Abstract
Lipoprotein lipase (LPL) is a principal enzyme responsible for the clearance of chylomicrons and very low density lipoproteins from the bloodstream. Two members of the Angptl (angiopoietin-like protein) family, namely Angptl3 and Angptl4, have been shown to inhibit LPL activity in vitro and in vivo. Here, we further investigated the structural basis underlying the LPL inhibition by Angptl3 and Angptl4. By multiple sequence alignment analysis, we have identified a highly conserved 12-amino acid consensus motif that is present within the coiled-coil domain (CCD) of both Angptl3 and Angptl4, but not other members of the Angptl family. Substitution of the three polar amino acid residues (His46, Gln50, and Gln53) within this motif with alanine abolishes the inhibitory effect of Angptl4 on LPL in vitro and also abrogates the ability of Angptl4 to elevate plasma triglyceride levels in mice. The CCD of Angptl4 interacts with LPL and converts the catalytically active dimers of LPL to its inactive monomers, whereas the mutant protein with the three polar amino acids being replaced by alanine loses such a property. Furthermore, a synthetic peptide consisting of the 12-amino acid consensus motif is sufficient to inhibit LPL activity, although the potency is much lower than the recombinant CCD of Angptl4. In summary, our data suggest that the 12-amino acid consensus motif within the CCD of Angptl4, especially the three polar residues within this motif, is responsible for its interaction with and inhibition of LPL by blocking the enzyme dimerization.
Lipoprotein lipase (LPL)3 is an endothelium-bound enzyme that catalyzes the hydrolysis of plasma triglyceride (TG) associated with chylomicrons and very low density lipoproteins (1, 2). This enzyme plays a major role in maintaining lipid homeostasis by promoting the clearance of TG-rich lipoproteins from the bloodstream. Abnormality in LPL functions has been associated with a number of pathological conditions, including atherosclerosis, dyslipidemia associated with diabetes, and Alzheimer disease (1).
LPL is expressed in a wide variety of cell types, particularly in adipocytes and myocytes (2). As a rate-limiting enzyme for clearance of TG-rich lipoproteins, the activity of LPL is tightly modulated by multiple mechanisms in a tissue-specific manner in response to nutritional changes (3, 4). The enzymatic activity of LPL in adipose tissue is enhanced after feeding to facilitate the storage of TG, whereas it is down-regulated during fasting to increase the utilization of TG by other tissues (5). The active form of LPL is a noncovalent homodimer with the subunits associated in a head-to-tail manner, and the dissociation of its dimeric form leads to the formation of a stable inactive monomeric conformation and irreversible enzyme inactivation (6). At the post-translational level, the LPL activity is regulated by numerous apolipoprotein co-factors. For instance, apoCII, a small apolipoprotein consisting of 79 amino acid residues in human, activates LPL by directly binding to the enzyme (7, 8). By contrast, several other apolipoproteins such as apoCI, apo-CIII, and apoE have been shown to inhibit the LPL activity in vitro (3).
Angiopoietin-like proteins (Angptl) are a family of secreted proteins consisting of seven members, Angptl1 to Angptl7 (9, 10). All the members of the Angptl family share a similar domain organization to those of angiopoietins, with an NH2-terminal coiled-coil domain (CCD) and a COOH-terminal fibrinogen-like domain. Among the seven family members, only Angptl3 and Angptl4 have been shown to be involved in regulating triglyceride metabolism (10, 11). The biological functions of Angptl3 in lipid metabolism were first discovered by Koishi et al. (12) in their positional cloning of the recessive mutation gene responsible for the hypolipidemia phenotype in a strain of obese mouse KK/snk. Subsequent studies have demonstrated that Angptl3 increases plasma TG levels by inhibiting the LPL enzymatic activity (13–15). Angptl4, also known as fasting-induced adipocyte factor, hepatic fibrinogen/angiopoietin-related protein, or peroxisome proliferator-activated receptor-γ angiopoietin-related, is a secreted glycoprotein abundantly expressed in adipocyte, liver, and placenta (16–18). In addition to its role in regulating angiogenesis, a growing body of evidence demonstrated that Angptl4 is an important player of lipid metabolism (10, 11). Elevation of circulating Angptl4 by transgenic or adenoviral overexpression, or by direct supplementation of recombinant protein, leads to a marked elevation in the levels of plasma TG and low density lipoprotein cholesterol in mice (19–22). By contrast, Angptl4 knock-out mice exhibit much lower plasma TG and cholesterol levels compared with the wild type littermates (19, 20). Notably, treatment of several mouse models (such as C57BL/6J, ApoE–/–, LDLR–/–, and db/db obese/diabetic mice) with a neutralizing antibody against Angptl4 recapitulate the lipid phenotype found in Angptl4 knock-out mice (19). The role of Angptl4 as a physiological inhibitor of LPL is also supported by the finding that its expression levels in adipose tissue change rapidly during the fed-to-fasting transitions and correlate inversely with LPL activity (23). In humans, a genetic variant of the ANGPTL4 gene (E40K) has been found to be associated with significantly lower plasma TG levels and higher high density lipoprotein cholesterol concentrations in several ethnic groups (24–26).
Angptl3 and Angptl4 share many common biochemical and functional properties (10). In both humans and rodents, Angptl3 and Angptl4 are proteolytically cleaved at the linker region and circulate in plasma as two truncated fragments, including NH2-terminal CCD and COOH-terminal fibrinogen-like domain (14, 27–29). The effects of both Angptl3 and Angptl4 on elevating plasma TG levels are mediated exclusively by their NH2-terminal CCDs (15, 22, 23, 27, 30). The CCDs of Angptl3 and Angptl4 have been shown to inhibit the LPL activity in vitro as well as in mice (23,30,31). Angptl4 inhibits LPL by promoting the conversion of the catalytically active LPL dimers into catalytically inactive LPL monomers, thereby leading to the inactivation of LPL (23, 31). However, the detailed structural and molecular basis underlying the LPL inhibition by Angptl3 and Angptl4 remain poorly characterized at this stage.
In this study, we analyzed all known amino acid sequences of Angptl3 and Angptl4 from various species and found a short motif, LAXGLLXLGXGL (where X represents polar amino acid residues), which corresponds to amino acid residues 46–57 and 44–55 of human Angptl3 and Angptl4, respectively, is highly conserved despite the low degree of their overall homology (∼30%). Using both in vitro and in vivo approaches, we demonstrated that this 12-amino acid sequence motif, in particular the three polar amino acid residue within this motif, is essential for mediating the interactions between LPL and Angpt4, which in turn disrupts the dimerization of the enzyme.
EXPERIMENTAL PROCEDURES
Materials—Restriction endonucleases were from New England Biolabs (Ipswich, MA). T4 DNA ligase and ImProm-II™ Reverse Transcription System were purchased from Promega (Madison, WI). Cell culture media, fetal bovine serum, and cloning vector pPROEx-Htb were obtained from Invitrogen. All oligonucleotide primers used for cloning and mutagenesis were synthesized by Techdragon Co. (Hong Kong, China). PS20 ProteinChip® array and all PCRs were performed with iProof High Fidelity PCR system from Bio-Rad. QIAprep™ miniprep kit, QIAquick™ gel extraction kit, and Ni2+-nitrilotriacetic acid affinity gel beads were from Qiagen GmbH (Hilden, Germany). Escherichia coli BL21(DE3)-codon plus RILP-competent cell was from Stratagene (La Jolla, CA). Human recombinant Angptl3-CCD was kindly provided by the Biovendor Laboratory Medicine, Inc. (Modrice, Czech Republic). Intralipid 10% emulsion (soybean triacylglycerol emulsified in egg yolk phospholipids) was from KABI-Fresenius (Uppsala, Sweden). [3H]Triolein was obtained from American Radiolabeled Chemicals Inc. (St. Louis, MO). Isopropyl β-d-thiogalactoside, purified bovine LPL, sinapinic acid, and the synthetic peptides were from Sigma. 3T3-L1 and HepG2 cells were from ATCC (Manassas, VA). Detoxi-Gel™ endotoxin removing gel was from Pierce.
