Abstract
SLAT (SWAP-70-like adaptor protein of T cells) is an adaptor protein expressed in cells of the hematopoietic system. SLAT interacts with and alters the function of small GTPase Rac1 in fibroblasts. In these nonhematopoietic models, the SLAT-Rac interaction leads to changes in F-actin and causes cytoskeletal reorganization. In T cells, SLAT expression regulates the development of T helper cells through Cdc42- and Rac1-mediated activation of the NF-AT transcription factor. Here we show that SLAT is expressed in macrophages. Overexpression of SLAT in a macrophage cell line inhibits the IgG Fcγ receptor-mediated phagocytic ability of THP1 cells. In bone marrow-derived macrophages, SLAT protein is recruited to the early phagosomes formed via Fcγ receptor engagement. SLAT recruitment to the phagosome was most efficient when the macrophages express at least one isoform of Rac (Rac1 or Rac2), because SLAT recruitment was reduced in macrophages of Rac-deficient mice. Macrophages derived from animals lacking SLAT show an elevation in the rate of Fcγ receptor-mediated phagocytosis. The absence of SLAT is associated with an increase in the amount of F-actin formed around these phagosomes as well as an increase in the amount of Rac1 protein recruited to the phagosome. Our results suggest that SLAT acts as a gatekeeper for the amount of Rac recruited to the phagosomes formed by Fcγ receptor engagement and thus is able to regulate F-actin re-organization and consequently phagocytosis.
Innate immune cells phagocytose IgG-opsonized particles through the immunoglobulin Fc receptors (FcγRs).2 The FcγR-bound particle is engulfed by filamentous actin (F-actin)-mediated extension of plasma membrane leading to internalization of the particle in a membrane-bound vesicle. Phagosomes, the vesicles containing the internalized particle, begin as a phagocytic cup, which is an actin-rich structure containing numerous actin remodeling proteins (1, 2). The phagosome subsequently matures by fusion with primary lysosomes containing hydrolytic enzymes (3). Digestion of the particle and its associated protein components leads to processing for subsequent antigen presentation on the cell surface (reviewed in Ref. 4).
Besides phagocytosis, FcγR stimulation induces other biological events in innate immune cells. FcγR stimulation triggers inflammatory cytokine mRNA and protein by the activation of transcription factors (5, 6). Inflammatory cytokine production also requires activation of the p38 module of mitogen-activated protein kinases, which promote mRNA stability (7). In neutrophils, FcγR activation also stimulates the production of reactive oxygen and nitrogen species through the activation of Vav3 (8) and the small GTPase Rac2 (9).
There are four distinct murine FcγR proteins as follows: FcγRI, FcγRII, FcγRIII, and FcγRIV (reviewed in Ref. 10). These four receptors are divided into two categories, depending on whether the receptor promotes or inhibits innate immune cell activation. The activating or the signal-promoting receptors include FcγRI, FcγRIII, and FcγRIV, all of which associate with a low molecular weight γ chain. The γ chain contains a sequence of amino acids characterized by dual YXX(I/L) motifs and termed the immunoreceptor tyrosine-based activation motif (ITAM) (11). The inhibitory receptor FcγRII does not associate with the γ chain, but it contains a single YXX(I/L) that functions to inhibit cell activation by recruiting phosphatases (11).
Upon phosphorylation of the tyrosine residues within the ITAM motif in the FcR-associated γ chain, the γ chain becomes a docking site for a variety of Src homology 2 domain-containing proteins. The ITAM tyrosines are phosphorylated by one of several members of the Src family of protein-tyrosine kinases (12). Then the Src homology 2 domain-containing protein-tyrosine kinases Syk and phosphatidylinositol 3-kinase (13) bind to the phosphorylated ITAMs. Both Syk (14) and phosphatidylinositol 3-kinase (15) are essential for FcγR-mediated phagocytosis. Besides these initial kinases, the phosphorylated ITAM recruits adaptor proteins (16–18) to activate other downstream effectors. Among these effectors are the guanine nucleotide exchange factors (GEFs), which activate the members of Rho GTPase family. Recent experiments indicate a role for a complex containing CrkII-DOCK180 as the Rac GEF and essential for phagocytosis (19, 20).
Rho GTPases, especially Rac and Cdc42, are established players in various F-actin-mediated cytoskeletal rearrangements, including the phagocytic cup formation, although their precise role is unclear. Activated (GTP-loaded) Cdc42 appears at actinrich sites within the phagocytic cup, whereas activated Rac1 and -2 appear uniformly around the cup, particularly during cup closure (2). Activated Cdc42 recruits Wiskott-Aldrich syndrome protein (21), a protein recently shown to be essential for FcγR-dependent phagocytic cup formation (22). Dominant-negative mutants of Rac block FcγR-dependent phagocytosis (23), and membrane-targeted Rac is sufficient to induce phagocytosis of cell-bound latex beads, bypassing the γ chain and FcγR signaling (24). Macrophages of Rac1/2–/– mice show defects in FcγR-mediated phagocytosis but are still able to form the initial phagocytic cup (25). Thus Rac is required for the late term actin rearrangements needed for FcγR-induced particle internalization but not for the early phase of FcγR-induced cup formation. Despite these findings, the molecular contribution of activated Rac is not understood. Rac (26, 27) and Rac1 in particular (28) is clearly required for other F-actin rearrangements such as chemotactic migration toward formylated peptides like fMLP.
SLAT (SWAP-70-like adaptor in T cells; also known as IBP and Def6) was originally identified in a screen of a myeloid cell line FDCP-Mix A4 for genes differentially expressed during myelopoiesis (29). SLAT/IBP/Def6 (hereafter referred to as SLAT) was found to be highly expressed in interleukin-4-secreting T cells of the Th2 phenotype (30). Structurally, SLAT has an EF hand at the N terminus, a central pleckstrin homology domain, and a Dbl homology-like domain at its C terminus. The pleckstrin homology domain of SLAT displayed affinity for the phosphatidylinositol 3-kinase product, phosphatidylinositol 3,4,5-trisphosphate, after SLAT became tyrosine-phosphorylated (31). SLAT appeared in the T cell synapse area of contact with antigen-presenting cells (30) and serves to activate the transcription factor NF-AT in a Cdc42- and/or Rac1-dependent manner (32). SLAT is a member of a structurally novel set of Rho family GTPase GEFs that includes SWAP-70 itself (33). Indeed, transfection and overexpression of SLAT in COS-7 fibroblasts were able to induce Rac, Cdc42, and RhoA activation, indicated by increased GTP binding of these small GTPases (31, 34). These findings suggest that SLAT acts upstream of Rac, Rho, and/or Cdc42. However, others show that SLAT was able to bind activated mutants of Rac, Cdc42, and RhoA (35) suggesting that SLAT acts distal to Rho GTPase family members.
