Abstract
Previously, we have demonstrated that human oxidative DNA glycosylase NEIL1 excises photoactivated psoralen-induced monoadducts but not genuine interstrand cross-links (ICLs) in duplex DNA. It has been postulated that the repair of ICLs in mammalian cells is mainly linked to DNA replication and proceeds via dual incisions in one DNA strand that bracket the cross-linked site. This process, known as “unhooking,” enables strand separation and translesion DNA synthesis through the gap, yielding a three-stranded DNA repair intermediate composed of a short unhooked oligomer covalently bound to the duplex. At present, the detailed molecular mechanism of ICL repair in mammalian cells remains unclear. Here, we constructed and characterized three-stranded DNA structures containing a single ICL as substrates for the base excision repair proteins. We show that NEIL1 excises with high efficiency the unhooked ICL fragment within a three-stranded DNA structure. Complete reconstitution of the repair of unhooked ICL shows that it can be processed in a short patch base excision repair pathway. The new substrate specificity of NEIL1 points to a preferential involvement in the replication-associated repair of ICLs. Based on these data, we propose a model for the mechanism of ICL repair in mammalian cells that implicates the DNA glycosylase activity of NEIL1 downstream of Xeroderma Pigmentosum group F/Excision Repair Cross-Complementing 1 endonuclease complex (XPF/ERCC1) and translesion DNA synthesis repair steps. Finally, our data demonstrate that Nei-like proteins from Escherichia coli to human cells can excise bulky unhooked psoralen-induced ICLs via hydrolysis of glycosidic bond between cross-linked base and deoxyribose sugar, thus providing an alternative heuristic solution for the removal of complex DNA lesions.
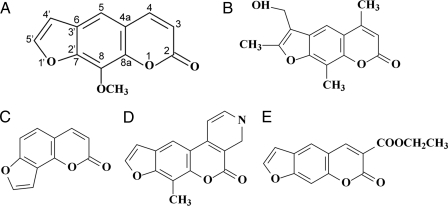
Interstrand cross-links (ICLs)4 are highly lethal DNA lesions that block DNA transcription, replication, and recombination by preventing strand separation. Due to their high cytotoxicity, DNA cross-linking agents such as mitomycin C, cisplatin, and psoralens are widely used against hyperplasic diseases such as cancer and psoriasis (1, 2). Furanocoumarins (psoralens), naturally occurring secondary metabolites in plants, are tricyclic compounds formed by the fusion of a furan ring with a coumarin (Fig. 1). Among other ICL-inducing agents, psoralens require UVA photoactivation following DNA intercalation to chemically react with both cellular DNA in vivo and naked DNA in vitro (3). 8-Methoxypsoralen (8-MOP) is an asymmetric, planar compound that intercalates into DNA duplex near pyrimidines, preferentially at 5′-TpA sites. Upon photoactivation, 8-MOP primarily photoalkylates DNA by cycloaddition to the 5,6-double bond of a thymidine generating monoadducts (MA) with either the 4′,5′-double bond of the furan (MAf) or the 3,4-double bond of the pyrone (MAp) side of the psoralen (4) (supplemental Fig. S1). A unique property of psoralen photochemistry is that the absorption of a second photon by the MAf leads to formation of a pyrone side 5,6-double bond adduct with a flanking thymine in the complementary strand, thus generating an ICL (5). An angular fusion of the two-ring systems forms isopsoralens such as angelicin (Ang), 3-carbethoxypsoralen (3CP), and 7-methylpyrido(3,4-c)psoralen (MePy). In contrast to psoralens, isopsoralens, due to their geometry, cannot generate cross-links and form only monoadducts with DNA and RNA. The structures of several psoralen and isopsoralen derivatives are shown in Fig. 1.
FIGURE 1.
Chemical structure and numbering system for the furanocoumarins. A, 8-MOP; B, HMT; C, angelicin; D, 3CP; and E, MePy.
The detailed structures of psoralen-DNA adducts including sites of covalent attachment of psoralen in DNA helix and chemical structure of psoralen-DNA adducts were well established (3). Crystal and NMR structures of psoralen-induced ICL revealed that psoralen can induce both minor and dramatic distortions into the DNA helix, depending on the sequence (6–8). This knowledge enabled geneticists to use photoactivated psoralen as a model agent to study ICL repair and mutagenesis. Importantly, up to 40% of induced DNA lesions by UVA+8-MOP exposure are ICLs, the remaining being two types of MAs (1). This is in striking contrast with other cross-linking agents such as mitomycin, cis-platin, and nitrogen mustard, which induce a plethora of different kinds of damage and only 1–8% of ICLs (1).
