ligand for galectin-3, on surface receptors oppose their constitutive endocytosis, promoting intracellular signaling and consequently cell migration and tumor metastasis.
FIGURE 1.
Potential N-glycosylation sites on the α5 subunit and its modification by GnT-III and GnT-V. A, schematic diagram of potential N-glycosylation sites on the α5 subunit. Putative N-glycosylation sites are indicated by triangles, and point mutations are indicated by crosses (N84Q, N182Q, N297Q, N307Q, N316Q, N524Q, N530Q, N593Q, N609Q, N675Q, N712Q, N724Q, N773Q, and N868Q). B, illustration of the reaction catalyzed by GnT-III and GnT-V. Square, GlcNAc; circle, mannose. TM, transmembrane domain.
In addition, sialylation on the non-reducing terminus of N-glycans of α5β1 integrin plays an important role in cell adhesion. Colon adenocarcinomas express elevated levels of α2,6 sialylation and increased activity of ST6GalI sialyltransferase. Elevated ST6GalI positively correlated with metastasis and poor survival. Therefore, ST6GalI-mediated hypersialylation likely plays a role in colorectal tumor invasion (16, 17). In fact, oncogenic ras up-regulated ST6GalI and, in turn, increased sialylation of β1 integrin adhesion receptors in colon epithelial cells (18). However, this is not always the case. The expression of hyposialylated integrin α5β1 was induced by phorbol esterstimulated differentiation in myeloid cells in which the expression of the ST6GalI was down-regulated by the treatment, increasing FN binding (19). A similar phenomenon was also observed in hematopoietic or other epithelial cells. In these cells, the increased sialylation of the β1 integrin subunit was correlated with reduced adhesiveness and metastatic potential (20–22). In contrast, the enzymatic removal of α2,8-linked oligosialic acids from the α5 integrin subunit inhibited cell adhesion to FN (23). Collectively these findings suggest that the interaction of integrin α5β1 with FN is dependent on its N-glycosylation and the processing status of N-glycans.
Because integrin α5β1 contains multipotential N-glycosylation sites, it is important to determine the sites that are crucial for its biological function and regulation. Recently we found that N-glycans on the β-propeller domain (sites 3, 4, and 5) of the integrin α5 subunit are essential for α5β1 heterodimerization, cell surface expression, and biological function (24). In this study, to further investigate the underlying molecular mechanism of GnT-III-regulated biological functions, we characterized the N-glycans on the α5 subunit in detail using genetic and biochemical approaches and found that site-4 is a key site that can be specifically modified by GnT-III.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies—A monoclonal antibody against human integrin α5 subunit (clone1) for Western blot analysis was obtained from BD Biosciences. For immunoprecipitation, the agarose-conjugated anti-green fluorescent protein (GFP) antibody (RQ2) was obtained from Medical & Biological Laboratories Co. Ltd. (Nagoya, Japan). Peroxidase-conjugated anti-mouse IgG was obtained from Cell Signaling Technology, Inc. (Danvers, MA). A VECTASTAIN ABC kit was purchased from Vector Laboratories, Inc. (Burlingame, CA). Antibodies against GnT-III (33A8) and GnT-V (24B11) were obtained from FUJIREBIO Inc. (Tokyo, Japan). Biotinylated erythroagglutinating phytohemagglutinin (E4-PHA), biotinylated leukoagglutinating phytohemagglutinin (L4-PHA), and biotinylated Datura stramonium lectin were purchased from Seikagaku Corp. (Tokyo, Japan). For fluorescence-activated cell sorting analysis, mouse anti-human α5β1 integrin monoclonal antibody (HA5, MAB1999) was purchased from Chemicon (Temecula, CA).
Cells and Cell Culture—The integrin α5 subunit-deficient CHO K1 cell line (CHO-B2) was a gift from Dr. Rudolf Juliano (School of Medicine, University of North Carolina, Chapel Hill, NC) (25). The CHO-B2 stable expression cells containing various integrin α5 with altered N-glycosylation sites were established in our laboratory (24). As shown in Fig. 1A, wild type (WT) indicates CHO-B2 expressing wild-type (full N-glycosylation sites) integrin α5; S-3,4,5, S-3,5, and S-4,5 show that all N-glycosylation sites were removed with site-directed mutagenesis except the indicated sites; and D-3 or D-4 represent single mutations at the indicated site. A HeLa cell line was purchased from RIKEN BioResource Center (Tsukuba, Japan). The stable expression of S-3,5 and S-4,5 mutants in HeLa cells was obtained by viral expression vector as mentioned below. These mutants and cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum, non-essential amino acids (Invitrogen), penicillin (100 units/ml), and streptomycin (100 μg/ml) (Nacalai Tesque, Inc., Kyoto, Japan) under a humidified atmosphere containing 5% CO2.
GnT-III, GnT-V, and α5 Mutant (S-3,5 and S-4,5) Expression with Viral Vectors—The cDNAs encoding human GnT-III and GnT-V were amplified for cloning into pENTR-D-Topo for the Gateway Conversion System (Invitrogen) according to the manufacturer's protocol. The cloned genes were inserted into the virus expression vector, pBABE-puro (Addgene, Inc. Cambridge, MA), accommodated into the Gateway Conversion System using LR Clonase reaction. The GnT-III and GnT-V constructs were transfected into Phoenix-Ampho cells with Lipofectamine 2000 (Invitrogen) for production of viral supernatants. The various α5 integrin mutants were infected with the resulting viral supernatant containing 10 μg/ml Polybrene (Sigma-Aldrich) and selected with 13 μg/ml puromycin for 2 weeks. In the case of HeLa cells expressing S-3,5 and S-4,5 mutants, after virus infection the infected cells were selected with 2.5 μg/ml puromycin. For mock transfection, the same protocol was performed using the empty virus expression vector only.