Plasmid Construction—The first strand cDNA for human Angptl4-CCD was prepared by reverse transcription PCR of total RNA extracted from HepG2 cells using primers as listed in supplemental Table 1. Restriction endonuclease cleavage sites SalI and XhoI were added to the 5′ and 3′ ends of the amplified fragment, respectively. The fragment was subcloned into pPROEX-HTb bacterial expression vector, which generated an in-frame fusion with an NH2-terminal His6 tag. The construct was verified by DNA sequencing and was used for the production of recombinant protein as well as the template for subsequent site-directed mutagenesis. All the mutants were generated by overlapping PCR mutagenesis using the oligonucleotide primers listed in supplemental Table 1. The restriction site at the 5′ end was changed to StylI instead of SalI after amplification using the 5′ primer “hA4-F-StyI” by the iProof High Fidelity PCR system following the manufacturer's instruction for the GC-rich conditions. The purified gene fragments were digested and cloned into pPROEX-HTb vector digested with StyI and XhoI, and the constructs were confirmed by DNA sequencing analysis as described above.
Expression and Purification of Recombinant Angptl4-CCD—The constructs encoding either wild type human Angptl4-CCD or its variants were transformed into E. coli BL21(DE3)-codon plus RILP-competent cells for protein expression as described (32). Cells were harvested and lysed by pulsed sonication in 50 mm sodium phosphate, 0.3 m NaCl, 5 mm imidazole, 20% glycerol (pH 6.5). After removing the debris by centrifugation, the lysates were loaded onto a Ni2+-nitrilotriacetic acid column pre-equilibrated with the same buffer. The wild type or mutated hAngptl4-CCD was eluted with 150 mm imidazole after extensive washing, concentrated by ultrafiltration, and dialyzed overnight at 4 °C against 50 mm sodium phosphate (pH 6.5). Endotoxin was removed by Detoxi-Gel™ according to the manufacturer's instructions.
Lipase Activity Assay—LPL activity was measured using a [3H]triolein-incorporated phospholipid emulsion of soybean triacylglycerols (10% Intralipid®) as described (33). All the reactions were performed using fetal calf serum as a source of apo-CII unless otherwise stated. The reaction mix (100 μl) was incubated at 30 °C for 1 h with gentle shaking. Free fatty acids, the main product of the reaction, were extracted by Dole's extraction method (34). Briefly, after the reaction was terminated by addition of 1 ml of Dole's solution (isopropyl alcohol:heptane: H2SO4, 40:48.3:1, v/v), 0.25 ml of H2O was added and mixed vigorously by vortexing. Following separation of the two phases by centrifugation at 1,500 × g for 5 min, 0.4 ml of upper layer was transferred to another fresh tube containing a 0.5-ml solution of alkaline ethanol (50% ethanol in 50 mm NaOH). Triacylglycerols were extracted twice by addition of 3 ml of heptane followed by vigorously mixing and phase separation. The amount of [3H]oleic acid in the lower aqueous phase was measured by determining the radioactivity using Liquid Scintillation Analyzer TRI-CARB 2900TR (PerkinElmer Life Sciences). One milliunit of lipase activity is equal to 1 nmol of fatty acids produced per min. All samples were measured at least in triplicate.
Cell Culture and Treatment—3T3-L1 preadipocytes were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and were induced for differentiation as described (35). Fully differentiated 3T3-L1 adipocytes were treated with different concentrations of wild type or mutant hAngptl4-CCD proteins for 4 h, and LPL activity in both secreted and surface-bound fractions was measured as above. The cell surface-bound LPL was released by incubating the sample with 0.5 ml of Dulbecco's modified Eagle's medium containing 0.2% BSA, 100 units/ml heparin for 30 min at 4 °C.
Mouse Serum TG Measurement—C57BL/6J mice were obtained from the Laboratory Animal Unit, and all animal experimental protocols were approved by the Animal Ethics Committee at the University of Hong Kong. Male C57BL/6J mice were housed at 25 °C under 12-h dark/12-h light cycle and had free access to water and standard rodent chows. The mice were fasted for 4 h prior to administration of recombinant wild type or mutant hAngptl4-CCD proteins (50 μg) through tail vein injection. The same volume of PBS was injected as the control. After injection, the mice were kept fasted throughout the experiment, and blood samples were collected from mice at various time points for measurement of serum TG using a commercial TG assay kit (Leadman Bioscience, Beijing, China).
Matrix-assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometric (MALDI-TOF MS) Analysis—MALDI-TOF MS experiments were performed using a 4800 MALDI TOF/TOF™ mass spectrometer (Applied Biosystems) equipped with a solid state OptiBeam laser (355 nm wavelength, 200 Hz). Briefly, bovine LPL (0.3 μg, Sigma) was incubated at room temperature with 1.5 μg of BSA, hAngptl4-CCD, or its mutant protein in a total volume of 10 μl of 20 mm Tris-HCl, 0.15 m NaCl (TBS) (pH 7.4), for 1 h. The protein samples were mixed with the sinapinic acid matrices (10 mg/ml in a 1:1 mixture of acetonitrile, 0.5% trifluoroacetic acid (v/v)) and applied immediately onto the 384-well specimen holder plates (Applied Biosystems). For MS analysis, positive ion spectra were acquired in linear mode using the following parameters: laser, 4000; laser shots, 25 × 20 subspectra; delayed time, 1500 ns; m/z range, 12,000–120,000. External calibration was performed with the Calmix3 provided by the manufacturer.
Surface-enhanced Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (SELDI-TOF MS)—A modified specimen holder plate that can hold three protein chips for analysis on 4800 MALDI TOF/TOF™ mass spectrometer was manufactured by Technology Support Centre of the University of Hong Kong with the permission from Applied Biosystems. All the protein-protein interaction experiments were performed using the SELDI protein chips with a pre-activated surface (PS20 Protein-Chip® array, Bio-Rad). Briefly, bovine LPL (5 μg) in 5 μl of PBS was immobilized onto spots of the PS20 protein chips as described before (36). After overnight coupling at 4 °C, free active sites on the spots were blocked with 0.5 m Tris-HCl (pH 8.0) for 2 h at room temperature before sequential washes with PBS containing 0.5% Triton X-100 followed by PBS alone. Subsequently, 500 ng of BSA, hAngptl4-CCD, or the mutant protein dissolved in PBS were applied onto the chip and incubated in a moist chamber at room temperature for 1 h. The unbound proteins were washed twice with PBS and rinsed with water. Saturated sinapinic acid in 50% acetonitrile and 0.5% trifluoroacetic acid was applied to the spot surface after air drying, and then the samples were analyzed on the 4800 MALDI TOF/TOF™ mass spectrometer using the settings described above. All of the spectra were externally calibrated using Calmix3.