Reconstitution of the repair of plasmids containing a single ICL in cell-free extract from Xenopus egg showed that ICL repair is coupled to DNA replication and involves convergence of two replication forks on the lesion (9). In mammalian cells, genetic evidence indicates that the repair of ICLs is also linked to DNA replication and proceeds via induction of a double strand break during DNA replication, probably due to replication fork collapse as a result of Mus81-Eme1 endonuclease cleavage of a template strand on the 3′-side of the lesion (10, 11). On the same strand, the XPF/ERCC1 nuclease might generate ICL-specific incision on the 5′-side of the damaged site to unhook a cross-linked DNA fragment from the template strand (12, 13). The resulting unhooked ICL swings free of the duplex, exposing a single-stranded gap. Translesion DNA synthesis (TLS) across the gap can be catalyzed by DNA polymerase κ (14) and/or by sequential action of Rev1 and polymerase ζ (15, 16), yielding a three-stranded DNA repair intermediate composed of a short oligomer covalently adducted to the duplex. This remaining cross-linked DNA fragment has to be removed from the template strand to unblock DNA replication. It has been proposed that in human cells, as in Escherichia coli and yeast, the unhooked ICLs are excised from the three-stranded DNA by the nucleotide excision repair (NER) machinery (17). However, with the exception of XPF/ERCC1-deficient cells, cells lacking critical NER components only show a moderate sensitivity to the cross-linking agents, which induce both MAs and ICLs (18, 19). These observations suggest that in human cells, either the NER pathway is only marginally responsible for the resolution of three-stranded DNA repair intermediates or an alternative pathway might exist.
In the base excision repair (BER) pathway, a DNA glycosylase binds to the abnormal base by flipping it out of the helix and catalyzes cleavage of the base-sugar bond, generating either an abasic site or a single strand break with 3′-blocking moiety (20). Human homologues of the E. coli oxidized base-specific bifunctional DNA glycosylase/endonuclease VIII (Nei) were identified by a data base search of the genome and named NEIL1, -2, and -3 (human Nei-like proteins 1, 2, and 3) (21, 22). Recently, we demonstrated that the oxidative DNA glycosylases, E. coli Fpg and Nei and human NEIL1, excise with a high efficiency psoralen-induced MAs in duplex DNA. Consistent with this result, HeLa cells lacking APE1 and/or NEIL1 became hypersensitive to 8-MOP+UVA exposure (23), suggesting that BER may provide an alternative pathway to classic NER to eliminate bulky DNA adducts. The next step of our investigation was to examine how a genuine ICL in an oligonucleotide covalently linked to another oligonucleotide is repaired. For this purpose, we constructed a three-stranded DNA structure with unhooked ICL, a physiologically relevant structure derived from the endonuclease incisions of the template strand containing ICL, and examined its repair by the BER proteins. The present study provides new biochemical and genetic insights into the molecular mechanism of ICL repair.
EXPERIMENTAL PROCEDURES
Reagents, Oligonucleotides, Plasmids, and Proteins—Chemical reagents including 8-MOP, 4′-hydroxy-methyl-4,5′,8-trimethylpsoralen (HMT), Ang, 3CP, and MePy were generously provided by Dr. Dietrich Averbeck (Institut Curie, Orsay, France). All oligonucleotides were purchased from Eurogentec (Seraing, Belgium), including regular oligonucleotides, small interfering RNAs, and those containing thymine glycol residue. 21-mer oligodeoxyribonucleotides C21, d(GCTCTCGTCTGXACACCGAAG), where X is either T or thymine glycol, and complementary D21, d(CTTCGGTGTACAGACGAGAGC), were hybridized to obtain duplexes referred to as D21·C21. These sequence contexts were previously used to study the repair of psoralen-induced ICLs in human cells (24). 21-mer chimeric RNA/DNA oligonucleotides R1 5′(gcucucgucugTacaccgaag)-3′, R2 5′(gcucucgucugTAcaccgaag)-3′, R3 5′(gcucucgucuGTAcaccgaag)-3′,R45′(gcucucgucTGTAcaccgaag)-3′, R4′ 5′(gcucucgucuGTACaccgaag)-3′,R2′ 5′(gcucucgucuGTacaccgaag)-3′, and D9 d(TCTGTACAC) consisting of ribonucleotides (lowercase italic font letters) and one or more deoxyribonucleotides (capital letters) were hybridized with the complementary oligodeoxyribonucleotide D21. To avoid potential sequence context effect for ICL repair, we switched the bottom and upper DNA strands in a cross-linked duplex. For this purpose, a 21-mer chimeric RNA/DNA oligonucleotide R1′ 5′-(cuucggugTacagacgagagc)-3′ and D9′ d(GGTGTACAG) were hybridized with the complementary oligodeoxyribonucleotide C21. The resulting oligonucleotide duplexes were used to generate MAs and ICLs as described (23). It should be emphasized that all psoralen-induced MAs used in this work were pyrone-sided (MAp) and obtained by incubation of purified cross-linked duplex in alkali as described (25).
The purified BER proteins including APE1 and NEIL1 were from laboratory stock. The activities of various DNA repair proteins were tested using their principal substrates and were checked just prior to use (data not shown). The bacterial expression vector phNEIL1 was generously provided by Dr. Hiroshi Ide (Hiroshima University, Japan), DNA polymerase β was generously provided by Dr. Grigory Dianov (Medical Research Council Harwell, Oxfordshire, UK), and the E. coli Nei protein was generously provided by Dr. Dmitry Zharkov (Institute of Chemical Biology and Fundamental Medicine, Novosibirsk, Russia).