Cell Adhesion Assay Using 96-well Plate—96-well plates (Corning Inc.) were coated with 3 μg/ml FN at 37 °C for 1 h and blocked with 1% bovine serum albumin (BSA) in DMEM at 37 °C for 1 h. The cells were detached with trypsin containing 1 mm EDTA, resuspended with 0.5 mg/ml trypsin inhibitor (Nacalai Tesque, Inc.) in DMEM. The suspended cells were centrifuged at 1,000 rpm for 3 min and diluted to 4 × 105 or 8 × 105 cells/ml with assay medium, 0.1% BSA in DMEM. One hundred-microliter aliquots of cell suspension were added to each well, and the plates were incubated at 37 °C for 20–25 min. After incubation, attached cells were fixed with 25% glutaraldehyde (Nacalai Tesque, Inc.) and stained with 0.5% crystal violet. The absorbance at 590 nm was measured using an automated microtiter plate spectrometer, Powerscan® HT (Dainippon Sumitomo Pharma Co., Ltd. Osaka, Japan) operated with Microplate Data Analysis Software, KC4™ (BioTek Instruments, Inc., Winooski, VT). Cell spreading assays were performed as described previously (12, 24). After a 20-min incubation, representative fields were observed using phase-contrast microscopy, and spread cells were counted. The rounded cells were not considered as spread cells.
Cell Adhesion Kinetics Assay Using the Real Time Cell Electronic Sensing (RT-CES™) System—The cell adhesion kinetics assay was performed using a RT-CES system (ACEA Biosciences, Inc.) (26). Briefly ACEA Biosciences, Inc. electrosensing 16-well plates were coated with 50 μl of 10 μg/ml FN (Sigma) at 37 °C for 1 h and then blocked with 1% BSA in DMEM at 37 °C for 1 h. The cells were detached with trypsin containing 1 mm EDTA and resuspended with 0.5 mg/ml trypsin inhibitor (Nacalai Tesque, Inc.) in DMEM. The suspended cells were centrifuged at 1,000 × g for 3 min and diluted to 1 × 105 or 8 × 105 cells/ml with assay medium (0.1% BSA in DMEM). Fifty-microliter aliquots of the cell suspension were added to each well, and the assay was performed using RT-CES SP software. The program was set up such that the cell index was measured every 2 min for 3 h.
Cell Migration—Transwells (BD BioCoat™ Control Inserts, 8.0-μm inserts; BD Biosciences) were coated only on the bottom side with 10 μg/ml FN at 37 °C for 1 h. Cells were starved in serum-free medium for 4 h, trypsinized, and suspended with 0.5 mg/ml trypsin inhibitor (Nacalai Tesque, Inc.) in DMEM. Suspended cells were centrifuged, and supernatants were removed. The cell pellets were resuspended with assay medium (0.1% BSA in DMEM containing 1% fetal bovine serum) and diluted to 4 × 105 cells/ml. One hundred-microliter aliquots of the cell suspension were added to each FN-coated Transwell; the cells were then incubated at 37 °C for 3 h. After incubation, cells on the upper side were removed by scraping with a cotton swab. The membranes in the Transwells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet for 30 min. Cells that had migrated to the lower side were counted using a phase-contrast microscope.
Immunoprecipitation, Western Blot, and Lectin Blot—Subconfluent cells were washed with phosphate-buffered saline twice and lysed with ice-cold lysis buffer (1% Triton in Tris-buffered saline containing protease inhibitor mixture (Nacalai Tesque, Inc.)). The cell lysates were centrifuged at 15,000 × g for 10 min at 4 °C. The supernatants were obtained, and protein concentrations were determined using a BCA™ Protein Assay kit (Pierce). Equal amounts of protein were incubated with 10 μl of agarose-conjugated anti-GFP antibody and 15 μl of Sepharose™ 4B at 4 °C for 1 h. The immunocomplexes were washed twice with ice-cold lysis buffer, then were eluted with SDS sample buffer, and boiled for 5 min. The immunoprecipitates were subjected to 6.0 or 7.5% SDS-PAGE and then were transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat milk or 5% BSA in Tris-buffered saline for Western blot and lectin blot, respectively. After blocking, the membranes were incubated with either primary antibody or lectin for 1 h. For Western blot, membranes were incubated with the secondary antibody conjugated with peroxidase for 1 h, and the immunoreactive proteins were visualized using an ECL kit (GE Healthcare). For lectin blot, the lectin-binding proteins were detected using a VECTASTAIN ABC kit and an ECL kit.
Flow Cytometry—Flow cytometry was performed as described previously (24). Briefly cells were detached by trypsinizing and incubated with mouse anti-human α5β1 integrin monoclonal antibody (HA5) followed by Alexa Fluor 647 anti-mouse IgG. Negative controls underwent the same procedure without primary antibody. The analyses were performed using a FACSCalibur instrument (BD Biosciences) equipped with CELLQuestPro software.
