Abstract
N-Linked glycosylation involves the ordered, stepwise synthesis of the unique lipid-linked oligosaccharide precursor Glc3Man9 GlcNAc2-PP-Dol on the endoplasmic reticulum (ER), catalyzed by a series of glycosyltransferases. Here we characterize Alg2 as a bifunctional enzyme that is required for both the transfer of the α1,3- and the α1,6-mannose-linked residue from GDP-mannose to Man1GlcNAc2-PP-Dol forming the Man3GlcNAc2-PP-Dol intermediate on the cytosolic side of the ER. Alg2 has a calculated mass of 58 kDa and is predicted to contain four transmembrane-spanning helices, two at the N terminus and two at the C terminus. Contradictory to topology predictions, we prove that only the two N-terminal domains fulfill this criterion, whereas the C-terminal hydrophobic sequences contribute to ER localization in a nontransmembrane manner. Surprisingly, none of the four domains is essential for transferase activity because truncated Alg2 variants can exert their function as long as Alg2 is associated with the ER by either its N- or C-terminal hydrophobic regions. By site-directed mutagenesis we demonstrate that an EX7E motif, conserved in a variety of glycosyltransferases, is not important for Alg2 function in vivo and in vitro. Instead, we identify a conserved lysine residue, Lys230, as being essential for activity, which could be involved in the binding of the phosphate of the glycosyl donor.
Asparagine-linked glycosylation is an essential protein modification highly conserved in eukaryotes (1–4), and several features of this pathway even occur in prokaryotes (5–7). In eukaryotes, biosynthesis of N-glycans starts with the assembly of the common core oligosaccharide precursor Glc3Man9 GlcNAc2-PP-Dol, the glycan moiety of which is subsequently transferred onto selected Asn-Xaa-(Ser/Thr) acceptor sites of the nascent polypeptide chain by the oligosaccharyl-transferase complex (8–10). The initial steps of the dolichol pathway up to Man5GlcNAc2-PP-Dol take place on the cytosolic site of the endoplasmic reticulum (ER),2 using sugar nucleotides as glycosyl donors. Upon translocation of the heptasaccharide to the luminal site, which is facilitated by Rft1 (11) and another not yet identified protein (12), it is extended by four mannose and three glucose residues deriving from Man-P-Dol and Glc-P-Dol. It has been demonstrated that the pathway operates sequentially in an ordered fashion based on differences in the substrate specificity of the various glycosyltransferases (13). In the yeast Saccharomyces cerevisiae, alg mutants (for asparagine-linked glycosylation) have been isolated, defective in lipid-linked oligosaccharide (LLO) assembly (14–17), and shown to be invaluable to define the pathway as well as to isolate the genes encoding the respective glycosyltransferases by complementing a particular phenotype characteristic of the respective mutant. Likewise various mutant cell lines from mammalian origin have been described that produce truncated lipid-linked oligosaccharides (18–20).
One of the temperature-sensitive alg mutants, alg2, was shown to accumulate lipid-linked Man2GlcNAc2 at the restrictive temperature (15), indicating that alg2 might have a defect in the glycosyltransferase catalyzing the transfer of the third, α1,6-linked mannose, i.e. in the formation of the branched pentasaccharide Man3GlcNAc2-PP-Dol (see Fig. 8). On the other hand, biochemical studies in human fibroblasts from a patient with a defect in the human ALG2 ortholog, causing congenital disorder of glycosylation type CDG1i, pointed to a role in the transfer of the second, α1,3-linked mannose residue, because no elongation of Man(1,6)ManGlcNAc2-PP-Dol occurred (21). In contrast, control fibroblasts were able to do so, albeit with reduced efficiency when compared with Man(1,3)ManGlcNAc2-PP-Dol as glycosyl acceptor. Because a bioinformatic approach of the yeast data base did not reveal an unknown open reading frame that might encode an additional putative mannosyltransferase being involved in LLO synthesis, we reasoned that ALG2 may have a dual function, i.e. synthesizing both Man2GlcNAc2-PP-Dol and Man3GlcNAc2-PP-Dol. While the current study was in progress, evidence was presented that a membrane fraction from Escherichia coli, expressing ALG2 from yeast, is able to carry out an α1,3- and α1,6-mannosylation to form the branched pentasaccharide intermediate (22). However, the contribution of native E. coli enzymes could not entirely be ruled out. So far Alg2 has not been studied biochemically in yeast. Here, we confirm and extent this finding by investigating Alg2 in yeast. We first established a radioactive in vitro assay and demonstrate that Alg2, immunoprecipitated from detergent extracts of yeast microsomal membranes, is indeed sufficient to catalyze both elongation of Man1GlcNAc2-PP-Dol to Man2GlcNAc2-PP-Dol and subsequently to Man3GlcNAc2-PP-Dol. Furthermore we investigated the membrane topology of Alg2 mannosyltransferase. Evidence will be presented that Alg2 is composed only of the two N-terminal of four predicted transmembrane domains (TMDs), whereas the C-terminal hydrophobic sequences contribute to ER localization merely in a nontransmembrane manner. Surprisingly, none of the four domains is essential for Alg2 activity because deletion of either the two N-terminal or C-terminal domains gives rise to an active transferase. Finally, we perform a mutational analysis of Alg2 and identify amino acids required for its activity.
FIGURE 8.
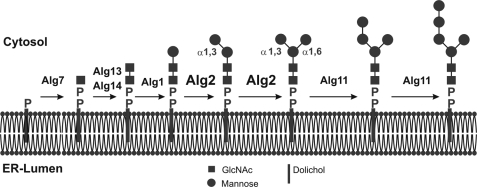
Early steps of lipid-linked oligosaccharide formation on the cytosolic side of the ER membrane. Biosynthesis starts with the transfer of a GlcNAc-phosphate to dolichol phosphate with formation of the pyrophosphate bond, catalyzed by Alg7. The second step is catalyzed be the dimeric Alg14/Alg13 complex, whereby membrane-bound Alg14 recruits cytosolic Alg13 to the membrane with formation of the active GlcNAc transferase. Following the addition of the β1,4-linked mannose by Alg1, Alg2 catalyzes, as demonstrated here, both the transfer of the α1,3- and α1,6-linked mannose. The two final α1,2-mannose residues are transferred by Alg11, before the Man5GlcNAc2-PP heptasaccharide is translocated across the ER membrane to the lumen, where further elongation takes place to the full-length core saccharide. All of the sugar residues are donated by sugar nucleotides.
MATERIALS AND METHODS
Yeast Strains, Media, and Genetic Methods—The following strains were used: SS328 (MATα ade2-101 ura3-52 his3Δ200 lys2-801), DBY747 (MATa his3-Δ1 leu2-3 leu2-112, ura3-52, trp1-289), W303-1A (MATa leu2-3 leu2-112, his3-11 his3-15 ura3-1 ade2-1 trp1-1), and alg2-1 (MATα ura3-52 ade2-101), STY50 (MATa his4-401 leu2-3, -112 trp1-1 ura3–52 HOL1-1 SUC2::LEU2). The strains were grown in YPD medium (1% yeast extract, 2% bacto-peptone, 2% glucose) or in selective YNB medium (0.67% YNB, 0.5% casamino acids, 2% glucose) supplemented with amino acids and nucleotide bases, as required.
To construct the expression plasmids pVT100-Alg2-ZZ and pVT100-Alg2-Bio, genomic ALG2 was amplified from W303-1A by PCR and engineered with HindIII and BamHI restriction sites at the 5′ and 3′ ends for cloning purpose and ligated into the HindIII/BamHI cut vectors pVT100-ZZ and pVT100-Bio, respectively, placing ALG2 under the control of the constitutive ADH1 promoter. The constructs were sequenced, and the functional expression of Alg2 containing two protein A (ZZ) epitopes or the Bio epitope in frame at the C terminus was verified by immunoblotting and complementation of the growth phenotype of the alg2 mutant. For PCR amplification the following primers were used: ALG2 fw (5′-CCCAAGCTTAAATGATTGAAAAGGATAAAAG-3′) and ALG2 rev (5′-CGGGATCCTATATTTCTTCATAAGGGTAG-3′). To generate the truncated Alg2-Bio reporter variants, internal ALG2-specific reverse primers annealing at the positions as indicated in the corresponding figures were used together with the above ALG2 fw primer (sequences available upon request). For constructing the Alg2-Suc2/His4C topology reporter, plasmid pJK90 was used to engineer the ALG2 variants into the SmaI restriction site (23), and the constructs were transformed into STY50 (24) for analysis. To generate site-specific alg2 mutants, the QuikChange® II XL site-directed mutagenesis kit (Stratagene) was used according to the manufacturer's instructions. Transformation into yeast and E. coli was carried out using standard techniques.