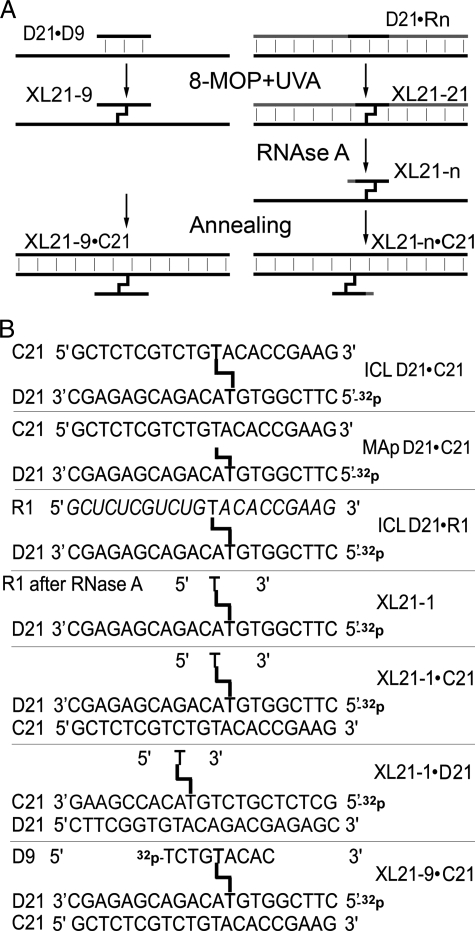
Preparation of Three-stranded DNA Substrate—Oligonucleotides containing psoralen mono- and diadducts were prepared as described previously (23). Shortly, oligonucleotide duplex was incubated for 15 min in the dark with 0.1 mm 8-MOP or other furanocoumarins in 50 μl of 100 mm Tris-HCl, pH 7.5, 5 mm EDTA, and 50 mm NaCl and then irradiated with 365 nm light at a dose of 240 kJ/m2 at room temperature. To prepare unhooked ICL substrate, the gel-purified cross-linked duplexes were incubated overnight at 37 °C with 20 μg/ml RNase A in 20 μl of Tris-EDTA buffer. The reaction was stopped by adding 0.1% SDS and 0.1 mg/ml protease K for 10 min at 50 °C and purified by denaturing PAGE. Oligonucleotides containing unhooked ICL referred to as XL21-n (n, number of remaining deoxyribonucleotides) were annealed with an excess of D21 or C21 oligonucleotide to obtain three-stranded DNA structures referred as XL21-n·C21 and XL21-n·D21, respectively, composed of the n-mer oligomer cross-linked to the 21-mer DNA duplex (see Fig. 2 and supplemental Fig. S2).
FIGURE 2.
Schematic presentation of the three-stranded DNA substrates used in the study. A, schematic presentation of the construction of three-stranded DNA structures with an unhooked adducted oligomer, which mimics ICL repair intermediate. RNA-containing strands are in gray color. B, schematic presentation of the primary structures of duplex and three-stranded DNA substrates used in the study.
DNA Repair Assay—The standard reaction mixture (20 μl) for ICL-specific DNA glycosylase activity contained 5 nm of 5′-32P-labeled XL21-1·C21, 25 mm HEPES-KOH, pH 7.6, 100 mm KCl, 1 mm EDTA, 5 mm 2-mercaptoethanol, 6% glycerol, and 20 nm NEIL1 for 10 min at 37 °C, unless otherwise stated. The incision assays with various BER proteins of different origins and the analysis of the reaction products were performed as described previously (23). For the reconstitution of the repair pathway for unhooked ICLs in vitro, 400 fmol of cold oligonucleotide containing unhooked ICL, XL21-1·C21, was incubated for 30 min at 25 °C with pure proteins, 5 nm APE1, 20 nm NEIL1, 10 nm polymerase β, and 2 units of T4 DNA ligase, in buffer (20 μl) containing 2 μCi of [α-32P]dTTP, 20 mm HEPES-KOH, pH 7.6, 100 mm KCl, 0.1 mg/ml bovine serum albumin, 1 mm dithiothreitol, 5 mm MgCl2, and 2 mm ATP. Reaction products were analyzed by electrophoresis at a constant temperature of 42 °C on denaturing 20% (w/v) polyacrylamide gels (29:1, 7 m urea, 0.5× Tris-borate-EDTA) for 2.5 h at 600 V. Gels were exposed to a Fuji FLA-3000 phosphor screen and analyzed using the Image Gauge Version 3.12 software.