Biotinylation and Separation of Bovine LPL by Sucrose Density Gradient Centrifugation—100 μg of bovine LPL was biotinylated with a Sulfo-NHS-Biotin kit (Pierce) and was then re-purified by sucrose density gradient ultracentrifugation to separate low density lipoprotein dimers and monomers as described elsewhere (23, 37). Briefly, 5–20% sucrose density gradient was made up in a buffer containing 20 mm Tris-HCl (pH 7.4), 2 m NaCl, and 2 mg/ml BSA. The total gradient volume was 3.6 ml. Biotinylated LPL was loaded into each tube, followed by centrifugation using a Beckman Coulter SW60 rotor at 270,000 × g for 16 h at 10 °C. After centrifugation, fractions of 0.24 ml were collected by puncturing the bottom of each centrifugation tube with a needle that was attached with tubing to a pump. Fractions that contain LPL dimers were collected as described (23, 37), pooled, and concentrated using a concentrator with molecular mass cutoff of 10,000 Da (Vivascience AG, Hannover, Germany). The protein samples were suspended in a buffer containing 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 2% BSA for further analysis.
Heparin-Sepharose Chromatography—Biotinylated LPL prepared as described above was subjected to heparin-Sepharose chromatography using an AKTA explorer fast protein chromatography system (GE Healthcare) as described (31, 37) with slight modifications. Briefly, 5 μg of biotinylated LPL was incubated with 1 ml of heparin-Sepharose beads in a capped tube with rotation for 1 h at 4 °C to allow the binding of LPL to the beads. The samples were then subjected to a brief centrifugation, followed by washing three times with 10 ml of buffer containing 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 2% BSA to remove unbound LPL. Afterward, the beads were incubated with 2 ml of the same buffer containing hAngptl4-CCD, Angptl3-CCD, or their mutants with shaking for another 2 h at 16 °C. The beads were collected by a brief centrifugation, washed three times as above, and placed into an empty chromatography column. LPL bound to the beads was eluted by a linear gradient of NaCl (from 0.20 to 1.8 m) in the same buffer. Proteins in each fraction were precipitated by 10% trichloroacetic acid, separated by SDS-PAGE, transferred to nylon membrane, and then probed with horseradish peroxidase-conjugated streptavidin to visualize biotinylated LPL.
RESULTS
NH2-terminal Coiled-coil Domains of Angptl3 and Angptl4 Differentially Inhibit the LPL Activity in Vitro—Recombinant human Angptl4-CCD with an NH2-terminal His6 tag was expressed in E. coli and purified as described under “Experimental Procedures.” SDS-PAGE analysis of the purified hAngptl4-CCD showed an apparent single band with a molecular mass of ∼21 kDa under a reducing condition (Fig. 1A, lane 3). In contrast, three bands with the molecular masses of ∼21, 42, and 85 kDa, which corresponded to the monomeric, dimeric, and tetrameric forms of hAngptl4-CCD, respectively, were separated by a nonreducing SDS-PAGE (Fig. 1A, lane 4) suggesting the formation of hAngptl4-CCD oligomers via disulfide bonds. On the other hand, SDS-PAGE analysis of the recombinant human Angptl3 CCD (hAngptl3-CCD) revealed an apparent single band under both reducing and nonreducing conditions (Fig. 1A, lanes 1 and 2).
FIGURE 1.
NH2-terminal CCDs of Angptl4 and Angptl3 inhibit the activities of LPL with different potencies. A, SDS-PAGE analysis of recombinant hAngptl3-CCD and hAngptl4-CCD. 3 μg of proteins per lane were separated under either reducing or nonreducing conditions by 12% SDS-PAGE. For DTT treatment, purified recombinant hAngptl4-CCD was incubated with 20 mm DTT for 1 h at room temperature to disrupt all the disulfide linkages. B, measurement of LPL activities in the presence of recombinant hAngptl3-CCD, hAngptl4-CCD, or DTT-treated hAngptl4-CCD. Note that DTT was completely removed by extensive dialysis before use. C, effects of hAngptl4-CCD on LPL activities stimulated with different concentrations of apoCIII. D, percentage of LPL activity remained after addition of different concentrations of hAngptl4-CCD. n = 4–5 in each group.
Both hAngptl4-CCD and hAngptl3-CCD showed dose-dependent inhibitory effects on LPL activities with IC50 values of 0.2 and 5 μg/ml, respectively (Fig. 1B). At a concentration of 1 μg/ml, hAngptl4-CCD almost completely inhibited the activities of LPL, whereas the same concentration of hAngptl3-CCD only caused a modest inhibition of this enzyme. Treatment with DTT (20 mm) completely eliminated the disulfide linkages of hAngptl4-CCD oligomers and converted the oligomeric complexes of hAngptl4-CCD into the monomers (Fig. 1A, lanes 5 and 6). Notably, disrupting the disulfide bond-mediated oligomerization of hAngptl4 greatly attenuated its inhibitory activity on LPL to a level comparable with that of hAngptl3-CCD (Fig. 1B). These results suggest that the disulfide bond formation facilitates the oligomeric formation of hAngptl4-CCD, which in turn enhance its inhibitory effects on the LPL activity.
We next determined whether the presence of apoCII, an activating co-factor of LPL, would interfere with the inhibitory effect mediated by hAngptl4-CCD. Increasing concentrations of human apoCII from 0 to 2 μg/ml in the assay mixture caused a dose-dependent elevation in the LPL activity (Fig. 1C). Addition of hAngptl4-CCD (0.125 to 1 μg/ml) inhibited both basal and apoCII-stimulated LPL activities. When the LPL activity was expressed as the percentage remaining in comparison with those without hAngptl4-CCD treatment (Fig. 1D), we found that the presence of apo-CII did not rescue hAngptl4-CCD-induced suppression of LPL activity. These data exclude the possibility that hAngptl4-CCD inhibits LPL activity through competing the activation site of apoCII.
A Highly Conserved Motif Is Present in the CCDs of Angptl3 and Angptl4—To determine the common structural features shared by the CCDs of Angptl3 and Angptl4 that are responsible for their LPL inhibitory effects, we aligned and analyzed the amino acid sequences of the CCDs of these two proteins from different species (supplemental Fig. 1). The overall homology of the CCDs from Angptl3 to that of Angptl4 was less than 30%. However, a highly conserved motif, LAXGLLXLGXGL, was identified near their NH2 termini, where X could be His or Gln. Most importantly, this sequence motif is not present in any other members of the Angptl family (data not shown). Therefore, it is intriguing to speculate that this region might be responsible for the LPL inhibitory activities shared by both Angptl3 and Angptl4. Prediction of the secondary structure for hAngptl4-CCD using PSIPRED showed that this motif was most likely localized within an α-helix (helix-1, Arg35 to Cys80, supplemental Fig. 1A). The amino acid sequence of helix-1 was submitted to the REPPER server for sequence periodicity prediction (38). This program predicted that helix-1 had a left-handed coiled-coil topology with periodicity at 3.54. Further analysis using HHrep (39) identified a 7-amino acid shifted self-alignment (p value = 2.8e–20; identities = 13%) with Val43 in the consensus motif aligned with Gln50 (data not shown), suggesting that helix-1 is possibly wound in a left-handed coiled-coil topology with periodicity of “7/2” (i.e. seven amino acid residues per two turns). A helical wheel projection model of helix-1 was built (supplemental Fig. 1B), which showed that the hydrophobic residues (positions 2 and 6) fall onto one face of the helix that is likely responsible for lipid interactions, whereas the polar residues (positions 1, 4, and 7) fall on the opposite face that is solvent-exposed and is probably involved in the protein-protein interactions. Interestingly, the three polar residues (His46, Gln50, and Gln53) of the motif are most notably situated in close proximity and solvent-accessible. These sequence and structural analyses suggest that the conserved motif might be important for the LPL inhibitory effects by both Angptl4 and Angptl3.