In Vivo Labeling of Lipid-linked Saccharides with [3H]Mannose—Labeling with [2-3H]mannose (15 Ci/mmol; GE Healthcare) and analysis of lipid-linked oligosaccharides by HPLC was performed as described previously (25). The cells were grown overnight at 25 °C. Subsequently the cells were labeled at 25 °C or at 34 °C as indicated.
Isolation of Microsomal Membranes and Preparation of Solubilized Enzyme Extract—Rough microsomal membranes were isolated as described (26). The membranes were resuspended in 30 mm Tris-HCl buffer, pH 7.5, containing 3 mm MgCl2, 1 mm dithiothreitol, 35% glycerol (v/v) at a concentration of 10 mg/ml protein. For membrane solubilization, Nonidet P-40 was added to a final concentration of 1%. After 20 min of incubation on ice, the solubilized extract was separated from insoluble material by centrifugation at 150,000 × g for 40 min. All of the steps were carried out at 4 °C, unless indicated otherwise.
Mannosyltransferase Assays—For assay I, activity of solubilized enzyme was determined with Man[14C]GlcNAc2-PP-Dol as acceptor and nonradioactive GDP-Man as glycosyl donor. The reaction contained the following in a final volume of 0.06 ml: 14 mm MES, pH 6.0, 0.1% Nonidet P-40, 10 mm MgCl2, 0.1 mm Na-EDTA, 1 mm dithiothreitol, 4 mm potassium citrate, 12% glycerol or 1 m sucrose, [14C]GlcNAc1-PP-Dol (3000 cpm), 1 mm GDP-Man and solubilized enzyme (equivalent to 70 μg of membrane protein). The reaction was started by the addition of GDP-Man and incubated at 26 °C, stopped with chloroform/methanol (3:2) at the times indicated. Labeled glycolipids were then extracted and washed, and oligosaccharides were released from the dolichol-PP moiety by mild acid and analyzed by HPLC as described (27). Man[14C]GlcNAc2-PP-Dol was prepared as described (21). For assay II, determination of enzyme activity by immunoprecipitation of Alg2-ZZ was carried out as follows. 0.2 ml of solubilized extract was incubated with 0.1 ml of IgG-Sepharose 6 Fast Flow (GE Healthcare) by gently tumbling for 60 min at 4 °C. IgG-Sepharose 6 was equilibrated before use with 40 mm PIPES, pH 6.8, 1% Nonidet P-40, 1 mm dithiothreitol, and 15% glycerol. After incubation the affinity matrix was washed five times with equilibration buffer and then three times with 20 mm MES, pH 6, containing 0.1% Nonidet P-40, 1 mm dithiothreitol, 10 mm MgCl2, 4 mm potassium citrate, and 1.5 m sucrose. The pellet was resuspended in 50 μl of mannosyltransferase assay I reaction mixture. The reaction was started by the addition of 1 mm GDP-Man (final concentration), incubated for 45 min, and processed as for assay I.
RESULTS
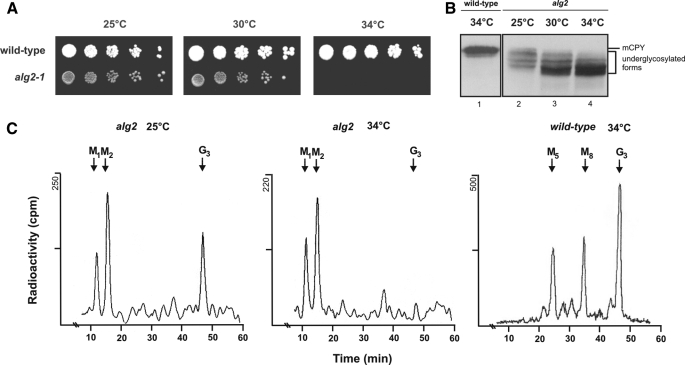
Characterization of the alg2 Mutant—alg2-1 is temperaturesensitive and displays a growth phenotype at 34 °C (Fig. 1A). Interestingly, its glycosylation defect occurs already to some extent at the permissive growth temperature of 25 °C, as demonstrated by Western blot analysis of the vacuolar model glycoprotein carboxypeptidase Y (CPY) (Fig. 1B), as well as by analysis of lipid-linked oligosaccharides metabolically labeled with [3H]mannose (Fig. 1C). CPY from wild-type cells contains four N-linked glycan chains and migrates as a distinct band, whereas the alg2 glycosylation defect gives rise to CPY species, lacking one or more chains, or having shortened chains and thus migrate with a higher mobility on SDS gels. It has been reported that the alg2-1 mutant accumulates Man2GlcNAc2-PP-Dol and Man1GlcNAc2-PP-Dol using Bio-Gel P4 chromatography (15), which is confirmed and extended in Fig. 1C by applying HPLC for separation. Whereas in wild-type yeast the fully assembled core oligosaccharide and larger intermediates occur, in the alg2 mutant Man2GlcNAc2-PP-Dol is dominating both at the permissive temperature and, even more pronounced, at the restrictive temperature. Sequencing of the alg2-1 mutant revealed at amino acid 377 an exchange of Gly to Arg in agreement with an earlier report (28). However, we could not detect the second mutation reported by this group for alg2-1, causing a Q386K exchange. Thus the alg2-1 defect is caused exclusively by the G377R mutation. Alg2 has four predicted potential glycosylation sites. However, these are not used, as demonstrated by endoglycosidase H treatment to cleave off N-glycan chains (supplemental Fig. S1), and hence Alg2 is nonglycosylated.
FIGURE 1.
Analysis of the growth and glycosylation defect of alg2. A, 3 μl of a serial 1:10 dilution of the indicated strains in liquid medium were plated on YEPD plates starting with 3 × 104 cells. The plates were incubated at 25, 30, or 34 °C for 72 h. B, Western blot analysis of CPY. Wild type (DBY747) (lane 1) and alg2 (lanes 2–4) were cultivated at the temperatures indicated, and the cytosolic fraction was obtained by lysis of cells with glass beads was analyzed. The equivalent of 107 cells was applied per lane and separated on an 8% SDS-polyacrylamide gel. The positions of the mature form of carboxypeptidase Y (mCPY), possessing four N-glycans and of the underglycosylated forms in alg2-1 cells are indicated.C, analysis of lipid-linked oligosaccharides in wild-type cells (SS328) and alg2 mutant cells. Lipid-linked oligosaccharides were metabolically labeled for 20 min with [2-3H]mannose at the temperature indicated, and the oligosaccharides released by mild acid hydrolysis were analyzed by HPLC. The positions of standards are indicated; M1, GlcNAc2Man1; M2, GlcNAc2Man2; M5, GlcNAc2Man5; M8, GlcNAc2Man8; G3, GlcNAc2Man9Glc3.