SLAT has thus far been linked to development and activation of T cells (30), although it was originally found in a screen of a myeloid cell line (29). SLAT has not been reported to play a role in macrophage biology. This situation prompted us to study SLAT function in FcR-mediated phagocytosis, which involves F-actin reorganization and is mediated by Rho GTPases. Here we show that SLAT is expressed in murine peritoneal and bone marrow-derived macrophages (BMM). Overexpression of SLAT in a human macrophage cell line caused a dramatic decrease in FcγR-mediated phagocytosis, suggesting that SLAT acts as a negative regulator of phagocytosis. In wild-type BMM, SLAT was recruited to the early phagosomes upon FcγR engagement. BMM from SLAT-deficient animals showed elevated phagocytosis relative to phagocytosis by wild-type-derived control macrophages. Furthermore, SLAT-deficient BMM showed increased levels of F-actin and increased Rac1 protein recruitment to the early phagosomes. SLAT recruitment to the phagosomes was partially reduced in macrophages of Rac1–/– or Rac2–/– animals and greatly reduced in macrophages of Rac1/2–/– double-deficient animals. Based on these findings, we hypothesize that SLAT modulates the amount of Rac recruited to the phagosomes, which in turn regulates the amount of F-actin reorganization. Consistent with this hypothesis, we found the SLAT-deficient macrophages showed increased migration toward formylated peptide (fMLP), an event that requires Rac and F-actin reorganization (26–28).
MATERIALS AND METHODS
Antibodies, Reagents, and Vector Constructs—SLAT rabbit antisera was described previously (30). Anti-GAPDH antibody, normal rabbit serum, normal mouse IgG, and normal rabbit IgG was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Rac1 mouse monoclonal antibody was purchased from Upstate Biotechnology, Inc. (Billerica, MA). Cy5-labeled anti-rabbit Ig and Cy5-labeled anti-mouse secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA). Alexafluor 660- and Alexafluor 488-conjugated phalloidin and prolong anti-fade solutions were purchased from Molecular Probes (Carlsbad, CA). PKH26 dye, rabbit anti sheep sera, fMLP and polystyrene latex beads were bought from Sigma. Sheep red blood cells were obtained from Colorado Serum Co. (Denver, CO). Recombinant mouse macrophage colony-stimulating factor (M-CSF) was obtained from R & D Systems (Minneapolis, MN). Transwell migration assay plates were obtained from Costar (Corning, NY).
Mig vector and Mig vector containing full-length SLAT were used for overexpression experiments in THP1 cells, as described previously (36). Phagocytosis measurements were done after sorting GFP-positive transfected cells.
Mice—SLAT–/– mice were mated as heterozygous animals and are on C57BL6 background (37). Wild-type mice with C57BL6 background were either bought from The Jackson Laboratory or were littermates of the heterozygous SLAT–/– breeders. Bones were from conditional Rac1–/– (28), Rac2–/– (38), and Rac1,2–/– (28) mice and their wild-type littermate controls, all on the C57Bl/6 background. The Rac deficiency is present only within the myeloid compartment, because of Cre expression driven by the LysM promoter (39).
Cell Culture—Bone marrow was flushed from 4- to 7-week-old wild-type or SLAT knock-out mice tibias and femurs and cultured in M-CSF containing RPMI 1640 media supplemented with 10% fetal bovine serum and penicillin/streptomycin/glutamine for 7–10 days. BMM were analyzed daily for the presence of cell surface markers by flow cytometry for the presence of mature macrophages in the culture. Cells in culture from day 7 onwards expressed mature macrophage markers F4/80, Mac-1, as well as Fc receptors. BMM from day 7 onwards were used for experiments and were cytokine- and serum-starved for at least 10 h prior to the experiment to reduce the biochemical events associated with signal transduction to resting levels.
Western Blots and PCR—Cell lysates were prepared as described (13), run on 12% SDS/PAGE, and transferred to nitrocellulose filters. The filters were blocked with 2.5% dry milk in Tris-buffered saline containing 0.1% Tween 20 (TBS/Tween), washed in TBS/Tween, and probed with various antibodies described in the text. The filters were washed with TBS/Tween and probed with horseradish peroxidase-conjugated secondary antibodies and developed and quantitated using ImageJ software as described (13).
PCR for SLAT used forward and reverse primers 5′-GGGCTCGAGATGGCCCTGCGC-3′ and 5′-GGGCCTAGGCTAATTCCCTGG-3′, respectively, and 30 cycles of 94 °C for 1 min, 55 °C for 2 min, and 72 °C for 2 min. GAPDH used forward and reverse primers 5′-AGTATGACTCCACTCACGGCAA-3′ and 5′-TCTCGCTCCTGGAAGATGGT-3′, respectively, and 35 cycles of 94 °C for 30 s, 58 °C for 45 s, and 72 °C for 45 s. PCR products were analyzed on 1% agarose and visualized by ethidium bromide.
Phagocytosis Assay—THP1 macrophage cell line or BMM from either wild-type or SLAT–/– mice were allowed to phagocytose sheep red blood cells (SRBCs) labeled with PKH26 dye and opsonized with rabbit antisera as described previously (13). Uninternalized SRBCs were lysed with NH4Cl buffer (13). Internalized SRBCs were counted under a fluorescent microscope to calculate percent phagocytic cells as the number of cells taking up at least one SRBC and phagocytic index as the total number of SRBCs per 100 cells, as described (13).
Confocal Microscopy—BMM from wild-type or the various knock-out strains of mice were grown on glass coverslips in 6-well plates overnight. After cytokine and serum starvation, the cells on the coverslips were washed with Hanks' balanced salt solution (HBSS) and allowed to phagocytose human IgG-coated, 3-μm diameter latex beads. Following phagocytosis, the coverslips were washed with HBSS to remove the uninternalized beads and immediately immersed in 2% methanol-free formaldehyde for 10 min at room temperature to fix the cells. Fixative was removed, and coverslips were washed with HBSS containing 3% fetal bovine serum. Washed coverslips were then immersed in saponin-containing permeabilization buffer for 10 min at room temperature. Primary antibody against the targeted protein as well as phalloidin were diluted in permeabilization buffer and added to the cells on coverslips in a fresh 6-well plate and incubated overnight at 4 °C. Following overnight incubation the coverslips were washed three times in permeabilization buffer and immersed for 1 h at 4 °C in fresh permeabilization buffer containing fluorescently conjugated secondary antibodies. After five washes the coverslips were mounted on to a glass slide using mounting media. Microscopy was done using ×100 objective lens for all the experiments.