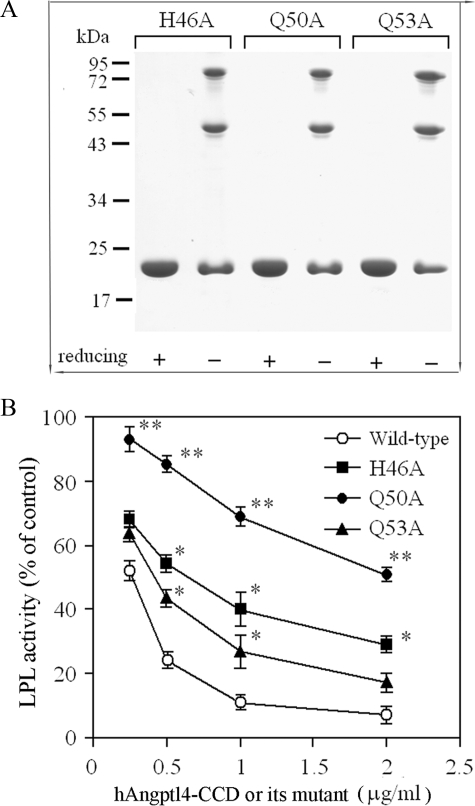
Substitution of the Three Polar Amino Acid Residues within the Conserved Motif Abolishes the Inhibitory Effects of Both Angptl4 and Angptl3 on LPL—To examine whether the three polar amino acid residues (His46, Gln50, and Gln53) within the consensus sequence motif are involved in mediating the inhibitory effects of human Angptl4 on LPL, we expressed and purified the recombinant human Angptl4-CCD mutants in which one of the polar amino acid residues was replaced by alanine (H46A, Q50A, or Q50A, respectively). Nonreducing SDS-PAGE analysis showed that all three mutants were present as several oligomeric complexes, a pattern similar to that of the wild type human Angptl4-CCD (Fig. 2A). Compared with wild type human Angptl4-CCD, the inhibitory activities of all these three mutants on LPL were significantly attenuated (Fig. 2B). Among these three mutants, the Q50A mutation caused the most dramatic effect, with a 75% drop in its inhibitory activity on LPL compared with wild type control. The effects of H46A and Q53A mutations were milder, with 33 and 44% decreases in the inhibitory activity on LPL, respectively. These results suggest that the three polar residues participated in the LPL inhibition. Gln50, which is completely conserved across all the species of Angptl3 and Angptl4, is apparently more crucial than His46 and Gln53 in conferring the LPL inhibition.
FIGURE 2.
Role of the three polar amino acid residues within the conserved motif in mediating the inhibitory activity of hAngptl4-CCD on LPL. A, SDS-PAGE analysis of the purified mutant hAngptl4-CCD with a single alanine substitution (H46A, Q50A, and Q53A). Each lane contains 3 μg of protein separated by 12% polyacrylamide gel under either reducing or non-reducing conditions. B, comparison of the inhibitory effects of wild type hAngptl4-CCD and the three mutants on LPL activity. Different concentrations of the wild type hAngptl4-CCD or the mutant proteins were included in the LPL activity assay mixture as indicated. The results are expressed as the percentage of LPL activity in the absence of hAngptl4-CCD or its mutants. *, p < 0.05; **, p < 0.01 versus wild type Angptl4-CCD treated group (n = 4–5).
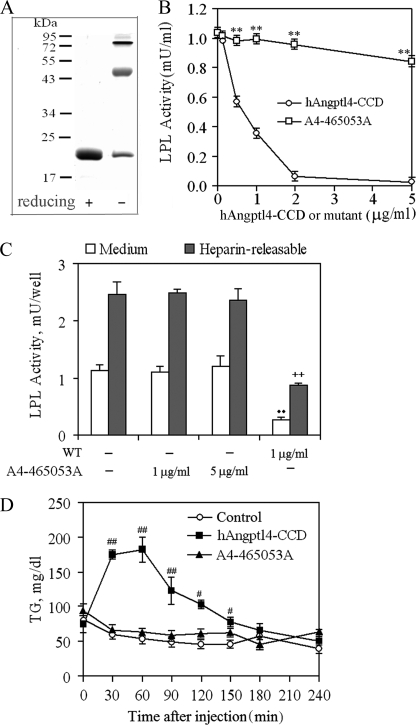
We next examined the LPL inhibitory effect of the hAngptl4-CCD mutant with all three polar amino acid residues replaced by alanines (designated as A4-212528A). As shown in Fig. 3A, simultaneous mutation of the three polar residues did not affect the oligomerization of this protein. However, the inhibitory activity of this Angptl4-CCD mutant on LPL was completely abolished (Fig. 3B). This observation was further confirmed in 3T3L1 adipocytes treated with hAngptl4-CCD or its mutant A4-212528A (Fig. 3C). 1 μg/ml hAngptl4-CCD inhibited the activity of LPL derived from both secreted and heparin-releasable cell surface-associated fractions by ∼64 and 57%, respectively, whereas the mutant A4-212528A at the same concentration or even 5-fold higher did not cause any significant effect. The inhibitory activity of Angptl3-CCD on LPL was also abrogated following substitution of the three polar amino acid residues with alanine (supplemental Fig. 2), suggesting that this conserved motif is indispensable for the LPL inhibition by both Angptl3 and Angptl4.
FIGURE 3.
Effects of simultaneous alanine substitutions at His46, Gln50, and Gln53 on LPL inhibition by hAngptl4-CCD. A, SDS-PAGE analysis of the purified hAngptl4-CCD mutant, A4-465053A, with His46, Gln50, and Gln53 being substituted with alanine residues. An equal amount of protein (3 μg) was loaded onto each lane and separated by 12% polyacrylamide gel under either reducing or nonreducing conditions. B, comparison of the effects between wild type hAngptl4-CCD and A4-465053A on purified bovine LPL activity. **, p < 0.01 versus wild type recombinant hAngptl4-CCD (n = 4). C, comparison of the effects between wild type hAngptl4-CCD and A4-465053A on LPL activity in differentiated 3T3-L1 adipocytes. Fully differentiated adipocytes were incubated with different concentrations of the recombinant proteins for 4 h at 37 °C. Free LPL in medium and heparin-releasable LPL fractions were collected to determine LPL activity. ••, p < 0.01 versus medium control; ++, p < 0.01 versus heparin-releasable control (n = 4–5). D, effects of wild type hAngptl4-CCD and A4-465053A on plasma TG levels of C57BL/6J mice. 50 μg of proteins were injected intravenously into C57BL/6J mice. Control mice were injected with PBS only. Blood samples were collected at the indicated time points and analyzed for TG contents. n = 4–5, #, p < 0.05; ##, p < 0.01 versus control mice (n = 4–5).
To further explore the physiological relevance of the above findings, the inhibitory activities of hAngptl4-CCD and its mutant A4-212528A on LPL were evaluated in C57/B6 mice (Fig. 3D). In control mice injected with PBS, the plasma TG levels gradually decreased throughout the course of the experiment (4 h) from 68.4 ± 4.4 to 43.2 ± 5.7 mg/dl. Intravenous injection of 50 μg of hAngptl4-CCD caused a marked elevation in the plasma TG levels in a time-dependent manner (Fig. 3D). The plasma TG levels were increased by 230% at 30 min after injection and reached the peak level at around 60 min, suggesting that bacterially expressed wild type hAngptl4-CCD functions properly in mice to inhibit the LPL activity and thus reduces the rate of TG hydrolysis. By contrast, injection of the same amount of its mutant form A4-465053A (50 μg) did not cause any significant change in the plasma TG level throughout the course of experiment compared with the control group, indicating that the mutations at the three polar amino acid residues abolish the LPL inactivating activity of Angptl4-CCD in vivo. Substitution of the three polar amino acid residues also led to a marked attenuation of Angptl3-induced elevation of plasma TG levels in mice (data not shown).