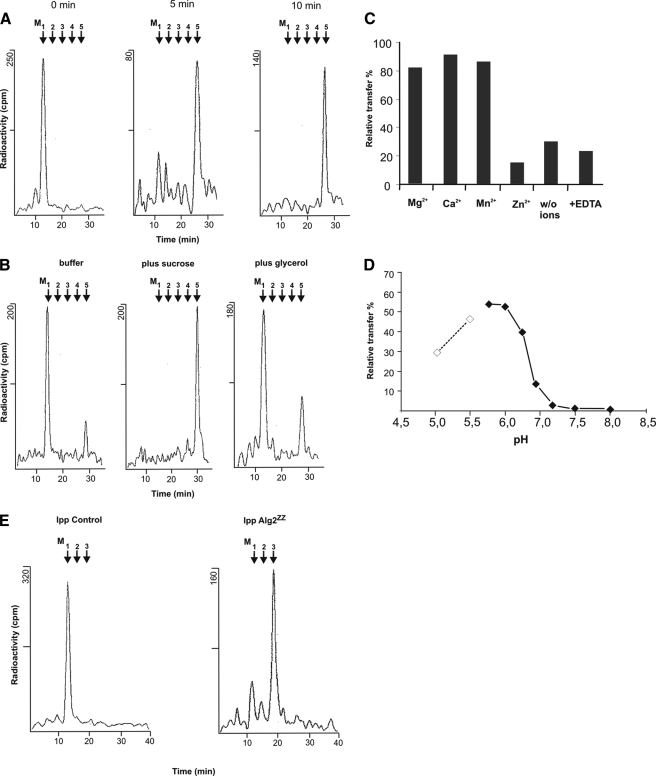
Alg2 Is a Bifunctional Transferase Catalyzing Both the Formation of Man2GlcNAc2-PP-Dol and Man3GlcNAc2-PP-Dol—To unravel the mannosylation defect of alg2 in more detail, we first established an enzymatic test that allowed us to measure early reactions of LLO synthesis from Man1GlcNAc2-PP-Dol to Man5GlcNAc2-PP-Dol involving the participation both of Alg2 and Alg11. So far in vitro assays for these reactions have not been demonstrated in yeast. As can be seen in Fig. 2A and supplemental Fig. S2, incubation of a detergent-solubilized extract from microsomal membranes of wild-type yeast with radioactive Man1[14C]GlcNAc2-PP-Dol as acceptor and unlabeled GDP-Man as glycosyl donor leads to rapid and time dependent elongation of the acceptor oligosaccharide up to Man5GlcNAc2-PP-Dol. Elongation was stimulated by glycerol in the assay or even more efficient by sucrose (Fig. 2B). Biosynthesis was also enhanced by divalent cations, such as Mg2+, Ca2+, or Mn2+, in a similar fashion, whereas Zn2+ ions were inhibitory (Fig. 2C). We also investigated the pH dependence of the elongation reactions and determined an optimum around pH 6 (Fig. 2D). This is somewhat surprising, because other glycosyltransferase of the N-glycosylation pathway in the ER are operative around pH 7.5 (27, 29–33). At this pH hardly any elongation of Man1GlcNAc2-PP-Dol to Man3GlcNAc2-PP-Dol was detectable in yeast. This finding is also in contrast to the experiments using a membrane fraction from E. coli expressing yeast Alg2, which were performed at pH 7.2 (22).
FIGURE 2.
Analysis of Alg2 mannosyltransferase activity. A, time dependence of lipid-linked oligosaccharide formation. Solubilized enzyme from wild-type membranes was incubated for the times indicated using assay I with Man1[14C]GlcNAc2-PP-Dol as glycosyl acceptor as described under “Experimental Procedures.” Lipid-linked oligosaccharides were extracted, and the oligosaccharide moiety was released by mild acid and separated by HPLC. B, influence of sucrose and glycerol on enzyme activity. Either 1 m sucrose or 15% glycerol was added to the standard assay. Incubation was 10 min. C, effect of metal ions on the reaction. Solubilized enzyme was incubated in the presence of 10 mm metal ions or 8 mm EDTA using assay I. D, pH dependence of the reaction. From pH 4.5 to 5.5 10 mm sodium acetate buffer was used (dotted line); from pH 5.7 to 8 10 mm MES buffer (solid line) was used under the conditions of assay I. E, Alg2 transferase activity in immunoprecipitates (Ipp). Alg2 with a C-terminal ZZ epitope was immunoprecipitated with IgG-Sepharose beads from solubilized extract, and the enzyme activity was determined using assay II (right panel). As control an extract from wild-type cells expressing a nontagged Alg2, which was not elongated, was immunoprecipitated (left panel).
To separate Alg2 activity from other mannosyltransferases of LLO synthesis and to measure exclusively Alg2 function, we introduced a protein A (ZZ) epitope at the C terminus of ALG2, which allowed us to immunoprecipitate Alg2-ZZ from the solubilized extract of wild-type cells with IgG-Sepharose as affinity matrix. In the experiment depicted in Fig. 2E (right panel) Man1[14C]GlcNAc2-PP-Dol was converted almost completely to Man3[14C]GlcNAc2-PP-Dol but without further elongation to Man5[14C]GlcNAc2-PP-Dol, as was the case in Fig. 2A without immunoprecipitation. No transfer onto Man1[14C] GlcNAc2-PP-Dol was measurable in a control experiment, in which a solubilized extract from membranes of wild-type cells expressing nontagged Alg2 was incubated with IgG-Sepharose beads (Fig. 2E, left panel). This means that immunoprecipitation of Alg2 is specific rather than that Alg2 binds nonspecifically to the affinity matrix. The results suggest that Alg2 is able to catalyze both the addition of the α1,3- and α1,6-linked mannose residue, mostly likely in this sequence. This conclusion is inferred from earlier studies dealing with the order of LLO assembly in mammalian cells in vivo (34, 35).
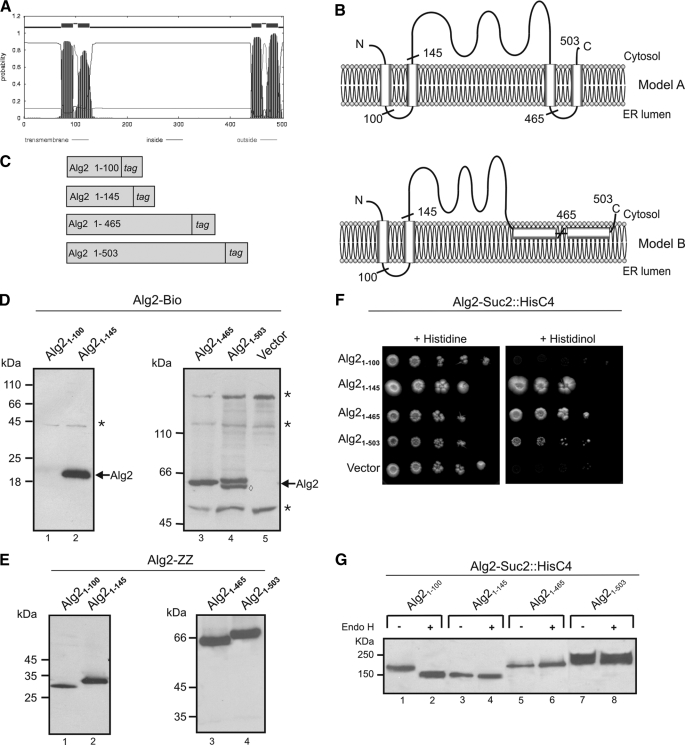
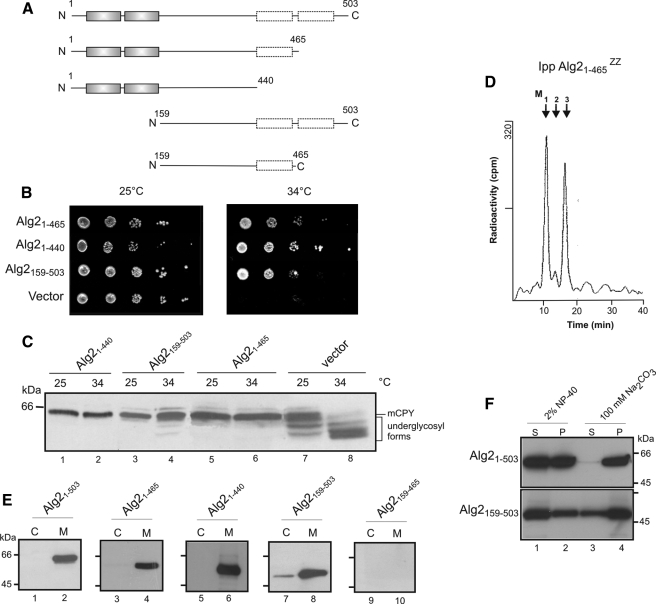
Determination of the Membrane Topology of Alg2 Reveals Two Membrane-spanning Domains—Alg2 is a 58-kDa protein and predicted by most algorithms to contain four transmembrane-spanning helices (TMDs). Two of them are closely spaced to the N terminus, whereas the other pair is grouped at the C terminus (36) (Fig. 3A). From this and the fact that the catalytic site must be localized to the cytosol, because the nucleotide sugar donor is only present in the cytosol, a topology can be proposed as depicted in Fig. 3B (Model A). To experimentally prove or disprove this prediction, we used two approaches. We designed various C-terminal fusions between Alg2 and a Bio tag or between Alg2 and a Suc2/His4C tag as topologysensitive reporters (Fig. 3C). In the case of the Bio epitope, this domain is a substrate for biotin ligase (37) and will be biotinylated only, when oriented to the cytosolic side, because this reaction takes place exclusively in the cytosol. His4C maintains histidinol dehydrogenase activity, the last step in histidine biosynthesis, and his4 mutants, expressing a Suc2/His4C fusion, are able to grow on minimal histidinol medium only when the His4C domain is on the cytoplasmic side of the ER but not when targeted to the ER lumen. In addition, in the latter case the chimeric protein becomes extensively glycosylated because of the presence of N-glycosylation sites (23, 38). As shown in Fig. 3D, Alg2 fusions with the Bio tag engineered downstream to the postulated second (Alg21–145), third (Alg21–465), and fourth (Alg21–503) TMD were all biotinylated, indicating that these fusions are oriented to the cytosol. Only the Alg21–100 construct was not labeled. To rule out the possibility that the lack of a signal for this construct is merely because it is not expressed, we replaced the Bio tag by a protein A epitope. However, as shown in Fig. 3E, this and also the other constructs are all expressed. As will be demonstrated below (Fig. 4), the two C-terminal hydrophobic regions, although not functioning as TMDs, nevertheless contribute to anchoring Alg2 to the ER in a nonperipheral manner, presumably integrating into the outer leaflet of the ER membrane. Therefore, model B is proposed for the topology of Alg2. This topology is also in agreement with the HisC4 technique. Apart from Alg1–100::Suc2/HisC4, all other fusions support growth on histidinol, implying that the C terminus of the respective constructs localizes to the cytosol (Fig. 3F). Moreover, the topology of the constructs has been validated by analyzing their glycosylation status. Treatment with endoglycosidase H results in a decrease of the molecular mass only in the case of the Alg1–100::Suc2/HisC4 construct (Fig. 3G, lanes 1 and 2), indicating an ER luminal orientation of the reporter. All other fusions were resistant and are not glycosylated and must therefore be cytosolic (Fig. 3G, lanes 3–8).