RESULTS
Construction and Characterization of the Three-stranded DNA Structure Containing Unhooked Adducted Oligomer—Although the yield of psoralen MAs to pyrimidine bases is 3-fold higher than that of ICLs, the latter class of damage appears to have a more severe biological effect (3). In vitro NEIL1 is not able to excise psoralen-induced ICLs within duplex DNA; however, the sensitivity of NEIL1-depleted human cells to UVA+8-MOP exposure suggests that NEIL1 may be directly involved in the removal of psoralen-induced ICLs (23). Previously, the DNA repair intermediate containing unhooked psoralen-induced ICL was constructed and used as a substrate for E. coli UV-endonuclease (UvrABC) nuclease (26, 27). Here, we constructed and characterized three-stranded DNA structures containing a single ICL that mimics DNA repair intermediates resulting after XPF/ERCC1-mediated unhooking and TLS through ICL (Fig. 2A and supplemental Fig. S2). The single-stranded 21-mer DNA oligonucleotide D21, containing a single 5′-TpA site at position 9, was annealed either to complementary 21-mer chimeric RNA/DNA oligonucleotides (R1–R4) or to a 9-mer DNA oligonucleotide, D9 (Fig. 2B and supplemental Fig. S2). R1–R4 fragments were used to obtain short unhooked ICL oligomers, whereas D9 was used to obtain a longer one. After annealing, the oligonucleotide duplexes were exposed to 8-MOP+UVA, and the resulting cross-linked DNA were purified by denaturing PAGE. Photoreaction of asymmetric psoralens with helix DNA yields two orientational isomers of the ICL that can be separated by denaturing PAGE (28). As expected, the cross-linked duplexes migrated as two bands in the gel, suggesting the presence of two orientational isomers of psoralen-induced ICL exhibiting different helix instabilities (Fig. 3A, lanes 2, 4, and 5) (25). The cross-linked RNA/DNA hybrid duplexes were treated by RNase A to remove ribonucleotides from the R strand generating a very short 1–4-mer DNA oligomer covalently bound to D21, XL21-n (Fig. 2 and supplemental Fig. S2). Extensive RNase A degradation of the 21-mer cross-linked duplexes (Fig. 3A, lanes 4 and 5) yields “fast migrating” XL21-2 and XL21-4 products (lanes 6 and 7), which still migrate slower than the 21-mer single-stranded oligonucleotide, indicating that they contain adducted DNA oligomers resistant to ribonuclease hydrolysis. Following this, the XL21-1, -2, -3, -4, and XL21-9 oligonucleotides containing an unhooked ICL were hybridized to C21, a complementary 21-mer DNA oligonucleotide, to obtain the three-stranded DNA substrate, XL21-n·C21 (Fig. 2 and supplemental Fig. S2).
FIGURE 3.
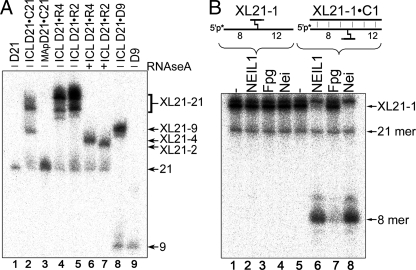
Construction and repair of the three-stranded DNA structures with an unhooked ICL adduct. A, electrophoretic mobility of the cross-linked DNA fragments. Lane 1, 21-mer single-stranded D21; lane 2, cross-linked D21·C21 duplex; lane 3, D21·C21 duplex containing single MAp; lanes 4 and 5, cross-linked D21·R4 and D21·R2 duplexes; lanes 6 and 7, as lanes 4 and 5 but treated with RNase A; lane 8, cross-linked D21·D9 duplex; lane 9, 9-mer single-stranded D9. B, DNA glycosylase activities on the duplex and three-stranded DNA structures containing unhooked ICL adduct. For details, see “Experimental Procedures.”
DNA Glycosylase-catalyzed Excision of Unhooked ICLs within Three-stranded DNA Structure—When using the 5′-32P-labeled XL21-1·C21 as a DNA substrate for E. coli Fpg, Nei, and human NEIL1 DNA glycosylases, we observed an 8-mer cleavage product suggesting excision of a cross-linked thymine at position 9 of D21 (Fig. 3B, lanes 6–8). Consequently, excision of adducted thymine residue within the ICL structure should also remove away the covalently bound short oligomer from XL21-1·C21. None of the DNA glycosylases tested were able to cleave XL21-1 when it was not annealed to C21 (lanes 2–4). It should be stressed that the appearance of the 5′-32P-labeled 8-mer product upon incubation with the DNA glycosylases was accompanied by the loss of XL21-1 (lanes 6–8), suggesting that Fpg, Nei, and NEIL1 specifically recognize an unhooked ICL within the three-stranded DNA structure but not in duplex DNA. Similar results were obtained with the other three-stranded DNA substrates containing unhooked oligomers of varying length and different sequence context (data not shown). Interestingly, when acting upon the three-stranded DNA substrate, in addition to the major 8-mer product, Nei and NEIL1 generate a minor slow migrating fragment (Fig. 3B, lanes 6 and 8). It was shown that Nei and NEIL1 can mediate β-elimination in addition to β,σ-lyase activity when acting on 5,6-dihydropyrimidines containing oligonucleotides. Subsequently, these bifunctional DNA glycosylases generate two cleavage products, one containing a 3′-α,β-unsaturated aldehyde and another containing 3′-phosphate, respectively (22). Based on this observation, we suggest that the minor slow migrating band in lanes 6 and 8 is a product of β-elimination reaction catalyzed by Nei-like DNA glycosylases.