Rac Assay—We used the procedure that applies GST-p21-activated kinase as described earlier (40). Briefly, cells were stimulated and lysed as above, and the lysates were incubated with GST-PAK prebound to glutathione-agarose beads. After washing, the beads were incubated with Laemmli SDS-PAGE sample buffer, and the eluted proteins were subjected to SDS-PAGE. After the transfer of proteins to a nitrocellulose membrane, Western blotting was performed using a monoclonal antibody to Rac1. The amount of GTP-loaded Rac1 was quantitated by measuring the mass of GTP-Rac1 in comparison to the mass of total Rac1. The value was then normalized to the GTP-Rac1/total Rac1 ratio of unstimulated cells.
Chemotaxis Assay—BMM from wild-type, SLAT–/–, or Rac–/– mice were placed in the upper chamber of transwell-permeable support system with 5-μm diameter pore size polycarbonate membrane inserts in a 24-well plate. The lower chamber/well contained either fMLP or M-CSF. The cells were allowed to migrate toward the chemokines for up to 60 min. The inserts were then taken out and immersed in fixative. After washing off the fixative, the cells were stained with Giemsa, and the number of cells having migrated toward the chemokines were counted under a light microscope. Data were represented as the average number of cells from five independent fields. The average number of cells from seven separate experiments are reported as an overall average and standard error.
Software—Confocal images were quantified using ipLab software (iVision from Biovision, Exton, PA) from Scanalytics, Inc. For quantification of confocal images, the area surrounding the phagosome was digitally magnified, and the pixels were selected to obtain the mean fluorescence intensity. Average mean fluorescence intensity of at least 20 phagosomes was obtained for each time point and for each genotype. The average fluorescence intensity of 20 phagosomes and standard error is shown in the figures. Band intensity quantification for Western blots was done using ImageJ 1.38X software (National Institutes of Health). Statistical analysis was done using Microsoft Excel software. The significance was determined with the help of Student's t test. p values <0.05 were considered as statistically significant.
RESULTS
SLAT was originally identified in a myeloid cell line, but most of the studies on SLAT to date have been done in lymphocytes. We first tested macrophages for expression of SLAT. For this experiment, we prepared macrophages from the bone marrow of C57Bl/6 animals by culturing whole marrow cells in media containing M-CSF. After 7 days the BMM expanded ∼10-fold and expressed Mac-1 (90–95%), F4/80 (90–95%), and FcγRII/III (75–80%) indicating the M-CSF-responsive population was largely macrophages. Additionally, mature ex vivo macrophages were collected by peritoneal lavage of wild-type mice. We prepared whole cell lysates by detergent extraction of the peritoneal and BMM macrophages. We also prepared detergent extracts of cell lines RAW 264.7 (mouse macrophage), THP1 (human macrophages), and Jurkat (human T cells). We used extracts of human embryonic kidney cells (HEK) as a negative control. The lysates were separated by SDS-PAGE, transferred to nitrocellulose filters, and probed with antibodies to SLAT and to GAPDH as a loading control. We found (Fig. 1A) that SLAT was expressed in all the hematopoietic cells (peritoneal macrophages, BMM, RAW 264.7, THP1, and Jurkat) but was absent in the HEK cell line. The minor band in HEK cells did not migrate with the SLAT protein, but to ensure its absence we analyzed cDNA by PCR from BMM, RAW cells, and HEK. We used the SLAT plasmid as a positive control. We found (Fig. 1B), like the Western blot result in Fig. 1A, that HEK lacked mRNA encoding SLAT. To quantitate the protein expression level, we determined the ratio of SLAT to GAPDH proteins (Fig. 1C) and found that BMM had approximately the same level of expression as the human T cell line Jurkat. We conclude that BMM and macrophage cell lines express SLAT at a level similar to T cells.
FIGURE 1.
SLAT is expressed in murine macrophages. A, SLAT protein expression in whole cell lysate of 25,000 cells of each cell type: peritoneal macrophages, bone marrow-derived macrophages, murine macrophage cell line RAW 264.1, human monocytic cell line THP1, human embryonic kidney cells, and human T cell line Jurkat. The immunoblot was stripped of anti-SLAT sera and re-probed with anti-GAPDH antibody. B, cDNA from HEK, BMM, and RAW 264 cells were analyzed for SLAT and GAPDH by 30 or 35 cycles of PCR as described under “Materials and Methods.” C, quantification of SLAT protein expression with respect to GAPDH expression in a given cell type. Data represented as the ratio of band intensity of SLAT to band intensity of GAPDH.
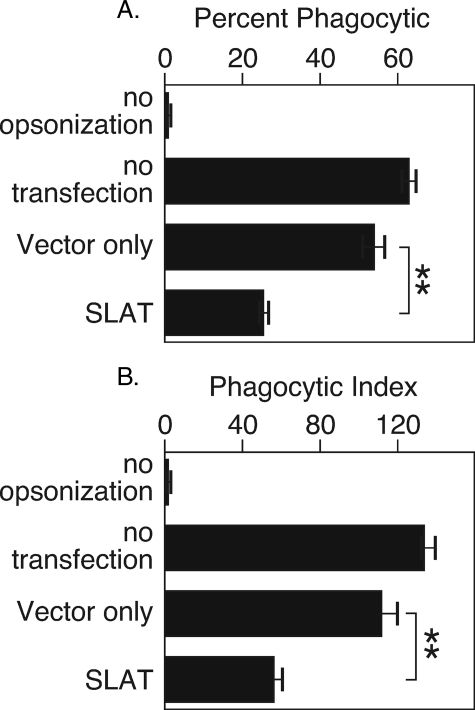
Macrophage function requires actin reorganization, and studies in fibroblasts suggested that SLAT regulates actin reorganization through Rac. Therefore, as an initial test of the function of SLAT, we transfected the human macrophage cell line THP1 with vector or with cDNA encoding SLAT. The vector encodes a bicistronic message that includes GFP; thus, the vector- or SLAT-transfected macrophages were sorted to collect the GFP-expressing populations. The cells are 100% viable at the end of the sort, and we observed no change in a propensity toward apoptosis when SLAT is overexpressed. The GFP+ fraction of transfectants was tested for phagocytic ability by incubation with IgG-opsonized sheep red blood cells, which stimulates all IgG receptors. We measured the percent of macrophages that took up at least one RBC (percent phagocytic) and the number of RBCs per 100 macrophages (phagocytic index). Controls were macrophages exposed to unopsonized RBCs and untransfected macrophages incubated with IgG-opsonized RBCs. We found that macrophages overexpressing SLAT had greatly reduced phagocytic ability as measured by percent phagocytic (Fig. 2A) or as phagocytic index (Fig. 2B). These findings suggest that SLAT acts to inhibit phagocytic function.
FIGURE 2.
SLAT overexpression in human monocytic cell line THP1 reduces phagocytosis. THP1 monocytes were transduced with SLAT or vector only and sorted to isolate the transduced cells. The SLAT- or vector-transduced THP1 cells were incubated with mouse anti-sheep RBC-opsonized sheep RBCs. The THP1 cells were analyzed for their phagocytic ability as described under “Materials and Methods.” A, percent phagocytic; for p values, ** is <0.05. B, phagocytic index; for p values, ** is <0.05. Data represent the average of three independent experiments.