FIGURE 3.
Investigation of the membrane topology of Alg2. A, hydrophobicity probability plot of Alg2 generated using the TMHMM server. B, models for the Alg2 membrane topology. The predicted topology of Alg2 suggests four TMD (model A); the experimental data using truncated Alg2 reporter fusions and truncated Alg2 variants (see Fig. 4) suggest model B, containing two TMD at the N terminus and two additional C-terminal hydrophobic regions that interact with, but do not cross the membrane. C, schematic representation of Alg2 reporter constructs. Either a Bio tag or a SUC2::HisC4 tag was fused in frame at the C-terminal side following amino acids 100, 145, 465, or 503 (see also B). D, analysis of Alg2 topology by Alg2-Bio fusions. The membranes from wild-type cells (SS328) expressing plasmid-borne Alg2 constructs, as indicated, were analyzed by Western blotting. The asterisks indicate biotinylated, endogenous yeast proteins; the band marked by a diamond in lane 4 is presumably a degradation product of the Alg2 construct. E, Western blot analysis of truncated Alg2-ZZ variants. To verify the protein expression of the truncated constructs, Alg2 fusions were made with ZZ instead of the Bio tag and probed for the ZZ tag. F, analysis of topology by Alg2-Suc2::HisC4 fusions. Strain STY50 containing a his4 mutation were transformed with truncated Alg2 variants, as indicated, and analyzed for growth on histidine or histidinol. Only cells with a cytosolic orientation of the epitope are able to grow on histidinol. G, analysis of glycosylation of Alg2-Suc2::HisC4 constructs. The membranes were isolated from wild-type cells (SS328), expressing the various Alg2-Suc2::HisC4 constructs, and incubated without (–) or with (+) endoglycosidase H (Endo H) for 3 h to release the glycan chains and analyzed by Western blotting. The constructs were probed with an antibody against the hemagglutinin epitope of the fusion.
FIGURE 4.
Alg2 protein lacking its two N-terminal transmembrane domains or its two C-terminal hydrophobic segments is functional. A, schematic representation of the Alg2 constructs. Constructs containing a C-terminal ZZ epitope for detection were engineered into expression vector pVT100 as indicated and transformed into alg2. B, growth of alg2 cells harboring an empty vector or the Alg2 constructs at the permissive temperature of 25 °C or at the restrictive temperature of 34 °C. C, glycosylation status of CPY analyzed by Western blot. Experimental conditions were as in Fig. 1. D, determination of mannosyltransferase activity of the Alg21–465-ZZ variant, immunoprecipitated from solubilized extract; assay II was used. E, localization of Alg2 constructs. Microsomal membranes (M) and cytosolic fraction (C) from cells expressing the various Alg2 constructs were analyzed by Western blot. Equivalent amounts of membranes and cytosol corresponding to 107 cells was applied. F, membrane association of Alg2. The membranes from cells expressing full-length Alg21–530 and Alg2 lacking both N-terminal TMDs, Alg2159–465, were subjected to treatment with 2% Nonidet P-40 or 100 mm Na2CO3 (pH 11) and further centrifuged at 100,000 × g. Equivalent amounts from the pellet (P) and supernatant (S) were analyzed by Western blot.
Functional Investigation of Truncated Alg2 Variants—The experimental approaches described above demonstrate that Alg2 consists of only two TMDs. We asked which function these two TMDs as well as the two other predicted, C-terminal hydrophobic segments could serve. Several Alg2 truncations, as illustrated in Fig. 4A, were engineered, and their role was investigated with respect to membrane anchoring, N-glycosylation, and growth. The constructs were cloned into a constitutive yeast expression vector, containing a C-terminal ZZ epitope for detection purpose, and their function was analyzed in the temperature-sensitive alg2 mutant. Quite unexpected, deletion of the two N-terminal TMDs in the Alg2159–503 variant led to a functionally active mannosyltransferase, which supported the temperature-sensitive growth defect of alg2 (Fig. 4B), rescued the underglycosylation of CPY (Fig. 4C, compare lanes 4 and 8) and bound to the membrane fraction (Fig. 4E, lane 8). The role of the additional deletion of the fourth hydrophobic membrane domain in the Alg159–465 construct could not be analyzed, because the truncated protein construct was not detectable, presumably because of instability (Fig. 4E, lanes 9 and 10). However, the lack of this domain per se is not the reason that Alg2159–465 is not detectable, because a construct comprising the two TMDs and the third hydrophobic region (Alg21–465) is active both in vivo (Fig. 4, B and C) and in vitro elongating Man1GlcNAc2-PP-Dol to Man3GlcNAc2-PP-Dol (Fig. 4D). Finally we investigated a construct (Alg21–440) containing both N-terminal TMDs but lacking the two C-terminal, nonspanning hydrophobic sequences. Alg21–440 was found to be functional because it complemented both the growth and glycosylation defect of alg2 at the restrictive temperature (Fig. 4, B and C). Altogether, these experiments imply that the cytosolic loop of amino acids 159–440 comprises the catalytic function of the transferase and can exert its role, when Alg2 is membrane-bound either via its N or C terminus. To address the contribution of the C-terminal hydrophobic domains to membrane anchoring, we performed an alkaline sodium carbonate extraction of membranes, known to release peripheral attached proteins, and compared it with a 2% Nonidet P-40 detergent treatment. As shown in Fig. 4F (lanes 3 and 4), in contrast to the full-length Alg21–503, the N-terminal truncated Alg2159–503 variant harboring the exclusively the C-terminal segments is somewhat extractable into the soluble fraction (S) but clearly not sufficient to suggest a peripheral attachment. Even extraction with 2% detergent did not completely solubilize the N-terminally truncated version. Hence, together with the results from the Bio and His4 localization experiments, a topology is proposed as depicted in model B positioning the hydrophobic helices in the leaflet of the membrane (for further discussion see below).