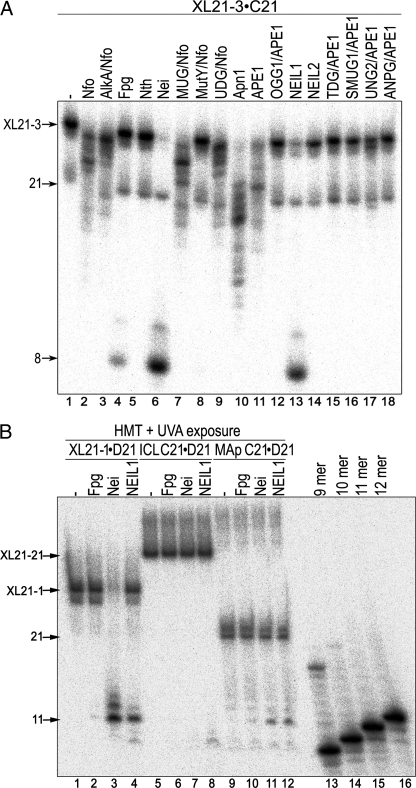
Next we investigated whether the unhooked ICLs were also substrates for previously characterized BER enzymes. We challenged the 5′-32P-labeled XL21-3·C21 containing an unhooked 3-mer DNA oligomer with the purified DNA glycosylases and apurinic/apyrimidinic (AP) endonucleases of E. coli, yeast, and human origins listed below. Because not all DNA glycosylases are endowed with AP-nicking activity, the assays were made in the presence of an AP endonuclease to cleave DNA duplex at the potential abasic sites generated after base excision. Among various DNA repair enzymes tested, only incubation with Nei and NEIL1, and with less efficiency, Fpg generated an 8-mer fragment (Fig. 4A, lanes 4, 6, and 13). Interestingly, excision of the unhooked DNA fragment by the Nei-like enzymes was more proficient than by Fpg (lanes 4 and 6 versus lane 13). Despite being used in an excess amount, Nfo, AlkA, Nth, MUG, MutY, UDG, Apn1, APE1, OGG1, NEIL2, TDG, SMUG1, hUNG, ANPG, and NTH1 proteins did not excise the unhooked ICL in XL21-3·C21 (Fig. 4A and data not shown). For quantitative comparison of the substrate specificity of NEIL1 toward an unhooked ICL, the amounts of incised oligonucleotides containing thymine glycol (canonical DNA substrate), MAp, and ICL were measured. Protein concentration-dependent cleavage reveals that all three DNA substrates were excised with high efficiency, showing that the unhooked ICL is one of the preferred substrates for NEIL1 (supplemental Fig. S3).
FIGURE 4.
DNA glycosylase-catalyzed excision of the unhooked ICL adducts when present in three-stranded DNA structure. A, activity of the various BER enzymes toward three-stranded oligonucleotide XL21-3·C21 containing single unhooked 8-MOP-induced ICL. 10 nm XL21-3·C21 32P-labeled at 5′ of 21-mer D21 was incubated with 20 nm of a respective enzyme at 37 °C for 10 min. Note that the D21 strand contains cross-linked thymine at position 9. Lane 1, control non-treated XL21-3·C21; lanes 2–18, as lane 1 but incubated with enzymes. B, DNA glycosylase activity of NEIL1 toward HMT-induced ICLs and MAs. 10 nm of 5′-32P-labeled oligonucleotides exposed to HMT+UVA were incubated with 20 nm DNA glycosylase at 37 °C for 10 min. In all DNA substrates, the 21-mer C21 strand was 32P-labeled at 5′. Note that the C21 strand contains cross-linked thymine at position 12. Lanes 1–4, three-stranded DNA construct XL21-1·D21; lanes 5–9, C21·D21 duplex oligonucleotide containing a single ICL; lanes 10–12, C21·D21 duplex oligonucleotide containing a single MAp; lanes 13–16, size markers. The products of the reaction were analyzed as described under “Experimental Procedures.”
The unhooked ICL fragment can be adducted to thymine in the duplex either via the furan side or via the pyrone side of a psoralen cross-link, resulting in two orientational isomers. Previously, it was shown that E. coli UvrABC nuclease selectively incises the pyrone side strand of the psoralen ICL within a three-stranded structure (29). The question arises whether Nei-like DNA glycosylases excise both orientational isomers in a three-stranded DNA substrate. When analyzing DNA glycosylase activities on the 5′-32P-labeled XL21-1·C21 and XL21-3·C21, we found that Nei and NEIL1 cleaves more than 90% of the substrate (Figs. 3B, lanes 6 and 8, and 4A, lanes 6 and 13). Assuming an equal probability for the formation of each isomer, these results suggest that the DNA glycosylases are able to excise both orientational isomers. However, we cannot exclude that Nei and NEIL1 might display some preference for one of the isomers.