RESULTS
Comparison of N-Glycosylation Patterns on S-3,4,5 α5 Subunit Mutant in GnT-III and GnT-V Transfectants—N-Glycosylation is essential for integrin α5β1 heterodimer formation and therefore plays an important role in the biological function of integrin. GnT-III-modified integrin α5β1 decreased cell adhesion and cell migration on FN (12). In contrast to GnT-III, GnT-V specifically modified only the β1 subunit and up-regulated integrin α5β1-mediated cell migration (14). Recently we found that three N-glycosylation sites, sites 3, 4, and 5 from the N terminus of the α5 subunit, were essential for the biological functions of integrin, such as cell adhesion and migration on FN and heterodimerization.
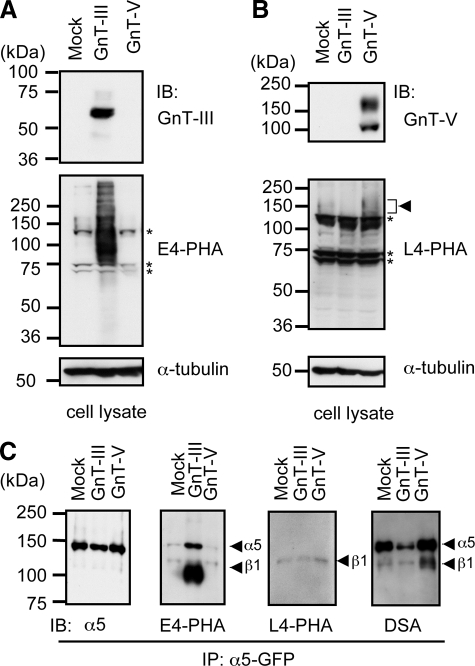
The purpose of the present study was to determine whether the S-3,4,5 mutant, which contained only three potential N-glycosylation sites (i.e. sites 3, 4, and 5), had characteristics similar to those of the wild-type α5 subunit, such as modification by GnT-III and GnT-V as described above. Various α5 subunit mutants were used in this study as shown in Fig. 1A. First the expression levels of GnT-III and GnT-V in S-3,4,5 mutant cells that had been transfected with a retrovirus system were examined by Western blotting (Fig. 2A). Their products were detected by E4-PHA lectin, which specifically recognizes bisecting GlcNAc, and by L4-PHA lectin, which selectively recognizes β1,6-branching GlcNAc, blots (Fig. 2B) (27, 28). As expected, bands corresponding to GnT-III and GnT-V as well as lectin reactivities of E4-PHA and L4-PHA were increased in the GnT-III and GnT-V transfectants, respectively (Fig. 2, A and B). Equal amounts of protein (20 μg) were loaded in each lane, and α-tubulin was used as the loading control. Next we immunoprecipitated α5 and detected N-glycans using E4-PHA and L4-PHA lectin blotting (Fig. 2C). The E4-PHA reactivities were much stronger in GnT-III transfectants than those in mock or GnT-V transfectants. This result indicates that both the α5 and β1 subunits are targets of GnT-III. In contrast, results of L4-PHA lectin staining indicated that only the β1 subunit could be modified by GnT-V, consistent with a previous study (14). The reactivities of D. stramonium lectin, which preferentially reacts with branched sugar chains (more than triantennary) (29), consistently showed a significant increase in the β1 subunit of GnT-V transfectants. A significant decrease in the α5 subunit of the GnT-III transfectants further supports the notion that introduction of GnT-III suppresses additional processing and branching formation of N-glycans catalyzed by other endogenous glycosyltransferases, such as GnT-V and GnT-IV. Although it is not clear whether the modification levels of branched N-glycans on α5 subunits were enhanced in GnT-V transfectants compared with the levels in mock transfectants, it could be argued that intrinsic GnT-V-mediated glycosylation is enough for occupation of some N-glycosylation sites on the α5 subunit, which may be specifically and exclusively modified by GnT-V.
FIGURE 2.
Comparison of N-glycosylation patterns on α5 subunits modified by GnT-III and GnT-V. GnT-III and GnT-V expression vectors were transfected into S-3,4,5 cells using the Phoenix retrovirus system, and stable expression cells were established as described under “Experimental Procedures.” Confluent cells were harvested and lysed for immunoblotting (IB). Equal amounts of protein (20 μg) were separated by 7.5% SDS-PAGE under reducing conditions, and the membranes were probed with antibodies against GnT-III (A, upper panel) and GnT-V (B, upper panel) or with the E4-PHA (A, middle panels) and L4-PHA lectins (B, middle panels) and reprobed with anti-α-tubulin, which was used as loading control (A and B, lower panels). Asterisks indicate nonspecific staining for E4-PHA or L4-PHA. C, cell lysates were subjected to immunoprecipitation (IP) using agarose-conjugated anti-GFP antibody. The immunoprecipitates were subjected to 6.0% SDS-PAGE under non-reducing conditions, blotted, and probed with E4-PHA, L4-PHA, and D. stramonium (DSA) lectins or probed with antibody against the α5 subunit.