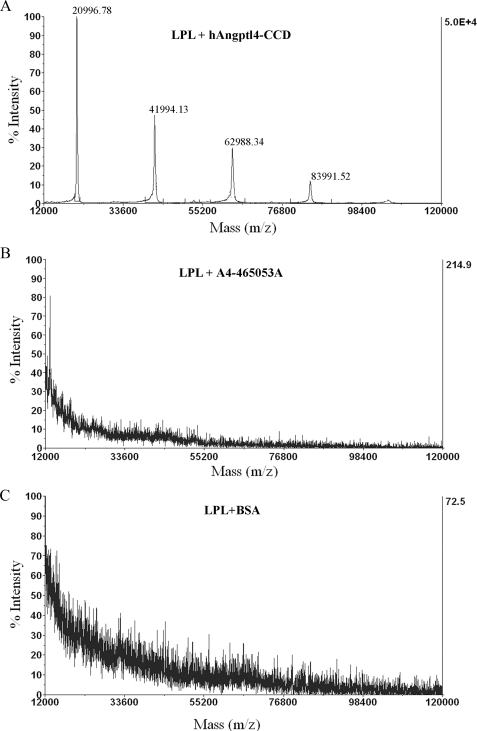
Three Polar Amino Acid Residues of Angptl4 Are Involved in the Direct Interaction with LPL and Conversion of LPL from Its Dimers to Monomers—Previous reports demonstrated that the CCD of Angptl4 binds to LPL and promotes the conversion of LPL from its catalytically active dimers to the inactive monomers in vitro and in vivo (23, 31). Here, we further characterized the structural basis underlying the interactions between Angptl4 and LPL using SELDI-TOF analysis. To this end, a biochip was pre-coated with bovine LPL as a binding ligand and was then incubated with wild type hAngptl4-CCD, the mutant protein A4-212528A, or BSA (as control). Following extensive washing, the biochip was subjected to mass spectrometric analysis. In the biochip incubated with wild type hAngptl4-CCD, mass spectrometric analysis detected four peptide species with molecular masses of 20,996, 41,994, 62,988, and 83,991 Da (Fig. 4), which corresponds to the monomeric, dimeric, trimeric, and tetrameric forms of human Angptl4-CCD, respectively, indicating that LPL interacts with several oligomeric forms of Angptl4. By contrast, in the biochip incubated with the mutant protein A4-212528A or BSA, no obvious mass peaks equivalent to these two proteins were retained and detected, indicating that the three polar amino acid residues within the conserved motif are critically involved in the interactions between LPL and Angptl4.
FIGURE 4.
Determination of the interactions between LPL and wild type or mutated hAngptl4-CCD using SELDI-TOF MS analysis. The protein chips were coated with bovine LPL and incubated with hAngptl4-CCD (A), its mutant A4-212528A (B), or BSA as control (C). The bound proteins were detected by mass spectrometry as described under “Experimental Procedures.” Note that the molecular mass of 20996.78, 42132.82, 63288.33 and 84419.64 represents the monomeric, dimeric, trimeric and tetrameric forms of hAngptl4-CCD, respectively.
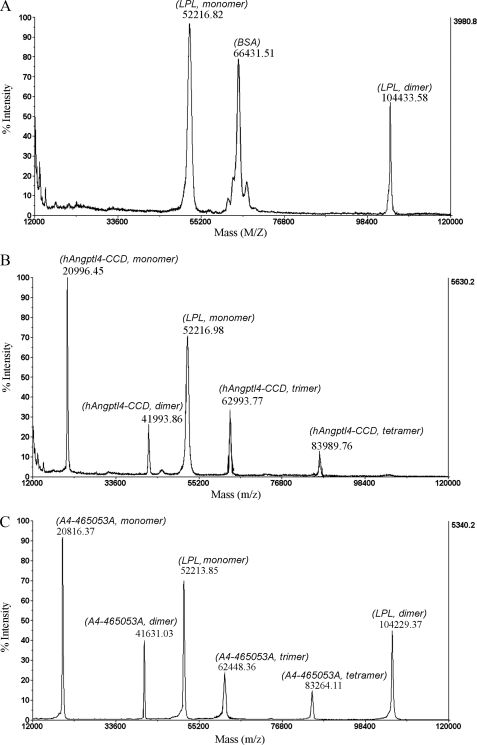
MALDI-TOF analysis revealed that bovine LPL formed both monomers and dimers with the molecular masses of 52 and 104 kDa, respectively (Fig. 5). Co-incubation with recombinant hAngptl4-CCD resulted in an almost complete absence of the LPL dimers, whereas both LPL monomers and dimers were detectable in the samples co-incubated with BSA or the mutant protein A4-212528A. These data suggest that the ability of Angptl4 to convert LPL from its catalytically active dimeric form to the inactive monomer is dependent on the three polar amino acid residues.
FIGURE 5.
MALDI-TOF MS analysis of LPL monomers and dimers. LPL was incubated with BSA (A), hAngptl4-CCD (B), or its A4-212528A (C) as described under “Experimental Procedures,” and the protein mixtures were analyzed by MALDI-TOF MS. Note that LPL dimeric form is not present in the sample treated with wild type hAngptl4-CCD.
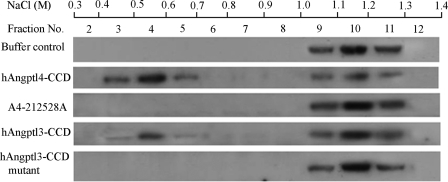
To further confirm the above findings, we next evaluated the effects of Angptl4-CCD and its mutant on oligomerization states of LPL using heparin-Sepharose chromatography. Because LPL monomers have much lower affinity for heparin than LPL dimers, this chromatographic method allows effective separation of monomeric and dimeric LPL by elution with a linear gradient of NaCl (23, 31, 37). To facilitate detection, we labeled bovine LPL with biotin. Afterward, biotinylated LPL was subjected to sucrose density gradient centrifugation. Fractions containing the dimeric LPL were collected, pooled, and subjected to further heparin-Sepharose chromatography. Immunoblot analysis showed that LPL was predominantly present in the fractions eluted by 1.0–1.3 m NaCl, which corresponds to the dimeric form of this enzyme (Fig. 6). After incubation with recombinant hAngptl4-CCD, a large portion of LPL was eluted by a much lower concentration of NaCl (0.4–0.7 m), which corresponds to LPL monomers. The increase in monomeric LPL by hAngptl4-CCD was accompanied by a marked reduction in LPL dimers eluted by 1.0–1.3 m NaCl. By contrast, LPL was still present predominantly as dimers eluted by 1.0–1.3 m NaCl after incubation with the mutant protein A4-212528A.
FIGURE 6.
Conversion of LPL dimers to monomers by both hAngptl4 and hAngptl3 are dependent on the three polar residues within the conserved motif of the coiled-coil domain. Bovine LPL was biotinylated and re-purified by sucrose density gradient ultracentrifugation to obtain LPL dimers. 5 μg of biotinylated LPL dimers were added to a tube containing 1 ml of heparin-Sepharose beads. The beads were collected by centrifugation, washed, and incubated further with 15 μg of hAngptl4-CCD or its mutant A4-212528A and 75 μg of hAngptl3-CCD or its mutant in which the three polar amino acid residues were replaced by alanine. Afterward, LPL bound to the beads were eluted by a linear gradient of NaCl (from 0.20 to 1.8 m) as described under “Experimental Procedures.” Protein in each fraction was precipitated by 10% trichloroacetic acid, transferred to nylon membrane, and then probed with horseradish peroxidase-conjugated streptavidin to detect biotinylated LPL. The representative blots were shown from three independent experiments. Note that LPL monomers and dimers were eluted by 0.4–0.7 m NaCl and 1.0–1.3 m NaCl, respectively.