Activities of Nei-like DNA Glycosylases toward Bulky DNA Adducts Induced by Various Psoralen and Isopsoralen Derivatives—An important issue is whether any bulky DNA adduct is a good substrate for the bifunctional DNA glycosylases when present in a three-stranded DNA construct or whether Nei and NEIL1 are highly specific to 8-MOP-induced mono- and di-DNA adducts and not to ICL as a general structure. To address this question, the specific activities of the DNA glycosylases were tested on DNA adducts induced by various furanocoumarins, distinct from 8-MOP: HMT and isopsoralens Ang, 3CP, and MePy. HMT is a structural analogue of 8-MOP containing in addition three methyl groups and one hydroxymethyl group. In contrast to 8-MOP, HMT has higher dark binding activity to DNA and forms less than 2% pyrone side monoadduct (30). Subsequently, HMT induces higher yield of ICLs in duplex DNA as compared with 8-MOP. Using the approach described above, we constructed a duplex C21·D21 containing a single ICL and three-stranded DNA structures XL21-1·D21 containing an unhooked ICL induced by HMT+UVA exposure. To obtain the HMT-derived MAp, cross-linked C21·D21 was incubated under hot alkali conditions. The primary structure of XL21-1·D21 with 5′-32P-labeled C21 strand containing cross-linked thymine at position 12 is shown in Fig. 2B. Nei and NEIL1 incise XL21-1· D21 and MAp-containing C21·D21 to generate an 11-mer cleavage products, indicating the excision of both HMT-thymine monoadduct (Fig. 4B, lanes 11–12) and unhooked HMT-ICL (lanes 3 and 4) at position 12 of C21 strand. Although we did not previously observe any activity on the duplex oligonucleotide containing 8-MOP-induced ICL (23), NEIL1 but not Nei incises with a very low efficiency HMT cross-linked duplex oligonucleotide C21·D21 and generates an ∼9-mer cleavage fragment, indicating the excision of a thymine at position 10 of the 21-mer C21 oligonucleotide (Fig. 4B, lane 8). Therefore, we may propose that in addition to thymine 12, a second minor photoreactive site is present in C21 oligonucleotide thymine 10. Because this thymine cannot form ICL with the opposite strand, it may form a monoadduct with HMT, which in turn becomes a substrate for NEIL1. These results suggest that substrate specificities of Nei-like DNA glycosylases are not limited to 8-MOP-generated ICLs but also include ICLs induced by compounds structurally distinct from 8-MOP and endowed with different properties.
We also investigated whether NEIL1 recognizes bulky DNA monoadducts generated by monofunctional isopsoralens Ang, 3CP, and MePy. For this, the isopsoralens+UVA-treated covalently closed circular plasmid DNA and the 21-mer D21·C21 duplex oligonucleotide were incubated with NEIL1. NEIL1 cleaves covalently closed circular DNA and converts it to open circular form, suggesting that it specifically excises bulky Ang-, 3CP-, and MePy-induced DNA adducts (supplemental Fig. S4A). Unexpectedly, 5′-labeled D21·C21 oligonucleotides treated by photoactivated isopsoralens were only weakly incised by NEIL1. This is likely due to inefficient formation of isopsoralen monoadducts in short oligonucleotides (supplemental Fig. S4B). Finally, altogether these results suggest that Nei-like DNA glycosylases recognize structurally diverse types of bulky DNA adducts and unhooked ICLs.
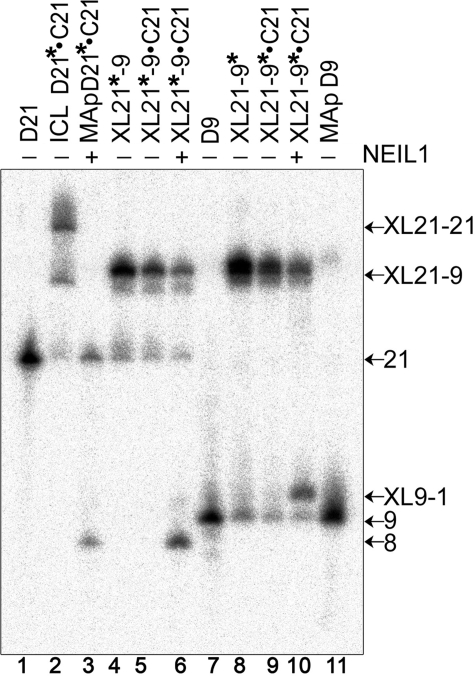
Mechanism of Action of NEIL1—To investigate the fine mechanism of action of NEIL1-mediated repair of ICLs, we used XL21-9·C21 containing the 9-mer D9 cross-linked to 21-mer D21·C21 duplex as a three-stranded DNA substrate (Fig. 2B). As with XL21-1·C21 and XL21-3·C21 substrates, NEIL1 generates an 8-mer product when acting upon XL21*-9·C21 containing 5′-32P-labeled D21 (Fig. 5, lane 6, asterisks indicate 32P-labeled strand). In contrast, when acting upon XL21-9·C21 containing 5′-32P-labeled D9, NEIL1 generates a short ∼9-mer DNA fragment (lane 10), which migrates slower than both the regular 9-mer (lane 7) and the 9-mer fragment containing MAp residue (lane 11). Based on these observations, we propose that the slow migrating ∼9-mer fragment carries a thymine residue adducted to D9. Again, appearance of a fast migrating band upon incubation with NEIL1 was concomitant with the reduction of the upper band, XL21-9, indicating the removal of the ICL in the three-stranded oligonucleotide, XL21-9·C21 (lane 10). Altogether, these results strongly suggest that NEIL1 cleaves the glycosidic bond between the psoralen-thymine adduct and the deoxyribose sugar to generate as reaction products thymine base cross-linked to the 9-mer oligonucleotide and the 21-mer DNA duplex with a single nucleotide gap.