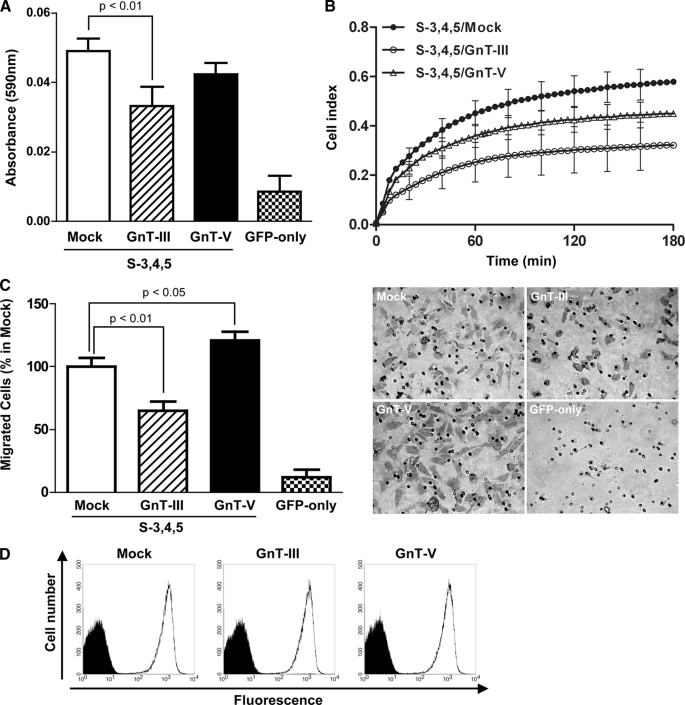
Effects of GnT-III and GnT-V on Integrin-mediated Cell Adhesion and Migration in S-3,4,5 Transfectants—It is well known that wild-type integrin α5β1-mediated cell migration can be positively and negatively regulated by GnT-V and GnT-III, respectively. Therefore, we determined whether modifications of S-3,4,5 mutants could mimic wild-type α5 to affect its biological functions, such as cell adhesion and cell migration. As shown in Fig. 3A, cell adhesion on FN was down-regulated in GnT-III transfectants compared with mock and GnT-V transfectants. The cell adhesion kinetics assay using RT-CES also showed the same tendency (Fig. 3B). On the other hand, cell migration was determined using the Transwell assay as described under “Experimental Procedures.” Interestingly overexpression of GnT-III significantly inhibited cell migration on FN, whereas GnT-V promoted cell migration relative to the mock transfectants (Fig. 3C). These results, taken together, suggest that the α5 subunit could be duplicated by the S-3,4,5 mutant. The up-regulation of cell migration in GnT-V transfectants could be ascribed to the N-glycans of the β1 subunit modified by GnT-V.
FIGURE 3.
Effects of overexpression of GnT-III and GnT-V on FN-mediated cell adhesion and migration in S-3,4,5 mutants. A, subconfluent cells were detached, and 40,000 cells were added to the 96-well plates coated with 3 μg/ml FN for the cell adhesion assay. The plates were incubated at 37 °C for 20 min and then washed twice with warmed phosphate-buffered saline to remove non-adherent cells. The adherent cells were fixed with 25% glutaraldehyde and stained with 0.5% crystal violet, and then the absorbance at 590 nm was measured. The bars represent the S.D. B, cell adhesion kinetics assay using the RT-CES system. Subconfluent cells were detached, and 10,000 cells were applied to wells of an electrosensing plate coated with 10 μg/ml FN. The device was operated with RT-CES SP software. The cell index represents the extent of cell adhesion. The bars represent the S.D. C, cell migration toward FN was determined using the Transwell assay as described under “Experimental Procedures.” Cells that migrated were stained with 0.5% crystal violet and counted under a microscope. The bars represent the S.D. A representative example is shown in the right panel. D, subconfluent cells were detached and labeled with primary antibody (mouse anti-human VLA5 antibody, HA5) for 30 min on ice. The labeled cells were washed with ice-cold phosphate-buffered saline and then incubated with Alexa Fluor 647 goat anti-mouse IgG for 30 min on ice. The expression levels of α5 integrin on the cell surface were measured using a FACSCalibur instrument (BD Biosciences). Negative controls were not treated with the primary antibody but underwent all other procedures.
To determine whether overexpression of GnT-III or GnT-V affected integrin α5β1 expression on the cell surface, we performed fluorescence-activated cell sorting analysis using anti α5β1 integrin antibody. As shown in Fig. 3D, there were no significant differences in the levels of cell surface expression among the three cell types, indicating that the functional alterations shown in Fig. 3 were due to N-glycosylation of the integrin modified by GnT-III or GnT-V.
GnT-III Selectively Modifies N-Glycosylation Site-4 on the α5 Subunit—As described above, the characteristics of the S-3,4,5 mutant are similar to those of wild-type α5. We therefore determined whether GnT-III could specifically modify the N-glycosylation site among site-3, site-4, and site-5. Because the N-glycosylation site-5 is essential for its expression on the cell surface, the mutant did not exhibit biological function such as cell adhesion (24). Thus, we chose the S-3,5 and S-4,5 mutants for use in further studies.