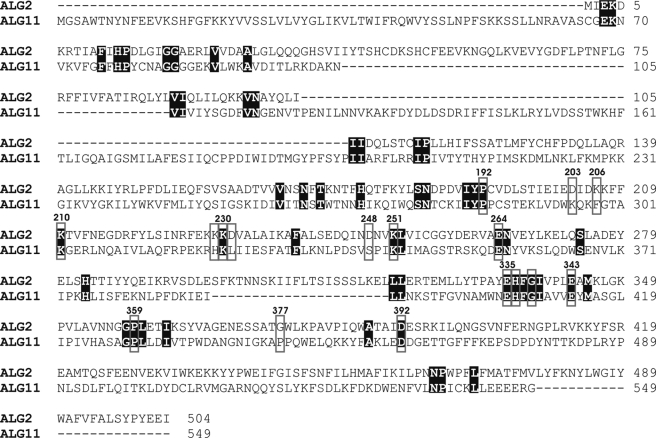
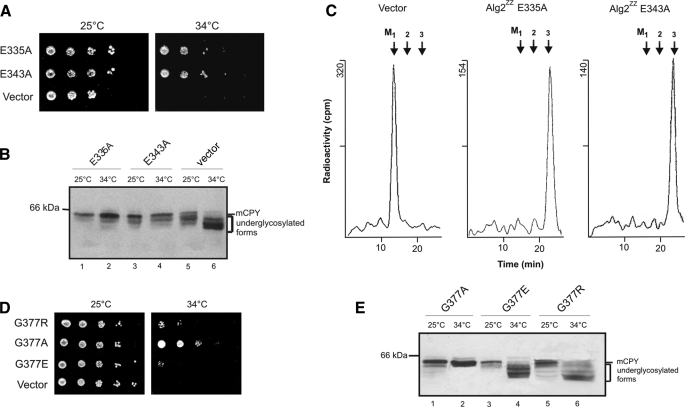
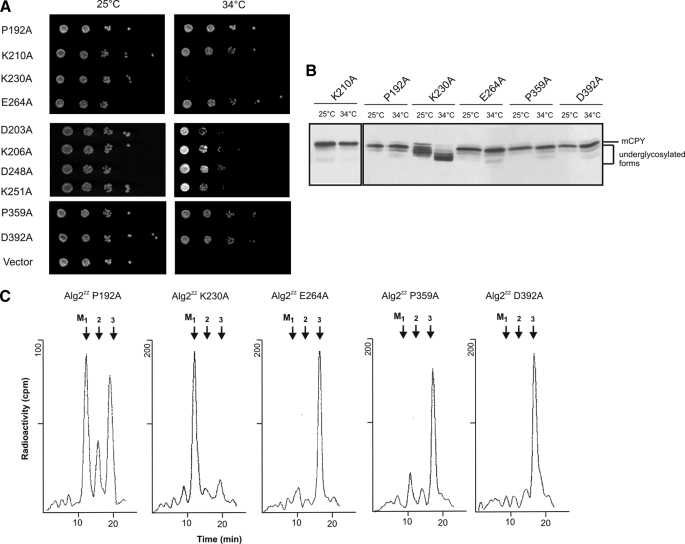
Analysis of Alg2 Function by Site-directed Mutagenesis—For identifying amino acids important for catalysis and eventually to discriminate between the α1,3 and α1,6 activity, we performed site-specific mutagenesis in conserved amino acid residues and motifs, respectively, discussed in the functional context of glycosyltransferases. A variety of Pfam GT1F glycosyltransferases contain the C-terminal sequence motif EX7E (39). The two glutamate residues have been proposed to be involved in catalysis, but their contributions have been discussed as controversial (40–42) and have yet to be identified by structural analysis. Fig. 5 shows an alignment of the Alg2 and Alg11 mannosyltransferases that are closely related and are grouped into family 4 of the CaZY data base. We have replaced both Glu335 and Glu343 by alanine and found that neither of these mutations abolished enzyme activity in vivo (Fig. 6B) or in vitro (Fig. 6C) and also complemented the Alg2 lethality at the restrictive temperature (Fig. 6A). This is in contrast to findings obtained with Alg2 expressed in E. coli, for which the E343A mutation was reported to be inactive and E335A had only some residual activity. The reason for this discrepancy is not known. Also two other alanine exchanges in this motif, H336A and G338A, had no influence on Alg2 function (data not shown). In addition we mutated residues that lie in conserved positions, such as Pro192, Lys210, Lys230, Glu264, Pro359, and Asp392 by alanine. In particular lysine residues and acidic amino acids such as Glu and Asp are proposed and have been shown to function in nucleotide sugar binding. From the six mutations only Lys230 was detrimental and caused a loss of Alg2 activity (Fig. 7 and supplemental Fig. S3) as revealed from the analysis of growth, glycosylation of CPY, and enzyme activity. The significance of residue Lys230 is also emphasized by the fact that exchanges in the immediate adjacent amino acids Lys229 and Asp231 to alanine did not effect growth and glycosylation (data not shown). As mentioned above, in the alg2-1 mutant amino acid Gly377 is mutated to an arginine residue (G377R). We have replaced in addition glycine 377 by a negatively charged amino acid such as glutamate. This exchange also led to a nonfunctional variant, whereas a G377A replacement was tolerated. Thus both a positive and a negative charge at this position are critical for activity (Fig. 6, D and E). Recently DXXK was identified as a novel motif, involved in catalysis of the oligosaccharyltransferase (43). We investigated two potential sequences of this kind and mutated Asp203, Lys206, Asp248, and Lys251 to alanine residues (Fig. 7). Because all mutations were able to complement the temperature growth defect of alg2, these amino acids do not seem to be important for the function of Alg2. Table 1 compiles the various mutations introduced.
FIGURE 5.
Sequence alignment of Alg2 and Alg11. Conserved amino acids are shaded, and amino acids altered by site-specific mutagenesis are framed by boxes.
FIGURE 6.
Site-specific mutagenesis of Alg2: exploring the EX7E motif and glycine 377 of the alg2 mutant. The mutations were introduced in the conserved EX7E motif of plasmid-borne Alg2ZZ as indicated and transformed into the alg2 mutant. Their role was investigated with respect to functional complementation on growth of alg2 cells (A), glycosylation of CPY monitored by Western blot (B), and Alg2 mannosyltransferase activity immunoprecipitated from solubilized extracts using assay II (C). In D and E mutations are shown at amino acid Gly377, which is altered to R in the alg2-1 mutant, and analyzed for growth (D) and glycosylation of CPY by Western blot analysis (E).
FIGURE 7.
Site-specific mutagenesis of Alg2: identification of Lys230 as essential amino acid for Alg2 function. Various amino acids, conserved in Alg2 and Alg11, with a potential role in Alg2 function were exchanged by site-specific mutagenesis as indicated. Plasmid-borne Alg2 was transformed into alg2 and tested for complementation of the growth defect of alg2 (A), of the underglycosylation of CPY analyzed by Western blot (B), and Alg2 mannosyltransferase activity, immunoprecipitated from solubilized extracts (C).
TABLE 1.