FIGURE 5.
Mechanism of action of NEIL1 on the three-stranded DNA substrate. Lane 1, 5′-32P-labeled 21-mer D21; lane 2, cross-linked D21·C21 duplex 32P-labeled at 5′ of D21; lane 3, as lane 2 but treated with alkali to generate MAp; lane 4, XL21-9 32P-labeled at 5′ of the 21-mer D21; lane 5, as lane 4 but hybridized to the 21-mer C21 to obtain three-stranded construct; lane 6, as lane 5 but incubated with NEIL1; lane 7, 5′-32P-labeled 9-mer D9; lane 8, XL21-9 32P-labeled at 5′ of the 9-mer D9; lane 9, as lane 8 but hybridized to the 21-mer C21 to obtain three-stranded construct; lane 10, as lane 9 but incubated with NEIL1; lane 11, XL21-9 32P-labeled at 5′ of the 9-mer D9 treated with alkali to obtain MAp. Asterisks indicate the 32P-labeled strand. For details, see “Experimental Procedures.”
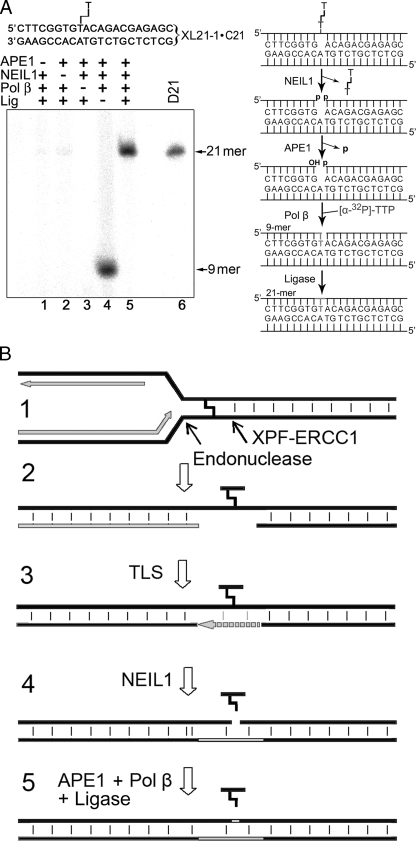
To examine whether NEIL1-catalyzed excision of an unhooked ICL in the three-stranded DNA substrate generates a gapped DNA duplex, we have reconstituted the BER pathway in vitro using purified proteins and non-labeled XL21-1·C21 oligonucleotide (Fig. 6A). As expected, incubation of XL21-1·C21 with NEIL1, APE1, DNA polymerase β, and [α-32P]TTP generates a labeled 9-mer fragment (lane 4). This result demonstrates that NEIL1 excises the adducted thymine residue in the 21-mer strand of XL21-1, generating a single nucleotide gap with 3′-P and 5′-P groups, and then APE1 removes the blocking 3′-P residue, allowing DNA polymerase β to incorporate one 32P-labeled thymidine nucleotide. The addition of a DNA ligase completes the restoration of the full-length 21-mer D21·C21 duplex (lane 5). In summary, these biochemical data obtained in vitro suggest that in vivo, the unhooked ICLs in the three-stranded DNA repair intermediate can be efficiently removed in the short patch BER pathway without recruitment of the NER machinery.
FIGURE 6.
NEIL1-initiated DNA repair pathway for ICLs. A, in vitro reconstitution of the ICL repair. Reactions were performed in presence of [α-32P]dTTP with non-labeled XL21-1·C21 containing a single unhooked ICL. Lanes 1–5, assays with various combinations of proteins; lane 6, 5′-32P-labeled 21 mer D21. Pol β, DNA polymerase β; Lig, T4 DNA ligase. B, a model for replication-associated repair of psoralen-induced ICLs. (1) ICL induces stalled replication fork by preventing strand separation. (2) Structure-specific endonucleases incise on the 3′-side of ICL to generate a double strand break and on the 5′-side to unhook the ICL. The unhooked cross-linked fragment swings out of the helix. (3) The single-stranded gap is bypassed by a TLS DNA polymerase. (4) NEIL1 excises the unhooked fragment in three-stranded DNA structure. (5) The single nucleotide gap is repaired by APE1 and DNA polymerase β.