We next examined the subcellular location of SLAT in BMM following an IgG-coated phagocytic stimulus. For these experiments, BMM were prepared by culturing with M-CSF for 7 days. The cells were removed from culture and grown overnight with M-CSF on glass coverslips. The attached cells were starved of M-CSF for 10 h and then incubated with latex beads coated with human IgG. The coverslip was washed at various periods of time to remove unbound beads, and the cells were fixed by the addition of formaldehyde. The samples were permeabilized with saponin and stained with antibodies to SLAT or antibodies to GAPDH, normal rabbit serum, or normal rabbit IgG as negative controls. The cells were counter-stained with labeled phalloidin to detect the actin-rich phagosomes. The results are shown in Fig. 3A (isotype, negative controls, and SLAT; each including a phalloidin stain for actin). We found that SLAT appeared at the same location as the actin-rich phagosomes, indicating recruitment of SLAT to the phagosome. To examine the recruitment in more detail, we prepared serial optical sections of a set of phagosomes; the sections are shown in Fig. 3B. The panels are labeled with the optical slice number and range from the bottom of the section (10th slice) to the top of the cell (13th slice). The co-localization of SLAT and F-actin-rich phagosomes is most obvious in the 12th and 13th optical slice and are indicated in Fig. 3B, arrow.
FIGURE 3.
SLAT is recruited to the phagosome in FcγR-stimulated macrophages. A, bone marrow-derived macrophages from wild-type mice were stimulated with human IgG-coated latex beads. The cells were fixed, permeabilized, and stained with antibodies to SLAT or GAPDH as indicated; NRS and normal rabbit IgG (NRIg) were used as negative controls. All cells were stained with phalloidin to detect F-actin. Representative optical slice from a confocal micrograph of BMM after 3 min of ingestion of human IgG-coated latex beads, stained with the indicated antibodies and with phalloidin. B, representative four serial optical slices from a confocal micrograph showing recruitment of SLAT on the phagosome along with F-actin indicated with arrows. C, mean fluorescence intensity of SLAT and F-actin stain around 15 phagosomes. Data represent average from three independent experiments.
To quantitate the amount of fluorescent signal from SLAT and actin at the phagosome, we used digital magnification of a ×100 image, focusing on isolated phagosomes as defined by the F-actin stain. The area containing the phagosome was electronically selected, and the mean fluorescence intensity of the pixels was calculated. The data in Fig. 3C shows the mean and standard error for each fluorescent signal (actin and SLAT, GAPDH, NRS, or normal rabbit IgG) in at least 20 isolated phagosomes. We found that the SLAT signal showed a high degree of co-localization with the F-actin signal at 3 min, and then declined. The amount of F-actin concomitantly declined, as shown previously (41). The signal from GAPDH, NRS, or normal rabbit IgG did not show significant co-localization with F-actin. These findings show that SLAT is recruited to the phagosome upon FcγR stimulation in bone marrow-derived macrophages.
The data in Figs. 2 and 3 suggest SLAT may regulate the phagocytic process. As an additional test of this possibility, we prepared BMM from wild-type and animals lacking SLAT (SLAT–/–). The bone marrow-derived macrophage progenitor cells from SLAT–/– animals responded to M-CSF and developed into macrophages essentially identical to those from wild-type animals (data not shown). Likewise, the normal complement of mature macrophages was found in the periphery of SLAT-deficient mice. Thus, SLAT does not appear to be required for proliferative responses to M-CSF nor does it appear to be required for macrophage development or differentiation. To test for function, BMM from 7-day M-CSF cultures of bone marrow from wild-type and SLAT–/– mice were starved of cytokine for 10 h and then provided IgG-coated RBCs to measure phagocytosis. The macrophages were incubated with the phagocytic stimulus for 5–30 min, and uninternalized RBCs were removed by exposure to ammonium chloride-containing buffer. The internalized RBCs were counted, and the data were expressed as percent phagocytic and phagocytic index. We found (Fig. 4A) that the percentage of macrophages that internalized at least one RBC was not significantly different between the two strains. Thus, macrophages from both strains were capable of phagocytosis. However, SLAT-deficient macrophages showed an increase in the phagocytic index, a measure of phagocytic rate. The rates of phagocytosis were such that SLAT-deficient macrophages from early time points (5 and 10 min) but not late time points (30 min) contained more RBCs than the wild-type counterparts. The increased number of internalized RBCs in the SLAT-deficient macrophages is consistent with the hypothesis mentioned above that SLAT is a negative regulator of FcγR-induced phagocytosis.
FIGURE 4.
Accelerated phagocytosis in macrophages of SLAT–/– mice. Bone marrow-derived macrophages from wild-type (open bars) or SLAT–/– mice (black bars) were prepared and provided IgG-opsonized sheep RBCs. Phagocytosis was quantitated as described under “Materials and Methods.” A, percent phagocytic cells; B, phagocytic index. For p values, * is <0.01 and ** is <0.05. Data represent average of four independent experiments.
To begin to address how SLAT affects macrophage phagocytosis, we examined the actin reorganization in macrophages from wild-type and SLAT–/– animals. Macrophages were prepared on glass coverslips as above and exposed to IgG-coated latex beads. The cells were fixed at various times with formaldehyde, permeabilized, and stained with Alexafluor 660-labeled phalloidin. We found (Fig. 5A) that the amount of F-actin around the phagosome was greatly elevated in the SLAT-deficient macrophages relative to the amount in wild-type macrophages. The increased F-actin was again particularly striking at early but not late time points, and it was coincident with the loss of F-actin around the phagosome that occurs in normal cells as the phagosome is internalized. We quantitated the amount of actin around 15–20 digitally isolated phagosomes as described above. The result is expressed as the mean fluorescent intensity of Alexafluor 660 and the standard error and examining 15–20 separate phagosomes from each condition. The data are shown in Fig. 5B and show a significant (p < 0.001; marked by *) increase in SLAT-deficient macrophages relative to wild-type macrophages after 3 min of incubation with the IgG-coated latex beads. The difference in F-actin amount declined with time and paralleled the decline in difference of phagocytosis shown in Fig. 4. The data indicate that SLAT-deficient macrophages recruit more F-actin to the phagosome, which might account for the elevations in phagocytic rates.
FIGURE 5.
Phagosomes in macrophages of SLAT–/– mice contain higher F-actin amounts. Bone marrow-derived macrophages from the indicated mouse strains were prepared and provided IgG-coated latex beads. The cells were fixed and stained with phalloidin. A shows a single optical slice from a confocal image of a representative field stained for F-actin after 3, 10, and 30 min of incubation with latex beads. Arrows indicate the F-actin formed around the phagocytosed latex beads. B, quantification of pixel intensity for F-actin stain around the phagosomes formed in macrophages from wild-type and SLAT knock-out mice from the experiment shown in A. For p values, * is <0.001. Average mean fluorescence intensity values around the phagosomes were plotted from four independent experiments.