Growth phenotype and glycosylation defects of ALG2 mutations
The table summarizes the results of site-specific mutagenesis. To test the complementation of the temperature-sensitive growth defect of alg2, the cells were transformed with the mutated ALG2 and incubated at the restrictive temperature of 34 °C. For the analysis of the glycosylation defect, the glycosylation status of CPY was investigated by Western blot of cells grown at 34 °C. C, conserved amino acid between Alg2 and Alg11; EX7E, signature motif of some glycosyltransferase; DXXK, motif of the Stt3 subunit of oligosaccharyltransferase, postulated to be related to binding of the lipid-linked oligosaccharide donor.
| Mutants | Conservation/motif | Complementation of temperature-sensitive growth defect of alg2 | Complementation of glycosylation defect of alg2 |
|---|---|---|---|
| P192A | C | Yes | Yes |
| D203A | DXXK | Yes | Yes |
| K206A | DXXK | Yes | Yes |
| K210A | C | Yes | Yes |
| K229A | Yes | Yes | |
| K230A | C | No | No |
| D231A | Yes | Yes | |
| D248A | DXXK | Yes | Yes |
| K251A | C and DXXK | Yes | Yes |
| E264A | C | Yes | Yes |
| P359A | C | Yes | Yes |
| D392A | C | Yes | Yes |
| E335A | EX7E | Yes | Yes |
| H336A | EX7E | Yes | Yes |
| G338A | EX7E | Yes | Yes |
| E343A | EX7E | Yes | Yes |
| G377R | alg2-1 defect | alg2-1 defect | |
| G377E | No | No | |
| G377A | Yes | Yes |
DISCUSSION
The results presented in this paper demonstrate that ALG2 encodes a bifunctional glycosyltransferase, catalyzing both the α1,3- and the α1,6-mannose linkage onto Man1GlcNAc2-PP-Dol with formation of the trimannosyl core Man3GlcNAc2-PP-Dol (Fig. 8). This finding closes a gap in the biosynthesis of the common lipid-linked oligosaccharide of protein N-glycosylation in yeast and because of the evolutionary conservation of the pathway presumably applies also to higher eukaryotes. It may explain previous failures to identify candidate transferases by bioinformatic or other means. We first established an in vitro assay that allowed measuring the cytosolic oriented mannosylation steps leading from Man1GlcNAc2-PP-Dol to Man5 GlcNAc2-PP-Dol in a membrane detergent extract. An important breakthrough thereby was the finding of an acidic pH requirement for the Alg2 reaction. At a slightly alkaline value normally applied for glycosyltransferases, hardly any elongation took place. Subsequently, we show that an Alg2 immunoprecipitate from this extract is able to catalyze only the steps from Man1GlcNAc2-PP-Dol to Man3GlcNAc2-PP-Dol. The present study is in agreement with and extends a recent investigation expressing Alg2 in E. coli, implicating a dual function for Alg2 (22), but is also in conflict with some of the results described there. Bifunctionality of glycosyl transferases forming two linkages is rather unusual but not without precedent. Examples are hyaluron synthase, which has an β1,3-N-acetylglucosamine and a β1,4-glucuronic acid transferase activity (44, 45), FT85 from Dictyostelium, a glycosyltransferase involved in Skp1 glycosylation transferring both β1,3-galactosyl and an α1,2-fucosyl residues at its core (46), the E. coli enzyme KfiC responsible for the alternating α1,4-N-acetylglucosamine and β1,4-glucuronic acid addition to the nonreducing end of the capsular polysaccharide chain (47), or chondroitin synthases having a β1,3-N-acetylglucosamine and a β1,4-glucuronic acid transferase activity (45, 48–51). Surveying these transferases it is noticeable, however, that they are larger than Alg2, and a common feature of them is having two separate domains, which is not obvious in case of Alg2. In conjunction with the discussion of the dual function of glycosyltransferases, Alg9 and Alg11 involved in LLO assembly should also be mentioned. But these enzymes carry out two subsequent mannosylation steps with the same α1,2-linkage (22, 52) and are not forming two different linkages as in the case of Alg2. Similarly, in the Pgl pathway of protein N-glycosylation in the bacteria Campylobacter jejuni three α1,4-linked GalNAc moieties are transferred iteratively to the glycan chain by PglH transferase (53).
The precise function of Alg2 was obscured for a long time by the fact that the temperature-sensitive alg2 accumulates Man2GlcNAc2 (15) (see also Fig. 1), suggesting that Alg2 encodes the transfer of the third mannosyl residue. On the other hand a later study identified an alg2 mutant from the zygomycete Rhizomucor pusillus, resulting in truncation of Alg2 in the middle of the codon region. This mutation led to accumulation of Man1GlcNAc2-PP-Dol, which was transferred to the protein as largest N-glycan chain, and surprisingly, the fungus was viable in contrast to the yeast null mutant (54). Together with the experimentally determined enzyme activity of Alg2 here, one can now postulate that the temperature-sensitive alg2 yeast mutant allows a small extent mannose addition to Man1GlcNAc2-PP-Dol, because of some residual activity, and thus also results in accumulation of Man2GlcNAc2-PP-Dol, which is not the case in the null mutant of R. pussilis. In this argumentation also, results seamlessly incorporate, deriving from the analysis of the enzyme activity from human skin fibroblasts of a patient with a mutation in hAlg2 causing a defect in elongating Man1GlcNAc2-PP-Dol (21). In this study it was also shown that Man(1,6)ManGlcNAc2-PP-Dol tetrasaccharide did not act as acceptor in the patient but was elongated when membranes from control fibroblasts were used as enzyme source, albeit less efficiently than with Man1GlcNAc2-PP-Dol as acceptor (21). This latter observation holds also true for the Alg2 transferase from yeast (supplemental Fig. S3). Presumably Alg2 has a relaxed specificity in vitro with respect to the acceptor substrate, whereas in vivo the reaction sequence is from α1,3-linkage followed by α1,6-linkage (34, 35).
Glycosyltransferases can be classified into a range of families based on amino acid sequence similarities and also on account of structural homology (39, 55–58). ALG2 and ALG11 belong to glycosyltransferase family 4 of the CAZY data base and share 22% overall homology. An alignment of these two transferases identifies conserved amino acids including the C-terminal sequence motif EX7E (Fig. 5). To test its importance and also of other amino acids for Alg2 function, we performed a series of site-specific mutagenesis. The two glutamate residues of the EX7E motif have been proposed to be involved in catalysis as nucleophile but with mixed results. Whereas studies of the human muscle glycogen synthase (40) and AceA mannosyltransferase from Acetobacter xylinum (41) indicate that the first Glu residue is critical for enzyme activity, Gpi3, involved in glycosylphosphatidylinositol biosynthesis of S. cerevisiae, showed that the second Glu residue is of greater importance (42). As depicted in Fig. 6, the exchange of both Glu335 and Glu343 in this motif to alanine was not deleterious to Alg2 and complemented the growth defect and the underglycosylation of CPY of the alg2 mutant and was also active in vitro in an Alg2 immunoprecipitate as enzyme source. This indicates that the glutamate residues in EX7E are not important for Alg2 activity (Fig. 6). Our results are in contrast to the finding expressing Alg2 in E. coli. In this study it was reported that E343A is inactive, and E335A had a much lower level of activity (22). The reason for the discrepancy is unclear. On the other hand we detect Lys230, also conserved in Alg11, as an essential amino acid required for Alg2 activity (Fig. 7). A lysine residue has been proposed to function in nucleotide sugar binding in the case of AceA and found to be critical for its activity (41). In addition we replaced several other lysine residues, e.g. Lys229, adjacent to the critical Lys230 as well as lysine residues Lys206 and Lys251 of a potential DXXK motif. This signature has been identified recently in the catalytic Stt3 subunit of oligosaccharyltransferase. Likewise conserved aspartate residues were mutagenized, which have also been discussed as nucleophiles by deprotonating the hydroxyl group of the acceptor, acting then as catalytic base and making a nucleophilic attack on the C1 atom of the sugar donor. None of these residues (Fig. 7 and Table 1) seems to be of importance for Alg2 function and thus underscoring the significance of Lys230 for function. It should be mentioned that the in vivo function of the various mutants described above was assessed in the temperaturesensitive alg2 mutant by rescuing the growth and underglycosylation defect at the restrictive temperature. Thus it is formally possible that an Δalg2 disruptant might not be rescued, because the various mutants may somehow enhance or stabilize the Alg2(G377R) protein of the alg2 mutant. Because for most of the site-directed engineered Alg2 variants the enzyme activity was also determined in vitro in Alg2 immunoprecipitates, this possibility is less likely for these cases.
Finally we had a closer look to residue Gly377, which is mutated to Arg377 in the yeast alg2 mutant. Interestingly, replacing Gly368 by arginine in the ALG2 from Rhizomucor (equivalent to the Gly377 of yeast Alg2) resulted in generation of a temperature-sensitive enzyme, too (28). We have replaced Gly377 also by the negatively charged amino acid glutamate and by a less detrimental alanine residue (Fig. 6). We find that Glu abrogates Alg2 activity similar to Arg, whereas an exchange to Ala is tolerated. The data suggest that Gly seems to be also critical for activity, and neither a negatively or positively charged amino acid is tolerated and most probably leads to structural perturbation. On the other hand exchange of the conserved Pro192 and Pro359 residues that are expected to have a strong effect on the protein architecture do not affect Alg2 function.