Incubation of the LPL-bound heparin-Sepharose beads with hAngptl3-CCD also caused conversion of LPL dimers to monomers (Fig. 6). However, compared with hAngptl4-CCD, the effect of hAngptl3-CCD was much weaker and was observed only when a much higher concentration of recombinant hAngptl3-CCD was used. Noticeably, the ability of hAngptl3-CCD was also abolished when the three polar residues within the conserved motif were mutated to alanine. Altogether, these findings highlight the critical role of the three polar amino acid residues in disruption of LPL dimerization by both Angptl4 and Angptl3.
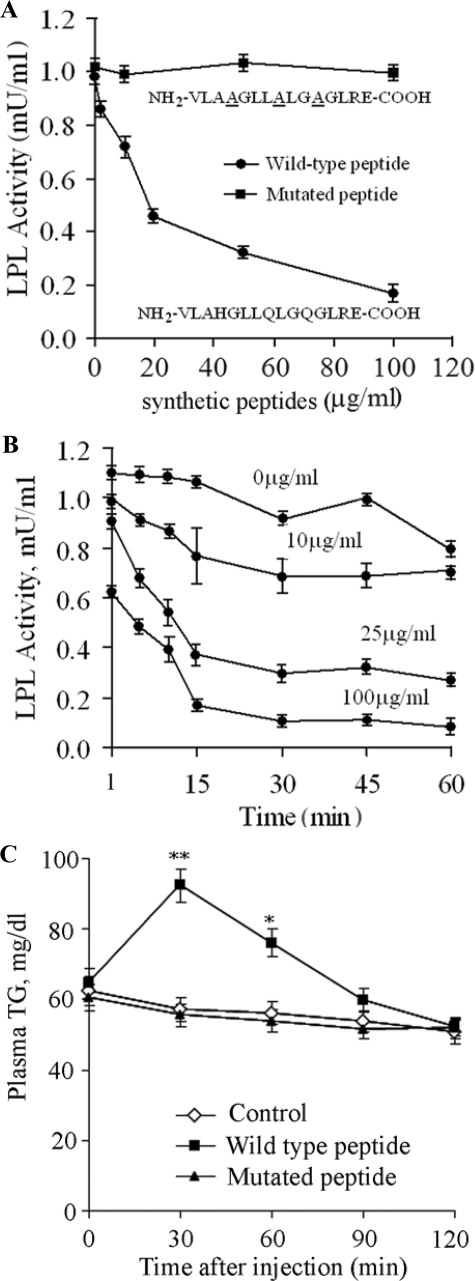
A Synthetic Peptide Containing the Conserved 12-Amino Acid Consensus Motif Mimics the Inhibitory Effects of Angptl4 on LPL Activities—Both our in vitro and in vivo findings described above highlight an essential role of the three polar amino acid residues within the conserved motif in mediating the inhibitory effect of Angptl4 on LPL activity. We next investigated whether this highly conserved motif alone is sufficient to inhibit LPL activity in vitro. To this end, a peptide of 15 amino acid residues containing the conserved motif, NH2-VLAHGLLQLGQGLRE-COOH, was synthesized. The last two amino acid residues, Arg56 and Glu57, were included to improve the solubility of this hydrophobic peptide. As shown in Fig. 7A, this synthetic peptide inhibited the activity of bovine LPL in a dose-dependent manner, with an IC50 value of 20 μg/ml. The molar ratio of the synthetic peptide relative to LPL dimer is estimated to be around 1500 to 1, which is ∼1,000-fold that for hAngptl4-CCD. The synthetic peptide inhibited the LPL activity in a time-dependent manner in the initial 15 min, whereas longer incubation did not result in further inhibition (Fig. 7B). On the other hand, the peptide with three polar residues being substituted by alanine, NH2-VLAAGLLALGAGLRECOOH, did not exhibit any inactivating effect on LPL even when 100 μg/ml concentration was used in the assay mixture, indicating that the inhibitory effect of the synthetic peptide on the LPL activity is also dependent on the three polar amino acid residues.
FIGURE 7.
Effect of the synthetic peptide containing the conserved sequence motif on LPL activity. A, synthetic peptides consisting of the wild type (NH2-VLAHGLLQLGQGLRE-COOH) or the mutated motif (NH2-VLAAGLLALGAGLRE-COOH) within the CCD of hAngptl4 were added to the LPL assay mixtures to a final concentration as indicated, and the reactions were performed at 30 °C for 1 h. B, time-dependent effect of the synthetic wild type peptide on the LPL activity. C, plasma triglyceride levels in C57/B6 mice at various time points after intravenous injection of wild type or mutated synthetic peptides (500 μg/mouse) or saline as control. *, p < 0.05; **, p < 0.01 versus control group (n = 5).
Consistent with the above in vitro findings, intravenous injection of the wild type synthetic peptide into C57/B6 mice also caused a significant elevation of plasma triglyceride levels (Fig. 7C). At 30 min after injection, plasma triglyceride levels were increased by 164% compared with the saline-treated controls, suggesting that the synthetic peptide alone is able to mimic the LPL inhibitory effect of hAngptl4-CCD in vivo. By contrast, the mutated synthetic peptide with the three polar residues being replaced by alanine did not have any effect on plasma triglyceride levels.
DISCUSSION
Although a large body of evidence has demonstrated the role of Angptl3 and Angptl4 as the endogenous inhibitors of LPL (3, 10, 11), the structural basis underlying the LPL inhibition by these two proteins remains poorly characterized. In this study, we have identified a highly conserved 12-amino acid consensus motif within the CCD of both Angptl3 and Angptl4, which mediate the LPL inhibition in vitro as well as in vivo. Notably, this consensus motif is present in all known species of Angptl3 and Angptl4, but not the other members of the Angptl family. The presence of this common motif in the CCD of both Angptl3 and Angptl4 might explain, at least in part, why the two proteins share a similar inhibitory property on LPL despite only ∼30% overall sequence homology.
The three-dimensional structures for either Angptl3 or Angptl4 have not been resolved so far. Our results from the secondary structural prediction suggest the presence of two α-helices located within the CCD of both Angptl3 and Angptl4 (supplemental Fig. 1). The highly conserved 12-amino acid motif is located at the central part of the first α-helix, with the three polar amino acid residues (His46, Gln50, and Gln53) in the consensus motif pointing to the same orientation in the hydrophilic face, which is likely to be solvent-exposed and is involved in the protein-protein interaction. Importantly, substitution of the three polar amino acid residues with alanine results in a complete abrogation of Angptl3- and Angptl4-mediated inhibition on LPL activity in vitro and abolishes the ability of Angptl4 to elevate plasma TG levels in mice (Fig. 3). Consistent with previous reports (23, 31), our SELDI-TOF analysis also demonstrates that the CCD of Angptl4 interacts with LPL and converts LPL from its catalytically active dimers to the inactive monomers (Fig. 5). By contrast, the mutant Angptl4 with the three polar amino acid residues being substituted by alanine loses such an activity. Taken in conjunction, these data suggest that the three polar amino acid residues within the conserved motif are directly involved in the interaction with LPL, thereby leading to the LPL inactivation by destabilizing the dimerization of the enzyme. In addition, we have prioritized the relative contributions of the three polar residues to the inhibitory effects of Angptl4 on LPL activities and found that alanine substitution of Gln50, which is completely conserved across all the species of Angptl3 and Angptl4, causes the most severe defect in inhibiting LPL activity (Fig. 2).