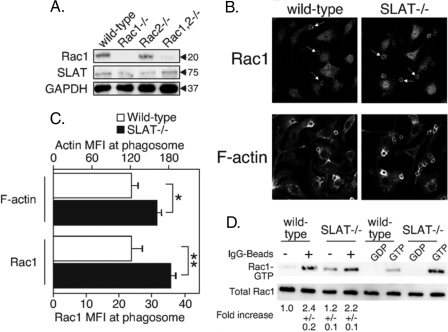
As mentioned in the Introduction, Rac is a small GTPase that is required for Fc receptor-mediated phagocytosis and is recruited to the phagosome following FcγR binding. Studies in fibroblasts suggested that SLAT can affect Rac, but it is unclear if SLAT acts proximal or distal to Rac activation. To address this issue, we stained wild-type and SLAT-deficient macrophages with antibodies to Rac isoforms. Unfortunately, we found that commercial antibodies to Rac2 are not specific and detect several bands in whole cell lysates, whereas Rac1 antibodies are very specific for Rac1. We show in Fig. 6A that antibodies to Rac1 detect the protein in wild-type and Rac2-deficient macrophages, but there is no detectable band in Rac1- or Rac1,2-double-deficient macrophages. SLAT is present in lysates from macrophages of all these mouse strains and at levels equal to that in macrophages from wild-type mice (Fig. 6A).
FIGURE 6.
Elevated levels of Rac1 recruitment to the phagosomes of SLAT-deficient macrophages. A, lysates of bone marrow-derived macrophages from wild-type and Rac-deficient mice were probed with antibodies to Rac1. B, bone marrow-derived macrophages from wild-type and SLAT–/– mice were incubated for 3 min with IgG-coated latex beads, fixed, and stained with antibodies to Rac1. The images are a single optical slice of a representative field co-stained for Rac1 protein and F-actin. Arrows indicate the actin-rich phagosomes containing the latex bead and co-localized Rac1 protein. C, quantification of pixel intensity for Rac1 and F-actin levels around the phagosomes. For p values, * is <0.05, and ** is <0.01. Average mean fluorescence intensity values around the phagosomes were plotted from three independent experiments. D, PAK-PBD domain was used to pull down Rac1 from lysates of bone marrow-derived macrophages from wild-type and SLAT–/– mice, stimulated with IgG-coated latex beads. The levels of Rac1 in the pulldown were determined by probing with antibodies to Rac1; band intensity was quantified using ImageJ software, and the numbers below the blot represent fold increase in active levels.
Using the specific Rac1 antibody, we measured the amount of Rac1 around the phagosome in BMM derived from wild-type and SLAT–/– animals as above by growing cells attached to glass coverslips, providing the cells IgG-coated latex beads and fixing and staining with anti-Rac1 and phalloidin to detect actin. The images are shown in Fig. 6B and reveal increased Rac1 in the phagosomes of SLAT-deficient macrophages. The amount of Rac1 was quantitated by digitally isolating the phagosome and quantitating the amount of Rac1 fluorescence around the phagosome. We found (Fig. 6C) that the amount of both F-actin and Rac1 was elevated in the SLAT-deficient macrophages relative to wild type (p < 0.05, marked by *; p < 0.01, marked by **). These observations suggest that SLAT functions to govern the amount of Rac1 recruitment to the site of actin reorganization and is consistent with observations in fibroblasts that SLAT acts downstream of activated, GTP-bound Rac. The increased recruitment of Rac1 accounts for the increased F-actin and increased phagocytosis in SLAT-deficient macrophages.
Experiments in nonhematopoietic cells suggested SLAT acts upstream of Rac by promoting formation of Rac-GTP. If this is also the case in FcγR-stimulated macrophages, we expect the activation of Rac would be defective in the SLAT-deficient cells. We measured FcγR-induced formation of Rac-GTP in macrophages of wild-type and SLAT–/– animals, using the PAK pulldown assay described previously (40). The macrophages were stimulated with latex beads coated with IgG, and lysates were subjected to the PAK pulldown. Rac-GTP that was bound to PAK was detected by immunoblotting the PAK-associated proteins with a monoclonal antibody to Rac1. We found (Fig. 6D) that formation of Rac1-GTP was induced in both wild-type and SLAT–/– macrophages, and the level of induction was unchanged. These findings indicate that SLAT is not an essential Rac1 GEF acting downstream of the FcγR.
The findings above show that there is more Rac1 associated with the phagosome when SLAT is absent. Others have shown that SLAT associated with activated, GTP-bound Rac in fibroblasts. Hence the recruitment of SLAT to the phagosome may be dependent on Rac. To test this possibility, we examined macrophages from wild-type or Rac1 or -2-deficient animals for the level of SLAT recruitment. For these experiments, BMM were grown on glass coverslips and provided IgG-coated latex beads. The cells were fixed at 3 min after stimulation, permeabilized, and stained for SLAT and F-actin. The images are shown in Fig. 7A, and a digitally magnified image of SLAT and F-actin for each condition is shown in Fig. 7A, right panels. SLAT was recruited to the F-actin-rich phagosome in wild-type cells. However, the amount of SLAT recruited in macrophages of Rac1 or Rac2 single-deficient animals was decreased, and more so in Rac1,2 double-deficient cells. The SLAT recruitment and F-actin levels were quantitated as above and are shown in Fig. 7B. The data show that SLAT recruitment to the phagosome is reduced by 34% in the macrophages lacking either Rac1 or Rac2 and reduced by more than 70% in the double-deficient macrophages. However, the formation of the phagocytic cup is intact in macrophages of all the Rac-deficient strains, consistent with an earlier report that phagocytic cup formation does not require Rac isoforms (25). These findings indicate that SLAT recruitment to the phagosome requires at least one isoform of Rac. Thus, as in fibroblasts, SLAT in macrophages may directly bind to Rac or a Rac-GTP effector protein after Rac activation and hence function downstream of Rac.
FIGURE 7.
SLAT recruitment to the phagosome requires Rac. A, bone marrow-derived macrophages from wild-type and the indicated Rac–/– strains of mice were stimulated with IgG-coated latex beads for 3 min and then fixed and stained with phalloidin and antibodies to SLAT. A single representative optical slice of the confocal images is shown. The boxed images at the right are digitally enhanced to ×300 magnification to show the amount of SLAT at the phagosome, indicated by the bright phalloidin staining. B, quantification of pixel intensity for SLAT and F-actin levels around 15 representative phagosomes. p value is as follows: * is <0.001; ** is 0.001, and # is <0.0001. Average mean fluorescence intensity values around the phagosomes were plotted from three independent experiments.