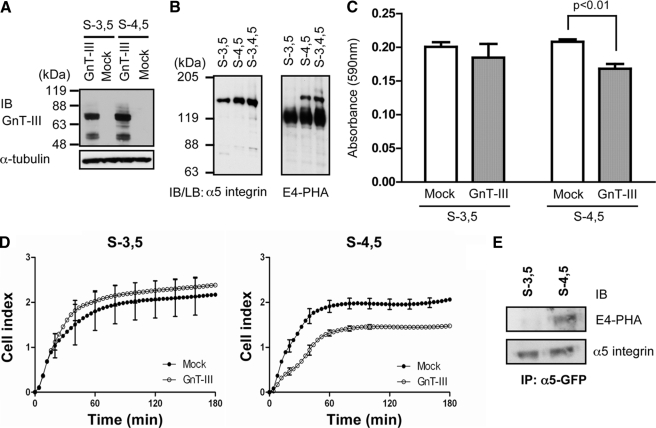
First GnT-III was overexpressed in both transfectants. The expression levels of GnT-III were almost the same in S-3,5 as in S-4,5 transfectants, which were examined by Western blot using anti-GnT-III antibody (Fig. 4A). It is of particular interest that the mutant S-4,5, but not the S-3,5 mutant, was clearly detected using E4-PHA lectin blot. The intensity of the lectin staining was comparable to that of S-3,4,5 (Fig. 4B). These results, taken together, suggest that site-3 may not be modified by GnT-III. Because introduction of bisecting GlcNAc into the α5 subunit down-regulates cell adhesion as described above, we checked whether this phenomenon occurred in these mutants. Overexpression of GnT-III in S-4,5 cells consistently inhibited cell adhesion on FN, whereas the inhibition of cell adhesion was not observed in S-3,5 cells overexpressing GnT-III (Fig. 4, C and D). It should be noted that cell adhesion activities of the S-3,5 mutant was similar to that of S-4,5 mutant because CHO-B2 cells do not express enough endogenous GnT-III to modify integrin as shown in Fig. 2.
FIGURE 4.
Comparison of effects of GnT-III on N-glycosylation and cell adhesion between S-3,5 and S-4,5 mutants. A, GnT-III was expressed in S-3,5 and S-4,5 mutants, and stable expression cells were established as described under “Experimental Procedures.” The expression levels of GnT-III were detected with an antibody against GnT-III. B, confluent cells were lysed and then subjected to immunoprecipitation (IP) using an agarose-conjugated antibody against GFP. The immunoprecipitates were subjected to 6.0% SDS-PAGE under non-reducing conditions, blotted, probed with E4-PHA (right panel), and then reprobed with antibody against the α5 subunit (left panel). C, the cell adhesion assay was carried out as described above (Fig. 3A). The bars represent the S.D. D, the cell adhesion kinetics assay using the RT-CES system was the same as that described in Fig. 3B. Subconfluent cells were detached, and 40,000 cells were added to the wells. The cell index reflects the extent of cell adhesion. The bars represent the S.D. E, the S-3,5 and S-4,5 expression vectors were transfected into HeLa cells using the Phoenix retrovirus system and selected with puromycin as described under “Experimental Procedures.” The cells were lysed, and the cell lysates (2 mg) were then subjected to immunoprecipitation using an agarose-conjugated antibody against GFP. The immunoprecipitates were subjected to 6.0% SDS-PAGE under non-reducing conditions, blotted, probed with E4-PHA (upper panel), and then reprobed with antibody against the α5 subunit (lower panel). IB, immunoblot; LB, lectin blot.
To confirm whether the site-specific modification also happens in endogenous conditions, we introduced the S-3,5 and S-4,5 mutants into HeLa cells that express a relatively higher level of endogenous GnT-III to examine the products of GnT-III as confirmed by E4-PHA lectin blot. Consistent with the results of overexpressing GnT-III, the E4-PHA lectin staining was clearly detected in S-4,5 but not in S-3,5 α5 subunit transfectants (Fig. 4E). Taken together, these results strongly suggest that GnT-III may specifically modify site-4 on the α5 subunit, which down-regulates its biological functions.
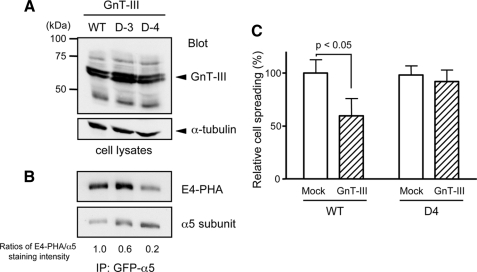
To further elucidate the importance of site-4 for GnT-III modification, we compared the E4-PHA staining patterns of WT with single mutants, such as site-3 (D-3) or site-4 (D-4), as shown in Fig. 1A. These three expression plasmids were co-transfected with GnT-III, and the α5 integrins were immunoprecipitated using an anti-GFP antibody. As shown in Fig. 5A, there were no significant differences in the GnT-III expression levels among the three transfectants. The intensity of E4-PHA staining in D-3 cells was less than that in WT cells, but they were comparable. However, the intensity of E4-PHA staining in D-4 cells was substantially less than that in WT or D-3 cells (Fig. 5B). The ratios of E4-PHA staining versus total α5 staining purified from WT, D-3, and D-4 cells were 1.0, 0.6, and 0.2, respectively. Furthermore to directly examine the effects of GnT-III on site-4 for cell adhesion, we compared cell spreading of the D-4 mutant with WT of α5 integrin. As expected, GnT-III significantly down-regulated cell spreading on FN in WT transfectants, whereas the deletion of site-4 abolished the suppression of cell spread induced by GnT-III in D-4 transfectants (Fig. 5C). Taken together, these results clearly show that N-glycosylation of site-4 is critical and effective for GnT-III modification.
FIGURE 5.