The indispensable role of the highly conserved 12-amino acid consensus motif in Angptl4- and Angptl3-mediated LPL inhibition is also supported by our observation that a synthetic peptide containing this motif alone is sufficient to suppress the enzyme activity in vitro, despite the fact that the IC50 value is 1000-fold higher than that of the recombinant-hAngptl4-CCD (Fig. 7). The key role of the three polar amino acid residues is further highlighted by our observation that a synthetic peptide with the three polar amino acid residues being replaced by alanine does not possess the inhibitory activity on LPL. Consistent with our finding, it is noteworthy that the synthetic peptide corresponding to apoCII Thr64–Val74 or Ser61–Glu79 has previously been shown to be capable of interacting with and activating LPL (40), although the degree of the activation is much less than the full-length apoCII. These findings suggest that the activity of LPL can be modulated by small molecules such as short peptides. Further characterization of the precise domains of LPL involved in its interaction with these short peptides will provide the structural basis for rational design of novel lipid-lowering drugs through enhancing the LPL activity.
Although our data show the 12-amino acid motif as a core sequence element for mediating the LPL suppression by Angptl4 and Angptl3, its much lower potency in comparison with the recombinant CCD of Angptl4 suggests that additional amino acid sequences and/or three-dimensional conformations are required to achieve its maximal bioactivity. Indeed, several recent genetic investigations in different ethnic groups have identified a genetic variant (E40K) of ANGPTL4 associated with significantly lower plasma TG levels and higher high density lipoprotein cholesterol levels (24–26). Site-directed mutagenesis analysis confirmed that this mutation causes a significant loss of the inactivating effects of Angptl4 on LPL in vitro (41). According to our secondary structural prediction, Glu40 is also located within the first α-helix and lies at the same solvent-exposed face with the three polar amino acids within the conserved motif. These findings support our hypothesis that the first α-helix of hAngptl4-CCD mediates its interaction with LPL through its solvent-exposed face. Whether or not Glu40 is also involved in the interaction between Angptl4 and LPL remains to be determined.
Despite the fact that both Angptl3 and Angptl4 inhibit LPL via their CCD, it is important to note that the two proteins exhibit distinct potency and expression patterns and might involve different mechanisms on LPL inhibition (42). Consistent with our observation, Angptl4 has been shown to be much more potent than Angptl3 on LPL inhibition in vitro and on elevation of plasma triglyceride in mice (15, 41). Angptl4 forms variably sized oligomers resulting from intermolecular disulfide bond formation mediated through two cysteine residues (76 and 80), although these two cysteine residues are not present in Angptl3 (43). The disulfide bond-mediated oligomerization has been shown to be important for mediating Angptl4-induced elevation of plasma triglyceride in mice (43). In line with this in vivo finding, this study demonstrated that disrupting disulfide bond-mediated oligomerization by DTT markedly attenuates the LPL inhibitory effects of Angptl4 to a level comparable with that of Angptl3 (Fig. 1), suggesting that the much higher potency of Angptl4 on LPL inhibition in comparison with Angptl3 is attributed, at least in part, to the disulfide bond-mediated oligomerization of Angptl4.
Another notable difference between the two proteins is the presence of a putative heparin-binding motif located within the CCD of Angptl3 ((61VHKTKG66) (14), whereas this motif cannot be found in any known sequence of Angptl4. Substitution of the basic amino acid residue within this motif decreases the ability of Angptl3 to elevate plasma TG levels in mice (14). However, whether or not heparin modulates the inhibitory effect of Angptl3 on LPL through binding with this motif is debatable. An earlier study by Ono et al. (14) showed that addition of heparin does not affect the inhibitory effect of Angptl3 on the LPL activity, suggesting that the putative heparin-binding site might interact with other factors instead of heparin. By contrast, a more recent report by Shan et al. (41) demonstrated that heparin can rescue LPL from the inhibition of Angptl3.
In summary, this study has identified a common structural basis that mediates the inhibitory effect of both Angptl3 and Angptl4 on the LPL activity. Even though the two proteins exhibit differential potencies and distinct regulation, we propose that the highly conserved 12-amino acid motif within the CCD of Angptl3 and Angptl4 acts as a core sequence element responsible for their LPL inhibition by the direct interaction with this enzyme, thereby converting the catalytically active dimers of the enzyme to its inactive monomers. Therefore, therapeutic intervention targeting this short consensus motif is expected to block the effect of both Angptl3 and Angptl4 on elevation of plasma triglyceride levels, and it might represent a promising strategy for treatment and prevention of dyslipidemia. Interestingly, a recent report by Desai et al. (19) demonstrated that treatment of several high fat-fed mouse lines with a monoclonal antibody against Angptl4 by tail vein injection potently alleviates hyperlipidemia in these mice, although it is unclear whether the 12-amino acid consensus motif is within the epitope recognized by this monoclonal antibody. Further studies are warranted to investigate whether an antibody specifically targeting this motif can block the effect of both Angptl3 and Angptl4 on LPL inhibition in vitro and on elevation of plasma triglyceride in mice.
Supplementary Material
This work was supported by the General Research Fund of Hong Kong Research Council Grant HKU 7609/05M, Outstanding Young Researcher award (to A. X.), a small project grant (to M. H. Y.) from the University of Hong Kong, and National 973 Program of China Grant 2006CB910202.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
Footnotes
The abbreviations used are: LPL, lipoprotein lipase; Angptl, angiopoietin-like protein; TG, triglyceride; CCD, coiled-coil domain; h, human; BSA, bovine serum albumin; PBS, phosphate-buffered saline; MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; SELDI-TOF, surface-enhanced laser desorption/ionization time-of-flight; DTT, dithiothreitol.