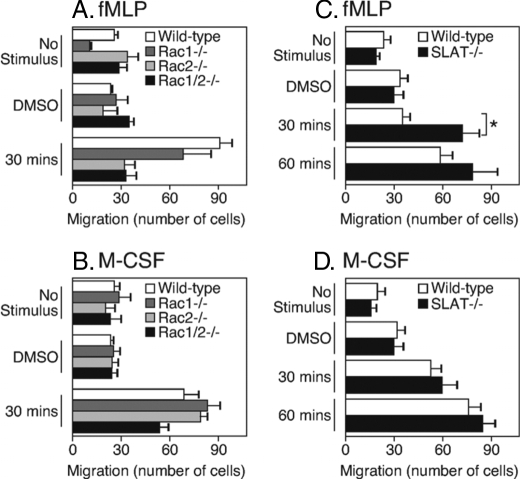
As mentioned in the Introduction, chemotactic migration toward fMLP requires F-actin rearrangement and both the Rac isoforms. In contrast, chemotactic migration to M-CSF appears to be Rac-independent, although the Rac dependence varies with the substrate (42, 43). With these earlier reports in mind, and because our findings here suggested that SLAT inhibits Rac-dependent events, we hypothesized that SLAT-deficient macrophages would exhibit defects in fMLP- but not M-CSF-induced chemotaxis. To test this hypothesis, we used a Transwell assay system to measure the number of migrating BMM toward fMLP or M-CSF in a 1-h assay. We first applied BMM obtained from Rac1–/– or Rac2–/– single-deficient or Rac1,2–/– double-deficient mice to the transwell system and measured responses to fMLP or M-CSF. We found that macrophages obtained from single Rac1- or Rac2-deficient mice showed a partial block in fMLP-induced migration, whereas macrophages from Rac1/2 double-deficient animals completely failed to migrate (Fig. 8A). Macrophages lacking any or both Rac isoforms were able to migrate toward M-CSF but the double-deficient macrophages showed a partial defect (Fig. 8B). These findings confirm the earlier conclusions that fMLP-induced migration is dependent on Rac.
FIGURE 8.
Elevated migration of macrophages from SLAT–/– mice to a Rac-dependent chemokine. Bone marrow-derived macrophages were generated from the indicated strains of mice. An equal number of macrophages were placed in Transwells (5-μm pore size), and the cells were stimulated with fMLP or M-CSF chemokines. At 30 or 60 min, the membranes were removed, and the number of macrophages that had migrated through were quantitated as described under “Materials and Methods.” The graphs show the average number of migrated cells at a given time point. For the p value, * is <0.05. Data represent seven independent experiments for SLAT-deficient macrophages and three independent experiments for Rac-deficient macrophages.
We used the same measurement to test the role of SLAT in fMLP- and M-CSF-induced migration, and we compared migrating cells obtained from wild-type or SLAT–/– mice. The results (Fig. 8C) show that the number of migrating macrophages in response to fMLP was about 2-fold higher when the cells were obtained from SLAT–/– mice. However, macrophage migration between wild-type and SLAT-deficient macrophages was equivalent in response to M-CSF (Fig. 8D). These findings are consistent with our hypothesis that SLAT affects F-actin-dependent events that require GTP-Rac.
DISCUSSION
Here we showed that SLAT is expressed in macrophages and is recruited to the F-actin-rich phagosome. SLAT function appears to be inhibitory toward phagocytosis because overexpression of SLAT reduces and knock-out of SLAT elevates macrophage phagocytosis. The elevated phagocytic function in SLAT-deficient macrophages is coincident with increased association of F-actin and increased Rac recruitment to the phagosome. SLAT also appeared to function in fMLP-triggered chemotaxis, an event likewise dependent on Rac and F-actin redistribution. The findings are consistent with earlier reports (34) that SLAT acts downstream of activated Rac and upstream of F-actin.
SLAT has been shown to function as a GEF for Rac and thus act as an upstream activator of Rac (34, 44). Other studies show that SLAT binds GTP-Rac and accordingly would act as a Rac effector (35). SLAT might be capable of acting as a GEF for Rac when overexpressed, but our data show that it is not an essential Rac1 GEF in macrophages stimulated by FcγR because Rac-GTP formation proceeds unimpeded in SLAT-deficient macrophages stimulated through FcγR. Our findings are consistent with the latter report, indicating SLAT is a Rac effector because of the following: (a) SLAT was recruited to the phagosome under conditions leading to formation of Rac-GTP; (b) SLAT recruitment to the phagosome was greatly reduced in macrophages of Rac1,2–/– animals; and (c) Rac recruitment to phagosomes was elevated in SLAT-deficient macrophages.
It is not clear how SLAT affects macrophage actin reorganization or how SLAT affects Rac-dependent functions. Experiments have established that the Rac GEF DOCK180 (19, 45) acts together in a signaling cascade with the adaptor protein CrkII (20) to induce Rac-GTP in Fc-dependent phagocytosis. DOCK180 is constitutively bound to a scaffold protein called Elmo, which supports the GEF activity of DOCK180 toward Rac (19, 45). The involvement of DOCK180/Elmo and CrkII in Rac activation during phagocytosis was originally identified in a Caenorhabditis elegans model (45). Mammalian cells lacking CrkII or DOCK180 are deficient in recruitment of Rac-GTP to the FcγR-induced phagocytic cup (20). Both CrkII and DOCK180 are recruited to the phagocytic cup, and short interfering RNA-mediated knockdown of CrkII protein expression blocks recruitment of Rac to the phagosome (20). Thus, one possible interpretation of these earlier findings is that inactive Rac is recruited to the phagocytic cup, where it encounters DOCK180 complexed with Elmo and CrkII. Rac then undergoes GDP/GTP exchange to become activated and stimulate downstream events.
How does SLAT regulate Rac recruitment and F-actin presence around the phagosome? One possibility is that SLAT regulates the CrkII-mediated recruitment of DOCK180/Elmo to the phagocytic cup. The reduced recruitment of DOCK180/Elmo by SLAT would cause less Rac to be present at the phagocytic cup. Alternatively, SLAT might regulate the amount of inactive Rac that is recruited to the cup by the complex CrkII-DOCK180-Elmo. In this possibility, the amount of CrkII-DOCK180-Elmo is unchanged, but the amount of Rac present at the phagosome would be decreased because the complex is modified by SLAT in its ability to recruit Rac. We showed that the amount of Rac1 recruited to the phagosome is elevated in macrophages lacking SLAT. Therefore, it is unlikely that SLAT regulates DOCK180 catalytic activity mediating the GDP/GTP exchange on Rac, because our measurements detect the presence of Rac protein and do not distinguish between active and inactive Rac. Experiments are in progress to test these possibilities.