Another major issue of the study was the investigation of the membrane topology of Alg2, which has not been investigated before. Contradictory to topology predictions, Alg2 was proven to consist of only two transmembrane-spanning domains located at the N terminus region between amino acid residues 1 and 145. However, these domains were demonstrated to be not essential for Alg2 function (Fig. 4) and could be deleted. Most surprisingly, this truncated Alg2 variant was still anchored to the membrane, obviously by the two predicted hydrophobic regions at the C-terminal end. Although not acting as TMD, as indicated from topology prediction, we experimentally demonstrate by alkaline carbonate and detergent extraction that they contribute to efficient membrane integration rather than to a merely peripheral association. We could not decide, whether only one or both of them are engaged, because construct Alg2159–465, in which the two TMDs and the fourth hydrophobic region are lacking (Fig. 4A), could not be detected by Western blot analysis. Obviously this truncated variant is not stable anymore. However, it was demonstrated that deletion of both the third and the fourth hydrophobic region (construct Alg21–440) is dispensable for activity, when Alg2 is anchored by the N-terminal TMDs (Fig. 4, B–D). These data suggest that the catalytic active part of the transferase must be in between amino acid residues 159 and 440.
The topology model B proposed is based on two different topology-sensitive reporter fusions of C-terminal truncated Alg2 variants. The results show that both the third and the fourth hydrophobic helix are accessible from the cytosolic face. Thus they are not likely to span the membrane, considering also the proven orientation of transmembrane domains 1 and 2. Although protein predictions have made an enormous progress during the last years, there is still not much known about the molecular details of how membrane helices are recognized and how they are oriented in the correct way in the membrane. Membrane-embedded helices can be long, be short, cross the membrane at oblique angles, lie flat on the surface, or even span only part of the bilayer and then turn back (59). Our experimental results do not allow qualifying the exact mode of localization of the two C-terminal domains. A helical wheel projection of membrane segments 3 and 4 (data not shown) reveals a concentration of hydrophobic and of more polar amino acid residues on two different sites of the helix. This may suggest an interaction with the surface and the nonpolar interior of the membrane. Association of hydrophobic segments with the membrane leaflet similar as described here have been discussed for Ost2, a subunit of the oligosaccharyltransferase (60), or the Sec61α and Sec61γ subunit of the translocon (61, 62). Moreover, insertion of monotopic proteins is often accomplished by hydrophobic helices interacting with one leaflet of the membrane (63–68).
Further work will be necessary to understand mechanistically how one and the same enzyme can accomplish catalyzing two different linkages and altering the specificity of the active site from the Man(β1,4)GlcNAc2 trisaccharide acceptor to the newly formed tetrasaccharide intermediate Man(α1,3) Man(β1,4)GlcNAc2. It is known already that small conformational changes can lead to dramatic alterations in the specificity. Thus an exchange of only one amino acid, Tyr289, in bovine β1,4-galactosyltransferase (or Tyr286 in human β1,4-galactosyltransferase) deter-mines enzyme specificity. Mutating Tyr289 to leucine, isoleucine, or asparagine made the Gal-transferase to a GalNAc-transferase (69). Furthermore, studies on several glycosyl transferases have shown that one or two flexible loops at the substrate binding site of the enzyme undergo marked conformational changes from an open to a closed conformation, in which the loop acts as a lid covering the bound donor (70). This flexibility may create a new acceptor-binding site or modulate it in combination with the conformational properties of the newly synthesized Man2GlcNAc2-PP-Dol oligosaccharide intermediate, thus altering Alg2 acceptor specificity.
Supplementary Material
Acknowledgments
We acknowledge the excellent technical assistance of Angelika Rechenmacher. We are also grateful to Verena Schmeiser for the contribution to the mutagenesis experiments and to Dr. J. Stolz for providing plasmids and helpful discussions. We also thank Dr. H. Kim for providing plasmid pJK90.
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Körber-Stiftung.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; CPY, carboxypeptidase Y; LLO, lipid-linked oligosaccharide; ZZ, protein A; TMD, transmembrane-spanning domain; HPLC, high pressure liquid chromatography; MES, 4-morpholineethanesulfonic acid; PIPES, 1,4-piperazinediethanesulfonic acid.
References
- 1.Burda, P., and Aebi, M. (1999) Biochim. Biophys. Acta 1426 239–257 [DOI] [PubMed] [Google Scholar]
- 2.Lehle, L., Strahl, S., and Tanner, W. (2006) Angew. Chem. Int. Ed. Engl. 45 6802–6818 [DOI] [PubMed] [Google Scholar]
- 3.Kornfeld, R., and Kornfeld, S. (1985) Annu. Rev. Biochem. 54 631–664 [DOI] [PubMed] [Google Scholar]
- 4.Helenius, A., and Aebi, M. (2004) Annu. Rev. Biochem. 73 1019–1049 [DOI] [PubMed] [Google Scholar]
- 5.Wacker, M., Linton, D., Hitchen, P. G., Nita-Lazar, M., Haslam, S. M., North, S. J., Panico, M., Morris, H. R., Dell, A., Wren, B. W., and Aebi, M. (2002) Science 298 1790–1793 [DOI] [PubMed] [Google Scholar]
- 6.Upreti, R. K., Kumar, M., and Shankar, V. (2003) Proteomics 3 363–379 [DOI] [PubMed] [Google Scholar]
- 7.Weerapana, E., and Imperiali, B. (2006) Glycobiology 16 91R–101R [DOI] [PubMed] [Google Scholar]
- 8.Kelleher, D. J., and Gilmore, R. (2006) Glycobiology 16 47R–62R [DOI] [PubMed] [Google Scholar]
- 9.Knauer, R., and Lehle, L. (1999) Biochim. Biophys. Acta 1426 259–273 [DOI] [PubMed] [Google Scholar]
- 10.Yan, A., and Lennarz, W. J. (2005) J. Biol. Chem. 280 3121–3124 [DOI] [PubMed] [Google Scholar]
- 11.Helenius, J., Ng, D. T., Marolda, C. L., Walter, P., Valvano, M. A., and Aebi, M. (2002) Nature 415 447–450 [DOI] [PubMed] [Google Scholar]
- 12.Sanyal, S., Frank, C. G., and Menon, A. K. (2008) Biochemistry 47 7937–7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burda, P., Jakob, C. A., Beinhauer, J., Hegemann, J. H., and Aebi, M. (1999) Glycobiology 9 617–625 [DOI] [PubMed] [Google Scholar]
- 14.Huffaker, T. C., and Robbins, P. W. (1982) J. Biol. Chem. 257 3203–3210 [PubMed] [Google Scholar]
- 15.Huffaker, T. C., and Robbins, P. W. (1983) Proc. Natl. Acad. Sci. U. S. A. 80 7466–7470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Runge, K. W., Huffaker, T. C., and Robbins, P. W. (1984) J. Biol. Chem. 259 412–417 [PubMed] [Google Scholar]
- 17.Runge, K. W., and Robbins, P. W. (1986) J. Biol. Chem. 261 15582–15590 [PubMed] [Google Scholar]