Comparison of GnT-III modification among wild-type (WT), site-3 (D-3) and site-4 (D-4) deletion mutants. GnT-III was expressed in WT and D-3 and D-4 deletion mutants, and stable expression cells were established as described under “Experimental Procedures.” The expression levels of the GnT-III and GnT-III products in total cell lysates were detected with antibodies against GnT-III (A, upper panel) and α-tubulin to ensure equal loading (A, lower panel), respectively. B, the cell lysates were subjected to immunoprecipitation (IP) using an agarose-conjugated antibody against GFP. The immunoprecipitates were separated by 6.0% SDS-PAGE under non-reducing conditions. The membrane blot was probed with E4-PHA lectin and then was reprobed with an antibody against the α5 subunit. The ratio of E4-PHA to total α5 staining in WT cells was set equal to 1.0. C, the percentages of spread cells were quantified and expressed as the mean ± S.D. from three independent experiments. The bars represent S.D. The rounded cells were not considered as spread cells. The ratio of spread cells versus total cells (∼300 cells) of WT transfectants was set as 100.
DISCUSSION
In the present study, we intensively investigated the effects of N-glycosylation on the β-propeller of the integrin α5 subunit on its biological functions such as cell adhesion and cell migration and found that site-4 is essential and effective for GnT-III modification among 14 potential N-glycosylation sites. To our knowledge, this is the first report to clearly demonstrate that a glycosyltransferase of N-glycosylation can specifically modify an N-glycosylation site among multiple potential sites and effectively regulate its biological functions.
Integrins can be activated by inside-out signaling mechanisms that trigger global conformational changes, which ultimately modulate integrin-ligand affinity. It is apparent that integrin activity can be regulated by other mechanisms, such as posttranscriptional modification, N-glycosylation. Altered integrin glycosylation has been associated with tumorigenesis, autoimmune disease, chronic inflammation, and cell adhesion events (11, 30). In particular, N-glycosylation of the integrin α5 and β1 subunits appears to be important for both structure and function. It has been reported that N-glycosylation of both the α5 and β1 subunits is necessary for α5β1 heterodimerization and its binding to FN. Moreover changes in integrin glycan composition, resulting from forced expression of selected glycosyltransferases, i.e. “remodeling,” reportedly modulate integrin functions as described above. However, most of these earlier studies examined only total changes without individual information. Therefore, the exact molecular mechanisms by which N-glycosylation of site(s) or glycan(s) occurs remain unknown. Recently we used site-directed mutagenesis to determine that N-glycosylation site-5 on the β-propeller plays an important role in the assembly of the integrin for its expression on the cell surface (24). These observations prompted us to determine whether there are specific N-glycosylation sites that regulate its biological functions. Here we clearly showed that site-4 is a key N-glycosylation site for the biological function of α5 subunit that is effectively modified by GnT-III. Taken together these results indicate that individual N-glycosylation sites may have unique functions.
Although the molecular mechanism by which bisecting GlcNAc is introduced into site-4, inhibiting its biological function, remain unknown, we speculate that the effect of altered glycosylation of site-4 may be related to conformational changes in the key functional regions of the β-propeller domain of the α5 subunit that are critical for integrin activation. In fact, the β-propeller domain has been postulated to be required for effective interaction between α5β1 integrin and its ligand (31). In contrast, the crystal structure of integrin αVβ3 has been successfully determined, and the main contact between the αV and β3 subunits is the β-propeller on the α and A domain on β3 with hydrophobic, ionic, and mixed contacts (32, 33). Because the α5 subunit has 47% homology to αV, Mould et al. (34) made a homologous modeling structure of α5β1. Based on the model, the α5 subunit seems to be surrounded by N-glycans, explaining the dissociation of the αβ heterodimer that occurs when α5β1 is deglycosylated by treatment with peptide-N-glycosidase F or removal of N-glycans on the β-propeller. Very recently, Liu et al. (35) used a molecular modeling approach to study the effects of altered glycosylation on the I-like domain of the β1 subunit, which is the partner of the β-propeller of the α subunit. These researchers found that α2,6 sialic acid affected the interactions between N-glycans and the I-like domain, which in turn altered the accessibility of the loop that determines specificity of ligand binding. In fact, the remodeling of N-glycans by GnT-III affects either the branching formations catalyzed by GnT-V and GnT-IV or the sialylation on the terminus of the N-glycans (11, 36). Therefore, a possible mechanism by which N-glycans are involved in the αβ interaction or conformational arrangement is that an unknown lectin domain may exist on the α or β subunit. The lectin domain of αMβ2 integrin is associated with GlcNAc on the non-reducing terminus of sugar chains on platelets, facilitating their phagocytosis (37, 38). These studies further support the observation that modification of bisecting GlcNAc on site-4 of the β-propeller may be critical for the regulation of its biological functions, which may shed light on the structural studies.
It is of interest to understand why GnT-III specifically and effectively modifies site-4 of the 14 putative N-glycosylation sites. There is currently no detailed information available regarding this observation, but several explanations have been proposed. First, N-glycosylation occurs on site-4 because it provides the easiest access for GnT-III. Because the integrin α5 crystal structure is currently unavailable this hypothesis cannot be proven. Second, GnT-III may associate with some other molecules, which define the specificities for protein or peptide substrates. Reportedly protein O-mannosyltransferase 1 (POMT1) and its homolog POMT2 are responsible for catalyzing the first step in O-mannosyl glycan, which is important for muscle and brain development (39). Interestingly Manya et al. (40) reported that formation of a POMT1-POMT2 complex is essential for POMT activity. Only two peptides derived from the mucin domain of α-dystroglycan are highly O-mannosylated by POMT, but no O-mannosylation occurs in mucin tandem repeat peptides (41). Similarly complex formation is also important for T-synthase (core 1 β1,3-galactosyl transferase) activity. Ju et al. (42, 43) reported that Tn syndrome, a rare autoimmune disease, in which subpopulations of blood cells of all lineages carry an incompletely glycosylated membrane glycoprotein, known as the Tn antigen, is associated with a somatic mutation in Cosmc, a gene on the X chromosome that encodes a molecular “chaperone” that is required for the proper folding and hence full activity of T-synthase. Indeed it has been reported that caveolin-1 may co-localize with GnT-III to regulate its localization and activity (44). Those results, taken together, suggest that glycosyltransferase complex formation may play a crucial role in determination of both activity and substrate specificity. The detailed molecular mechanism requires further study.