References
- 1.Mead, J. R., Irvine, S. A., and Ramji, D. P. (2002) J. Mol. Med. 80 753–769 [DOI] [PubMed] [Google Scholar]
- 2.Merkel, M., Eckel, R. H., and Goldberg, I. J. (2002) J. Lipid Res. 43 1997–2006 [DOI] [PubMed] [Google Scholar]
- 3.Otarod, J. K., and Goldberg, I. J. (2004) Curr. Atheroscler. Rep. 6 335–342 [DOI] [PubMed] [Google Scholar]
- 4.Enerback, S., Semb, H., Tavernier, J., Bjursell, G., and Olivecrona, T. (1988) Gene (Amst.) 64 97–106 [DOI] [PubMed] [Google Scholar]
- 5.Doolittle, M. H., Ben-Zeev, O., Elovson, J., Martin, D., and Kirchgessner, T. G. (1990) J. Biol. Chem. 265 4570–4577 [PubMed] [Google Scholar]
- 6.Osborne, J. C., Jr., Bengtsson-Olivecrona, G., Lee, N. S., and Olivecrona, T. (1985) Biochemistry 24 5606–5611 [DOI] [PubMed] [Google Scholar]
- 7.Vainio, P., Virtanen, J. A., Kinnunen, P. K., Voyta, J. C., Smith, L. C., Gotto, A. M., Jr., Sparrow, J. T., Pattus, F., and Verger, R. (1983) Biochemistry 22 2270–2275 [DOI] [PubMed] [Google Scholar]
- 8.MacPhee, C. E., Hatters, D. M., Sawyer, W. H., and Howlett, G. J. (2000) Biochemistry 39 3433–3440 [DOI] [PubMed] [Google Scholar]
- 9.Katoh, Y., and Katoh, M. (2006) Int. J. Mol. Med. 17 1145–1149 [PubMed] [Google Scholar]
- 10.Li, C. (2006) Curr. Opin. Lipidol. 17 152–156 [DOI] [PubMed] [Google Scholar]
- 11.Kersten, S. (2005) Biochem. Soc. Trans. 33 1059–1062 [DOI] [PubMed] [Google Scholar]
- 12.Koishi, R., Ando, Y., Ono, M., Shimamura, M., Yasumo, H., Fujiwara, T., Horikoshi, H., and Furukawa, H. (2002) Nat. Genet. 30 151–157 [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto, K., Koishi, R., Shimizugawa, T., and Ando, Y. (2006) Exp. Anim. 55 27–34 [DOI] [PubMed] [Google Scholar]
- 14.Ono, M., Shimizugawa, T., Shimamura, M., Yoshida, K., Noji-Sakikawa, C., Ando, Y., Koishi, R., and Furukawa, H. (2003) J. Biol. Chem. 278 41804–41809 [DOI] [PubMed] [Google Scholar]
- 15.Yoshida, K., Shimizugawa, T., Ono, M., and Furukawa, H. (2002) J. Lipid Res. 43 1770–1772 [DOI] [PubMed] [Google Scholar]
- 16.Kersten, S., Mandard, S., Tan, N. S., Escher, P., Metzger, D., Chambon, P., Gonzalez, F. J., Desvergne, B., and Wahli, W. (2000) J. Biol. Chem. 275 28488–28493 [DOI] [PubMed] [Google Scholar]
- 17.Kim, I., Kim, H. G., Kim, H., Kim, H. H., Park, S. K., Uhm, C. S., Lee, Z. H., and Koh, G. Y. (2000) Biochem. J. 346 603–610 [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon, J. C., Chickering, T. W., Rosen, E. D., Dussault, B., Qin, Y., Soukas, A., Friedman, J. M., Holmes, W. E., and Spiegelman, B. M. (2000) Mol. Cell. Biol. 20 5343–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desai, U., Lee, E. C., Chung, K., Gao, C., Gay, J., Key, B., Hansen, G., Machajewski, D., Platt, K. A., Sands, A. T., Schneider, M., Van Sligtenhorst, I., Suwanichkul, A., Vogel, P., Wilganowski, N., Wingert, J., Zambrowicz, B. P., Landes, G., and Powell, D. R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11766–11771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koster, A., Chao, Y. B., Mosior, M., Ford, A., Gonzalez-DeWhitt, P. A., Hale, J. E., Li, D., Qiu, Y., Fraser, C. C., Yang, D. D., Heuer, J. G., Jaskunas, S. R., and Eacho, P. (2005) Endocrinology 146 4943–4950 [DOI] [PubMed] [Google Scholar]
- 21.Mandard, S., Zandbergen, F., van Straten, E., Wahli, W., Kuipers, F., Muller, M., and Kersten, S. (2006) J. Biol. Chem. 281 934–944 [DOI] [PubMed] [Google Scholar]
- 22.Yu, X., Burgess, S. C., Ge, H., Wong, K. K., Nassem, R. H., Garry, D. J., Sherry, A. D., Malloy, C. R., Berger, J. P., and Li, C. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 1767–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sukonina, V., Lookene, A., Olivecrona, T., and Olivecrona, G. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 17450–17455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Folsom, A. R., Peacock, J. M., Demerath, E., and Boerwinkle, E. (2008) Metabolism 57 1591–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romeo, S., Pennacchio, L. A., Fu, Y., Boerwinkle, E., Tybjaerg-Hansen, A., Hobbs, H. H., and Cohen, J. C. (2007) Nat. Genet. 39 513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talmud, P. J., Smart, M., Presswood, E., Cooper, J. A., Nicaud, V., Drenos, F., Palmen, J., Marmot, M. G., Boekholdt, S. M., Wareham, N. J., Khaw, K. T., Kumari, M., and Humphries, S. E. (2008) Arterioscler. Thromb. Vasc. Biol. 28 2319–2325 [DOI] [PubMed] [Google Scholar]
- 27.Ge, H., Yang, G., Huang, L., Motola, D. L., Pourbahrami, T., and Li, C. (2004) J. Biol. Chem. 279 2038–2045 [DOI] [PubMed] [Google Scholar]
- 28.Mandard, S., Zandbergen, F., Tan, N. S., Escher, P., Patsouris, D., Koenig, W., Kleemann, R., Bakker, A., Veenman, F., Wahli, W., Muller, M., and Kersten, S. (2004) J. Biol. Chem. 279 34411–34420 [DOI] [PubMed] [Google Scholar]
- 29.Yang, Y. H., Wang, Y., Lam, K. S., Yau, M. H., Cheng, K. K., Zhang, J., Zhu, W., Wu, D., and Xu, A. (2008) Arterioscler. Thromb. Vasc. Biol. 28 835–840 [DOI] [PubMed] [Google Scholar]
- 30.Shimizugawa, T., Ono, M., Shimamura, M., Yoshida, K., Ando, Y., Koishi, R., Ueda, K., Inaba, T., Minekura, H., Kohama, T., and Furukawa, H. (2002) J. Biol. Chem. 277 33742–33748 [DOI] [PubMed] [Google Scholar]
- 31.Lichtenstein, L., Berbee, J. F., van Dijk, S. J., van Dijk, K. W., Bensadoun, A., Kema, I. P., Voshol, P. J., Muller, M., Rensen, P. C., and Kersten, S. (2007) Arterioscler. Thromb. Vasc. Biol. 27 2420–2427 [DOI] [PubMed] [Google Scholar]
- 32.Xu, A., Lam, M. C., Chan, K. W., Wang, Y., Zhang, J., Hoo, R. L., Xu, J. Y., Chen, B., Chow, W. S., Tso, A. W., and Lam, K. S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 6086–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergo, M., Olivecrona, G., and Olivecrona, T. (1996) Biochem. J. 313 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rimmer, A. D., Schonbaum, E., and Carroll, K. K. (1962) Clin. Chim. Acta 7 877–880 [DOI] [PubMed] [Google Scholar]
- 35.Chen, B., Lam, K. S., Wang, Y., Wu, D., Lam, M. C., Shen, J., Wong, L., Hoo, R. L., Zhang, J., and Xu, A. (2006) Biochem. Biophys. Res. Commun. 341 549–556 [DOI] [PubMed] [Google Scholar]
- 36.Wang, Y., Lam, K. S., Xu, J. Y., Lu, G., Xu, L. Y., Cooper, G. J., and Xu, A. (2005) J. Biol. Chem. 280 18341–18347 [DOI] [PubMed] [Google Scholar]
- 37.Zhang, L., Lookene, A., Wu, G., and Olivecrona, G. (2005) J. Biol. Chem. 280 42580–42591 [DOI] [PubMed] [Google Scholar]
- 38.Gruber, M., Soding, J., and Lupas, A. N. (2005) Nucleic Acids Res. 33 W239–W243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soding, J., Remmert, M., and Biegert, A. (2006) Nucleic Acids Res. 34 W137–W142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng, Q., Blackett, P., Jackson, K. W., McConathy, W. J., and Wang, C. S. (1990) Biochem. J. 269 403–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shan, L., Yu, X. C., Liu, Z., Hu, Y., Sturgis, L. T., Miranda, M. L., and Liu, Q. (2009) J. Biol. Chem. 284 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ge, H., Cha, J. Y., Gopal, H., Harp, C., Yu, X., Repa, J. J., and Li, C. (2005) J. Lipid Res. 46 1484–1490 [DOI] [PubMed] [Google Scholar]
- 43.Ge, H., Yang, G., Yu, X., Pourbahrami, T., and Li, C. (2004) J. Lipid Res. 45 2071–2079 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.