- 18.Beck, P. J., Gething, M. J., Sambrook, J., and Lehrman, M. A. (1990) Somat. Cell Mol. Genet. 16 539–548 [DOI] [PubMed] [Google Scholar]
- 19.Hall, C. W., McLachlan, K. R., Krag, S. S., and Robbins, A. R. (1997) J. Cell. Biochem. 67 201–215 [PubMed] [Google Scholar]
- 20.Zeng, Y., and Lehrman, M. A. (1991) Anal. Biochem. 193 266–271 [DOI] [PubMed] [Google Scholar]
- 21.Thiel, C., Schwarz, M., Peng, J., Grzmil, M., Hasilik, M., Braulke, T., Kohlschutter, A., von Figura, K., Lehle, L., and Korner, C. (2003) J. Biol. Chem. 278 22498–22505 [DOI] [PubMed] [Google Scholar]
- 22.O'Reilly, M. K., Zhang, G., and Imperiali, B. (2006) Biochemistry 45 9593–9603 [DOI] [PubMed] [Google Scholar]
- 23.Kim, H., Yan, Q., Von Heijne, G., Caputo, G. A., and Lennarz, W. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7460–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strahl-Bolsinger, S., and Scheinost, A. (1999) J. Biol. Chem. 274 9068–9075 [DOI] [PubMed] [Google Scholar]
- 25.Knauer, R., and Lehle, L. (1999) J. Biol. Chem. 274 17249–17256 [DOI] [PubMed] [Google Scholar]
- 26.Knauer, R., and Lehle, L. (1994) FEBS Lett. 344 83–86 [DOI] [PubMed] [Google Scholar]
- 27.Sharma, C. B., Knauer, R., and Lehle, L. (2001) Biol. Chem. 382 321–328 [DOI] [PubMed] [Google Scholar]
- 28.Yamazaki, H., Shiraishi, N., Takeuchi, K., Ohnishi, Y., and Horinouchi, S. (1998) Gene (Amst.) 221 179–184 [DOI] [PubMed] [Google Scholar]
- 29.Sharma, C. B., Lehle, L., and Tanner, W. (1982) Eur. J. Biochem. 126 319–325 [DOI] [PubMed] [Google Scholar]
- 30.Bickel, T., Lehle, L., Schwarz, M., Aebi, M., and Jakob, C. A. (2005) J. Biol. Chem. 280 34500–34506 [DOI] [PubMed] [Google Scholar]
- 31.Kean, E. L. (1983) Biochim. Biophys. Acta 750 268–273 [DOI] [PubMed] [Google Scholar]
- 32.Kean, E. L., Wei, Z., Anderson, V. E., Zhang, N., and Sayre, L. M. (1999) J. Biol. Chem. 274 34072–34082 [DOI] [PubMed] [Google Scholar]
- 33.Heifetz, A., and Elbein, A. D. (1977) J. Biol. Chem. 252 3057–3063 [PubMed] [Google Scholar]
- 34.Chapman, A., Li, E., and Kornfeld, S. (1979) J. Biol. Chem. 254 10243–10249 [PubMed] [Google Scholar]
- 35.Vijay, I. K., and Perdew, G. H. (1982) Eur. J. Biochem. 126 167–172 [DOI] [PubMed] [Google Scholar]
- 36.Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001) J. Mol. Biol. 305 567–580 [DOI] [PubMed] [Google Scholar]
- 37.Stolz, J., Ludwig, A., and Sauer, N. (1998) FEBS Lett. 440 213–217 [DOI] [PubMed] [Google Scholar]
- 38.Sengstag, C., Stirling, C., Schekman, R., and Rine, J. (1990) Mol. Cell. Biol. 10 672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bourne, Y., and Henrissat, B. (2001) Curr. Opin. Struct. Biol. 11 593–600 [DOI] [PubMed] [Google Scholar]
- 40.Cid, E., Gomis, R. R., Geremia, R. A., Guinovart, J. J., and Ferrer, J. C. (2000) J. Biol. Chem. 275 33614–33621 [DOI] [PubMed] [Google Scholar]
- 41.Abdian, P. L., Lellouch, A. C., Gautier, C., Ielpi, L., and Geremia, R. A. (2000) J. Biol. Chem. 275 40568–40575 [DOI] [PubMed] [Google Scholar]
- 42.Kostova, Z., Yan, B. C., Vainauskas, S., Schwartz, R., Menon, A. K., and Orlean, P. (2003) Eur. J. Biochem. 270 4507–4514 [DOI] [PubMed] [Google Scholar]
- 43.Igura, M., Maita, N., Kamishikiryo, J., Yamada, M., Obita, T., Maenaka, K., and Kohda, D. (2008) EMBO J. 27 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jing, W., and DeAngelis, P. L. (2000) Glycobiology 10 883–889 [DOI] [PubMed] [Google Scholar]
- 45.Jing, W., and DeAngelis, P. L. (2003) Glycobiology 13 661–671 [DOI] [PubMed] [Google Scholar]
- 46.Van Der Wel, H., Fisher, S. Z., and West, C. M. (2002) J. Biol. Chem. 277 46527–46534 [DOI] [PubMed] [Google Scholar]
- 47.Griffiths, G., Cook, N. J., Gottfridson, E., Lind, T., Lidholt, K., and Roberts, I. S. (1998) J. Biol. Chem. 273 11752–11757 [DOI] [PubMed] [Google Scholar]
- 48.Yada, T., Gotoh, M., Sato, T., Shionyu, M., Go, M., Kaseyama, H., Iwasaki, H., Kikuchi, N., Kwon, Y. D., Togayachi, A., Kudo, T., Watanabe, H., Narimatsu, H., and Kimata, K. (2003) J. Biol. Chem. 278 30235–30247 [DOI] [PubMed] [Google Scholar]
- 49.Yada, T., Sato, T., Kaseyama, H., Gotoh, M., Iwasaki, H., Kikuchi, N., Kwon, Y. D., Togayachi, A., Kudo, T., Watanabe, H., Narimatsu, H., and Kimata, K. (2003) J. Biol. Chem. 278 39711–39725 [DOI] [PubMed] [Google Scholar]
- 50.Ninomiya, T., Sugiura, N., Tawada, A., Sugimoto, K., Watanabe, H., and Kimata, K. (2002) J. Biol. Chem. 277 21567–21575 [DOI] [PubMed] [Google Scholar]
- 51.Hwang, H. Y., Olson, S. K., Esko, J. D., and Horvitz, H. R. (2003) Nature 423 439–443 [DOI] [PubMed] [Google Scholar]
- 52.Frank, C. G., and Aebi, M. (2005) Glycobiology 15 1156–1163 [DOI] [PubMed] [Google Scholar]
- 53.Glover, K. J., Weerapana, E., and Imperiali, B. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14255–14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takeuchi, K., Yamazaki, H., Shiraishi, N., Ohnishi, Y., Nishikawa, Y., and Horinouchi, S. (1999) Glycobiology 9 1287–1293 [DOI] [PubMed] [Google Scholar]
- 55.Breton, C., Snajdrova, L., Jeanneau, C., Koca, J., and Imberty, A. (2006) Glycobiology 16 29R–37R [DOI] [PubMed] [Google Scholar]
- 56.Kapitonov, D., and Yu, R. K. (1999) Glycobiology 9 961–978 [DOI] [PubMed] [Google Scholar]
- 57.Coutinho, P. M., Deleury, E., Davies, G. J., and Henrissat, B. (2003) J. Mol. Biol. 328 307–317 [DOI] [PubMed] [Google Scholar]
- 58.Campbell, J. A., Davies, G. J., Bulone, V., and Henrissat, B. (1997) Biochem. J. 326 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Heijne, G. (2006) Nat. Rev. Mol. Cell Biol. 7 909–918 [DOI] [PubMed] [Google Scholar]
- 60.Silberstein, S., Collins, P. G., Kelleher, D. J., and Gilmore, R. (1995) J. Cell Biol. 131 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilkinson, B. M., Critchley, A. J., and Stirling, C. J. (1996) J. Biol. Chem. 271 25590–25597 [DOI] [PubMed] [Google Scholar]
- 62.Blobel, G. (2000) Chembiochem. 1 86–102 [DOI] [PubMed] [Google Scholar]
- 63.Zakharov, S. D., and Cramer, W. A. (2002) Biochimie (Paris) 84 465–475 [DOI] [PubMed] [Google Scholar]
- 64.Zhang, J., Frerman, F. E., and Kim, J. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 16212–16217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Picot, D., Loll, P. J., and Garavito, R. M. (1994) Nature 367 243–249 [DOI] [PubMed] [Google Scholar]
- 66.Bracey, M. H., Hanson, M. A., Masuda, K. R., Stevens, R. C., and Cravatt, B. F. (2002) Science 298 1793–1796 [DOI] [PubMed] [Google Scholar]
- 67.Partridge, A. W., Melnyk, R. A., Yang, D., Bowie, J. U., and Deber, C. M. (2003) J. Biol. Chem. 278 22056–22060 [DOI] [PubMed] [Google Scholar]
- 68.Blobel, G. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 1496–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramakrishnan, B., and Qasba, P. K. (2002) J. Biol. Chem. 277 20833–20839 [DOI] [PubMed] [Google Scholar]
- 70.Qasba, P. K., Ramakrishnan, B., and Boeggeman, E. (2005) Trends Biochem. Sci. 30 53–62 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.