This study specifically focused on N-glycosylation of the integrin α5 subunit. To fully understand the effects of the N-glycans on integrin structure and function, it will be necessary, in future studies, to investigate the interaction of glycans with glycans or peptides of integrin. The current study also has implications for engineering α5 that contains the glycans necessary for its activation that may facilitate the study of its crystal structure.
This work was supported in part by Core Research for Evolutional Science and Technology; the Japan Science and Technology Agency; the “Academic Frontier” Project for Private Universities from the Ministry of Education, Culture, Sports, Science and Technology of Japan; the core to core program (Japan Society for the Promotion of Science); and Takeda Science Foundation, Japan.
Footnotes
The abbreviations used are: FN, fibronectin; BSA, bovine serum albumin; E4-PHA, erythroagglutinating phytohemagglutinin; GFP, green fluorescent protein; GlcNAc, N-acetylglucosamine; GnT-III, N-acetylglucosaminyltransferase III; GnT-V, N-acetylglucosaminyltransferase V; L4-PHA, leukoagglutinating phytohemagglutinin; CHO, Chinese hamster ovary; DMEM, Dulbecco's modified Eagle's medium; RT-CES, real time cell electronic sensing; WT, wild type; POMT, protein O-mannosyltransferase.
References
- 1.Apweiler, R., Hermjakob, H., and Sharon, N. (1999) Biochim. Biophys. Acta 1473 4–8 [DOI] [PubMed] [Google Scholar]
- 2.Dwek, R. A. (1995) Biochem. Soc. Trans. 23 1–25 [DOI] [PubMed] [Google Scholar]
- 3.Saxon, E., and Bertozzi, C. R. (2001) Annu. Rev. Cell Dev. Biol. 17 1–23 [DOI] [PubMed] [Google Scholar]
- 4.Hynes, R. O. (2002) Cell 110 673–687 [DOI] [PubMed] [Google Scholar]
- 5.George, E. L., Georges-Labouesse, E. N., Patel-King, R. S., Rayburn, H., and Hynes, R. O. (1993) Development 119 1079–1091 [DOI] [PubMed] [Google Scholar]
- 6.Goh, K. L., Yang, J. T., and Hynes, R. O. (1997) Development 124 4309–4319 [DOI] [PubMed] [Google Scholar]
- 7.Watt, F. M., and Hodivala, K. J. (1994) Curr. Biol. 4 270–272 [DOI] [PubMed] [Google Scholar]
- 8.Yang, J. T., Rayburn, H., and Hynes, R. O. (1993) Development 119 1093–1105 [DOI] [PubMed] [Google Scholar]
- 9.Akiyama, S. K., Yamada, S. S., and Yamada, K. M. (1989) J. Biol. Chem. 264 18011–18018 [PubMed] [Google Scholar]
- 10.Zheng, M., Fang, H., and Hakomori, S. (1994) J. Biol. Chem. 269 12325–12331 [PubMed] [Google Scholar]
- 11.Gu, J., and Taniguchi, N. (2004) Glycoconj. J. 21 9–15 [DOI] [PubMed] [Google Scholar]
- 12.Isaji, T., Gu, J., Nishiuchi, R., Zhao, Y., Takahashi, M., Miyoshi, E., Honke, K., Sekiguchi, K., and Taniguchi, N. (2004) J. Biol. Chem. 279 19747–19754 [DOI] [PubMed] [Google Scholar]
- 13.Zhao, Y., Nakagawa, T., Itoh, S., Inamori, K., Isaji, T., Kariya, Y., Kondo, A., Miyoshi, E., Miyazaki, K., Kawasaki, N., Taniguchi, N., and Gu, J. (2006) J. Biol. Chem. 281 32122–32130 [DOI] [PubMed] [Google Scholar]
- 14.Guo, H. B., Lee, I., Kamar, M., Akiyama, S. K., and Pierce, M. (2002) Cancer Res. 62 6837–6845 [PubMed] [Google Scholar]
- 15.Partridge, E. A., Le Roy, C., Di Guglielmo, G. M., Pawling, J., Cheung, P., Granovsky, M., Nabi, I. R., Wrana, J. L., and Dennis, J. W. (2004) Science 306 120–124 [DOI] [PubMed] [Google Scholar]
- 16.Seales, E. C., Jurado, G. A., Brunson, B. A., Wakefield, J. K., Frost, A. R., and Bellis, S. L. (2005) Cancer Res. 65 4645–4652 [DOI] [PubMed] [Google Scholar]
- 17.Shaikh, F. M., Seales, E. C., Clem, W. C., Hennessy, K. M., Zhuo, Y., and Bellis, S. L. (2008) Exp. Cell Res. 314 2941–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seales, E. C., Jurado, G. A., Singhal, A., and Bellis, S. L. (2003) Oncogene 22 7137–7145 [DOI] [PubMed] [Google Scholar]