The function of SLAT appears to be inhibitory with regard to phagocytosis, because phagocytosis is increased in macrophages from SLAT-deficient animals. Other FcγR-dependent functions like cytokine production (we measured tumor necrosis factor-α and interleukin-6) were not different from those of wild-type macrophages (data not shown). This finding suggests that signal transduction stimulated by FcγR and leading to actin reorganization is regulated by SLAT, whereas FcγR-stimulated signaling and leading to activation of transcription factors are not regulated by SLAT. This is not the case in T cells, where SLAT clearly regulates NF-AT transcriptional activation through Cdc42 and Rac (32). NF-AT is an important transcriptional regulator of macrophage gene expression triggered by the pattern recognition receptor dectin-1 (46), but there is no information on FcγR-induced cytokine responses. Although we did not test transcription factor activity in macrophages, we did not find any difference in the induction of proinflammatory cytokines in the SLAT-deficient macrophages.
SLAT-deficient animals on a mixed 129 and B6 background ultimately develop a systemic lupus erythematosus-like disease (47). The pathology could be the result of elevated antigen presentation, which is one of the biological consequences of phagocytosed protein (reviewed in Ref. 48). Some of the cargo from an apoptotic cell would include nuclear self-antigens. Autoantibodies in systemic lupus erythematosus patients characteristically (49, 50) and diagnostically (51) target nuclear self-antigens. It is therefore possible that the autoimmune disease characterizing the SLAT-deficient animal is because of the elevated phagocytosis by macrophages of naturally occurring apoptotic cells and inappropriate presentation of the self-antigens to T lymphocytes. Our laboratory is studying the antigen-presenting capacity of wild-type and SLAT-deficient macrophages to test this possibility.
Acknowledgments
We are grateful to Drs. Bill Rodgers (Oklahoma Medical Research Foundation) and Brian Ceresa (University of Oklahoma) for critical reading of the manuscript. We thank Dr. Ute Hochgeschwender (Duke University) for input into the experiments with SLAT knock-out animals and Drs. Linda Thompson (Oklahoma Medical Research Foundation) and James Tomasek and Eric Howard (University of Oklahoma) for advice.
This work was supported, in whole or in part, by National Institutes of Health Grant AI068320 (to A. A).
Footnotes
The abbreviations used are: FcγR, Fcγ receptor; BMM, bone marrow-derived macrophages; F-actin, filamentous actin; fMLP, formyl-Met-Leu-Phe; GEF, guanine nucleotide exchange factor; GFP, green fluorescence protein; HBSS, Hanks' buffered salt solution; ITAM, immunotyrosine-based activation motif; M-CSF, macrophage colony-stimulating factor; NRS, normal rabbit serum; SLAT, SWAP-70-like adaptor of T cells; SRBC, sheep red blood cell; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RBC, red blood cell; PAK, p21-activated kinase; HEK, human embryonic kidney cell.
References
- 1.Coppolino, M. G., Krause, M., Hagendorff, P., Monner, D. A., Trimble, W., Grinstein, S., Wehland, J., and Sechi, A. S. (2001) J. Cell Sci. 114 4307–4318 [DOI] [PubMed] [Google Scholar]
- 2.Hoppe, A. D., and Swanson, J. A. (2004) Mol. Biol. Cell 15 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvier, G., Benoliel, A. M., Foa, C., and Bongrand, P. (1994) J. Leukocyte Biol. 55 729–734 [DOI] [PubMed] [Google Scholar]
- 4.Niedergang, F., and Chavrier, P. (2004) Curr. Opin. Cell Biol. 16 422–428 [DOI] [PubMed] [Google Scholar]
- 5.Lucas, M., Zhang, X., Prasanna, V., and Mosser, D. M. (2005) J. Immunol. 175 469–477 [DOI] [PubMed] [Google Scholar]
- 6.Ganesan, L. P., Joshi, T., Fang, H., Kutala, V. K., Roda, J., Trotta, R., Lehman, A., Kuppusamy, P., Byrd, J. C., Carson, W. E., Caligiuri, M. A., and Tridandapani, S. (2006) Blood 108 718–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winzen, R., Kracht, M., Ritter, B., Wilhelm, A., Chen, C. Y., Shyu, A. B., Muller, M., Gaestel, M., Resch, K., and Holtmann, H. (1999) EMBO J. 18 4969–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Utomo, A., Cullere, X., Glogauer, M., Swat, W., and Mayadas, T. N. (2006) J. Immunol. 177 6388–6397 [DOI] [PubMed] [Google Scholar]
- 9.Kim, C., and Dinauer, M. C. (2001) J. Immunol. 166 1223–1232 [DOI] [PubMed] [Google Scholar]
- 10.Nimmerjahn, F., and Ravetch, J. V. (2006) Immunity 24 19–28 [DOI] [PubMed] [Google Scholar]
- 11.Taylor, L. S., Paul, S. P., and McVicar, D. W. (2000) Rev. Immunogenet. 2 204–219 [PubMed] [Google Scholar]
- 12.Duchemin, A. M., and Anderson, C. L. (1997) J. Immunol. 158 865–871 [PubMed] [Google Scholar]
- 13.Cooney, D. S., Phee, H., Jacob, A., and Coggeshall, K. M. (2001) J. Immunol. 167 844–854 [DOI] [PubMed] [Google Scholar]
- 14.Crowley, M. T., Costello, P. S., Fitzer-Attas, C. J., Turner, M., Meng, F., Lowell, C., Tybulewicz, V. L., and DeFranco, A. L. (1997) J. Exp. Med. 186 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox, D., Tseng, C. C., Bjekic, G., and Greenberg, S. (1999) J. Biol. Chem. 274 1240–1247 [DOI] [PubMed] [Google Scholar]
- 16.Tridandapani, S., Lyden, T. W., Smith, J. L., Carter, J. E., Coggeshall, K. M., and Anderson, C. L. (2000) J. Biol. Chem. 275 20480–20487 [DOI] [PubMed] [Google Scholar]