- 19.Semel, A. C., Seales, E. C., Singhal, A., Eklund, E. A., Colley, K. J., and Bellis, S. L. (2002) J. Biol. Chem. 277 32830–32836 [DOI] [PubMed] [Google Scholar]
- 20.Pretzlaff, R. K., Xue, V. W., and Rowin, M. E. (2000) Cell Adhes. Commun. 7 491–500 [DOI] [PubMed] [Google Scholar]
- 21.Kawano, T., Takasaki, S., Tao, T. W., and Kobata, A. (1993) Int. J. Cancer 53 91–96 [DOI] [PubMed] [Google Scholar]
- 22.Dennis, J., Waller, C., Timpl, R., and Schirrmacher, V. (1982) Nature 300 274–276 [DOI] [PubMed] [Google Scholar]
- 23.Nadanaka, S., Sato, C., Kitajima, K., Katagiri, K., Irie, S., and Yamagata, T. (2001) J. Biol. Chem. 276 33657–33664 [DOI] [PubMed] [Google Scholar]
- 24.Isaji, T., Sato, Y., Zhao, Y., Miyoshi, E., Wada, Y., Taniguchi, N., and Gu, J. (2006) J. Biol. Chem. 281 33258–33267 [DOI] [PubMed] [Google Scholar]
- 25.Schreiner, C. L., Bauer, J. S., Danilov, Y. N., Hussein, S., Sczekan, M. M., and Juliano, R. L. (1989) J. Cell Biol. 109 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solly, K., Wang, X., Xu, X., Strulovici, B., and Zheng, W. (2004) Assay Drug Dev. Technol. 2 363–372 [DOI] [PubMed] [Google Scholar]
- 27.Cummings, R., and Kornfeld, S. (1982) J. Biol. Chem. 257 11230–11234 [PubMed] [Google Scholar]
- 28.Yamashita, K., Hitoi, A., and Kobata, A. (1983) J. Biol. Chem. 258 14753–14755 [PubMed] [Google Scholar]
- 29.Yamashita, K., Totani, K., Ohkura, T., Takasaki, S., Goldstein, I. J., and Kobata, A. (1987) J. Biol. Chem. 262 1602–1607 [PubMed] [Google Scholar]
- 30.Bellis, S. L. (2004) Biochim. Biophys. Acta 1663 52–60 [DOI] [PubMed] [Google Scholar]
- 31.Mould, A. P., Askari, J. A., and Humphries, M. J. (2000) J. Biol. Chem. 275 20324–20336 [DOI] [PubMed] [Google Scholar]
- 32.Xiong, J. P., Stehle, T., Zhang, R., Joachimiak, A., Frech, M., Goodman, S. L., and Arnaout, M. A. (2002) Science 296 151–155 [DOI] [PubMed] [Google Scholar]
- 33.Xiong, J. P., Stehle, T., Diefenbach, B., Zhang, R., Dunker, R., Scott, D. L., Joachimiak, A., Goodman, S. L., and Arnaout, M. A. (2001) Science 294 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mould, A. P., Symonds, E. J., Buckley, P. A., Grossmann, J. G., McEwan, P. A., Barton, S. J., Askari, J. A., Craig, S. E., Bella, J., and Humphries, M. J. (2003) J. Biol. Chem. 278 39993–39999 [DOI] [PubMed] [Google Scholar]
- 35.Liu, Y., Pan, D., Bellis, S. L., and Song, Y. (2008) Proteins 73 989–1000 [DOI] [PubMed] [Google Scholar]
- 36.Koyota, S., Ikeda, Y., Miyagawa, S., Ihara, H., Koma, M., Honke, K., Shirakura, R., and Taniguchi, N. (2001) J. Biol. Chem. 276 32867–32874 [DOI] [PubMed] [Google Scholar]
- 37.Hoffmeister, K. M., Josefsson, E. C., Isaac, N. A., Clausen, H., Hartwig, J. H., and Stossel, T. P. (2003) Science 301 1531–1534 [DOI] [PubMed] [Google Scholar]
- 38.Josefsson, E. C., Gebhard, H. H., Stossel, T. P., Hartwig, J. H., and Hoffmeister, K. M. (2005) J. Biol. Chem. 280 18025–18032 [DOI] [PubMed] [Google Scholar]
- 39.Endo, T., and Manya, H. (2006) Methods Enzymol. 417 137–152 [DOI] [PubMed] [Google Scholar]
- 40.Manya, H., Chiba, A., Yoshida, A., Wang, X., Chiba, Y., Jigami, Y., Margolis, R. U., and Endo, T. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 500–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manya, H., Suzuki, T., Akasaka-Manya, K., Ishida, H. K., Mizuno, M., Suzuki, Y., Inazu, T., Dohmae, N., and Endo, T. (2007) J. Biol. Chem. 282 20200–20206 [DOI] [PubMed] [Google Scholar]
- 42.Ju, T., and Cummings, R. D. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 16613–16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ju, T., and Cummings, R. D. (2005) Nature 437 1252. [DOI] [PubMed] [Google Scholar]
- 44.Sasai, K., Ikeda, Y., Ihara, H., Honke, K., and Taniguchi, N. (2003) J. Biol. Chem. 278 25295–25301 [DOI] [PubMed] [Google Scholar]