- 17.Bonilla, F. A., Fujita, R. M., Pivniouk, V. I., Chan, A. C., and Geha, R. S. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1725–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu, H., Botelho, R. J., Yu, M., Grinstein, S., and Neel, B. G. (2003) J. Cell Biol. 161 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gumienny, T. L., Brugnera, E., Tosello-Trampont, A. C., Kinchen, J. M., Haney, L. B., Nishiwaki, K., Walk, S. F., Nemergut, M. E., Macara, I. G., Francis, R., Schedl, T., Qin, Y., Van Aelst, L., Hengartner, M. O., and Ravichandran, K. S. (2001) Cell 107 27–41 [DOI] [PubMed] [Google Scholar]
- 20.Lee, W. L., Cosio, G., Ireton, K., and Grinstein, S. (2007) J. Biol. Chem. 282 11135–11143 [DOI] [PubMed] [Google Scholar]
- 21.Symons, M., Derry, J. M., Karlak, B., Jiang, S., Lemahieu, V., McCormick, F., Francke, U., and Abo, A. (1996) Cell 84 723–734 [DOI] [PubMed] [Google Scholar]
- 22.Tsuboi, S., and Meerloo, J. (2007) J. Biol. Chem. 282 34194–34203 [DOI] [PubMed] [Google Scholar]
- 23.Cox, D., Chang, P., Zhang, Q., Gopal Reddy, P., Bokoch, G. M., and Greenberg, S. (1997) J. Exp. Med. 186 1487–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castellano, F., Montcourrier, P., and P. Chavrier, P. (2000) J. Cell Sci. 113 2955–2961 [DOI] [PubMed] [Google Scholar]
- 25.Hall, A. B., Gakidis, M. A., Glogauer, M., Wilsbacher, J. L., Gao, S., Swat, W., and Brugge, J. S. (2006) Immunity 24 305–316 [DOI] [PubMed] [Google Scholar]
- 26.Glogauer, M., Hartwig, J., and Stossel, T. (2000) J. Cell Biol. 150 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, C. X., Downey, G. P., Zhu, F., Koh, A. L., Thang, H., and Glogauer, M. (2004) Blood 104 3758–3765 [DOI] [PubMed] [Google Scholar]
- 28.Glogauer, M., Marchal, C. C., Zhu, F., Worku, A., Clausen, B. E., Foerster, I., Marks, P., Downey, G. P., Dinauer, M., and Kwiatkowski, D. J. (2003) J. Immunol. 170 5652–5657 [DOI] [PubMed] [Google Scholar]
- 29.Hotfilder, M., Baxendale, S., Cross, M. A., and Sablitzky, F. (1999) Br. J. Haematol. 106 335–344 [DOI] [PubMed] [Google Scholar]
- 30.Tanaka, Y., Bi, K., Kitamura, R., Hong, S., Altman, Y., Matsumoto, A., Tabata, H., Lebedeva, S., Bushway, P. J., and Altman, A. (2003) Immunity 18 403–414 [DOI] [PubMed] [Google Scholar]
- 31.Gupta, S., Fanzo, J. C., Hu, C., Cox, D., Jang, S. Y., Lee, A. E., Greenberg, S., and Pernis, A. B. (2003) J. Biol. Chem. 278 43541–43549 [DOI] [PubMed] [Google Scholar]
- 32.Becart, S., Balancio, A. J., Charvet, C., Feau, S., Sedwick, C. E., and Altman, A. (2008) Immunity 29 704–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinohara, M., Terada, Y., Iwamatsu, A., Shinohara, A., Mochizuki, N., Higuchi, M., Gotoh, Y., Ihara, S., Nagata, S., Itoh, H., Fukui, Y., and Jessberger, R. (2002) Nature 416 759–763 [DOI] [PubMed] [Google Scholar]
- 34.Mavrakis, K. J., McKinlay, K. J., Jones, P., and Sablitzky, F. (2004) Exp. Cell Res. 294 335–344 [DOI] [PubMed] [Google Scholar]
- 35.Oka, T., Ihara, S., and Fukui, Y. (2007) J. Biol. Chem. 282 2011–2018 [DOI] [PubMed] [Google Scholar]
- 36.Vedham, V., Phee, H., and Coggeshall, K. M. (2005) Mol. Cell. Biol. 25 4211–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becart, S., Charvet, C., Canonigo Balancio, A. J., De Trez, C., Tanaka, Y., Duan, W., Ware, C., Croft, M., and Altman, A. (2007) J. Clin. Investig. 117 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts, A. W., Kim, C., Zhen, L., Lowe, J. B., Kapur, R., Petryniak, B., Spaetti, A., Pollock, J. D., Borneo, J. B., Bradford, G. B., Atkinson, S. J., Dinauer, M. C., and Williams, D. A. (1999) Immunity 10 183–196 [DOI] [PubMed] [Google Scholar]
- 39.Clausen, B. E., Burkhardt, C., Reith, W., Renkawitz, R., and Forster, I. (1999) Transgenic Res. 8 265–277 [DOI] [PubMed] [Google Scholar]
- 40.Benard, V., Bohl, B. P., and Bokoch, G. M. (1999) J. Biol. Chem. 274 13198–13204 [DOI] [PubMed] [Google Scholar]
- 41.Diakonova, M., Bokoch, G., and Swanson, J. A. (2002) Mol. Biol. Cell 13 402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wells, C. M., Walmsley, M., Ooi, S., Tybulewicz, V., and Ridley, A. J. (2004) J. Cell Sci. 117 1259–1268 [DOI] [PubMed] [Google Scholar]
- 43.Weiss-Haljiti, C., Pasquali, C., Ji, H., Gillieron, C., Chabert, C., Curchod, M. L., Hirsch, E., Ridley, A. J., van Huijsduijnen, R. H., Camps, M., and Rommel, C. (2004) J. Biol. Chem. 279 43273–43284 [DOI] [PubMed] [Google Scholar]
- 44.Gupta, S., Lee, A., Hu, C., Fanzo, J., Goldberg, I., Cattoretti, G., and Pernis, A. B. (2003) Hum. Immunol. 64 389–401 [DOI] [PubMed] [Google Scholar]
- 45.Brugnera, E., Haney, L., Grimsley, C., Lu, M., Walk, S. F., Tosello-Trampont, A. C., Macara, I. G., Madhani, H., Fink, G. R., and Ravichandran, K. S. (2002) Nat. Cell Biol. 4 574–582 [DOI] [PubMed] [Google Scholar]
- 46.Goodridge, H. S., Simmons, R. M., and Underhill, D. M. (2007) J. Immunol. 178 3107–3115 [DOI] [PubMed] [Google Scholar]
- 47.Fanzo, J. C., Yang, W., Jang, S. Y., Gupta, S., Chen, Q., Siddiq, A., Greenberg, S., and Pernis, A. B. (2006) J. Clin. Investig. 116 703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blander, J. M., and Medzhitov, R. (2006) Nat. Immunol. 7 1029–1035 [DOI] [PubMed] [Google Scholar]
- 49.Munoz, L. E., van Bavel, C., Franz, S., Berden, J., Herrmann, M., and van der Vlag, J. (2008) Lupus 17 371–375 [DOI] [PubMed] [Google Scholar]
- 50.Kalaaji, M., Fenton, K. A., Mortensen, E. S., Olsen, R., Sturfelt, G., Alm, P., and Rekvig, O. P. (2007) Kidney Int. 71 664–672 [DOI] [PubMed] [Google Scholar]
- 51.Zandman-Goddard, G., Gilburd, B., Shovman, O., Blank, M., Berdichevski, S., Langevitz, P., and Shoenfeld, Y. (2005) Clin. Dev. Immunol. 12 107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]