DISCUSSION
Putative Model for Psoralen-induced ICL Repair in Human Cells—Several arguments point to the preferential involvement of NEIL1 in the replication-associated repair of ICLs. (i) NEIL1 can act toward the three-stranded DNA repair intermediate generated during DNA replication; (ii) NEIL1 is an S phase regulated protein (21); (iii) it excises base lesions from single-stranded DNA regions, and (iv) it interacts with proliferating cell nuclear antigen (PCNA) and flap endonuclease 1 (FEN-1) (31, 32). Based on these data, we propose a model for the mechanism of ICL repair in mammalian cells that implicates the DNA glycosylase activity of NEIL1 downstream of XPF/ERCC1 and TLS repair steps (Fig. 6B). Several studies have demonstrated that an ICL at the stalled replication fork is converted to a double strand break and unhooked cross-linked fragment via dual incisions that bracket the lesion site (steps 1 and 2) (9, 11–13). Once the ICL is released from one strand, it may swing out of the helix to allow bypass by a TLS DNA polymerase to generate a postulated three-stranded DNA repair intermediate (step 3) (9, 14). NEIL1 excises the unhooked cross-linked fragment (step 4), resulting in the single nucleotide gap, which is then filled in the short patch BER pathway (step 5). Besides the repair of bulky DNA adducts, NEIL1 was initially characterized as a DNA glycosylase specific for oxidized, saturated, and ring-fragmented bases (21, 22). Similar to ICLs, clustered base damage can also block replicative DNA polymerases on both strands, resulting in the stalled replication fork, which is in turn prone to mechanical or nuclease-induced breakage. Earlier, it has been demonstrated that NEIL1 is specialized in excision of oxidized bases located in the proximity of single strand breaks (33, 34). Taken together, these observations suggest that NEIL1 may also repair base lesions left after replication fork collapse.
Structural Implications for DNA Damage Recognition in BER—In general, substrate specificities of DNA glycosylases are limited to small non-bulky DNA base damage. Three-dimensional crystal structures of the Nei-DNA complex and free NEIL1 protein showed that both proteins consist of two domains forming a DNA binding cleft (35, 36). Based on molecular modeling, it was suggested that NEIL1 binds to DNA and flips out damaged bases in a shallow and comparatively cramped recognition pocket (37). However, the psoralen MAs and three-stranded DNA structure containing unhooked ICL fragments with varying length from 1 to 17 nucleotides constitute very large substrates that could present topological constraint for their accommodation in the active site of Nei-like DNA glycosylases. Several lines of evidence argue that DNA glycosylases are able to accommodate bulky DNA adducts despite their relatively small active sites. It was demonstrated that Fpg and T4 endonuclease V recognize very bulky DNA adducts such as an imidazole ring opened form of N-hydroxy-2-aminofluorene, a C8 guanine adduct, and a cyclobutane dimer, respectively (38, 39). Interestingly, human O6-alkylguanine-DNA alkyltransferase (hAGT) can repair an oligonucleotide containing a heptane (seven-carbon) cross-link via formation of a hAGT-oligonucleotide complex and further reaction of a second hAGT molecule yielding a hAGT dimer and free DNA (40). Three-dimensional crystal structures of the complex of T4 endonuclease V and Fpg bound to their bulky DNA substrates provide insights to the structural basis of bulky lesion accommodation in the DNA glycosylase active sites (39, 41). T4 endonuclease V kinks the DNA helix by about 60° and flips out the opposing adenine base complementary to thymine dimer out of the DNA base stack, thus avoiding steric problems (39), whereas Fpg flips the bulky N7-substituted FapydG derivative guanine lesion out of the DNA helix to the binding pocket but enables the N7-bulky group to stay outside (41). Based on these observations, we may hypothesize that translesion synthesis across unhooked ICL results in ejection of the base with adducted oligomer from duplex and formation of a three-stranded DNA repair intermediate. This might enable recognition and accommodation of the psoralen-thymine adduct in a “flipped out” conformation by Nei and NEIL1. Furthermore, we propose that the DNA glycosylases accommodate only thymine base in their active site, whereas the bulky cross-linked DNA oligomer stays outside, thus avoiding severe steric hindrance. It is tempting to speculate that in the protein-DNA cross-links, expulsion of the DNA base adducted to protein out of the duplex may be also recognized by NEIL1. However, a detailed structural model of Neilike DNA glycosylase interaction with bulky and ICL DNA adducts will have to await further investigations.
Conclusion
DNA glycosylase-initiated BER is a major pathway for the removal of non-bulky oxidative DNA base damage, whereas it was thought that bulky DNA lesions, such as psoralen-induced MAs and ICLs, are eliminated only in the NER pathway (17, 42). In this study, we demonstrated that the Nei-like proteins from E. coli to human cells can remove the unhooked psoralen-derived ICLs via hydrolysis of a glycosidic bond between adducted base and deoxyribose sugar, thus providing an alternative pathway to the classic NER. Consistent with these results, HeLa cells with reduced expression of the NEIL1 and APE1 proteins are sensitive to the 8-MOP+UVA exposure (23). However, the biochemical experiments use only photoactivated psoralen-induced MAs and ICLs as DNA substrates, raising a question whether NEIL1-mediated repair makes a contribution only to cells challenged with these agents. Given the wide variety of the unhooked ICL substrates tested, we propose to extrapolate these results to a more general case. Because NEIL1 is involved in the repair of both MA and ICL, our results may explain why xeroderma pigmentosum cells defective in the NER pathway are only moderately sensitive to the cross-linking agents. The DNA replication-linked BER pathway removes unhooked ICLs and oxidized bases occurring at a broken replication fork. Otherwise, persistence of the unrepaired adducted template DNA strand leads to the inability to accurately repair replication-induced double strand break, which in turn results in a high frequency of DNA damage-induced chromosomal aberrations in cells exposed to cross-linking agents. The biological role of NEIL1 in the repair of DNA damage is further supported by the fact that NEIL1 deficiency and/or low DNA glycosylase activities might be involved in the pathogenesis of a subset of gastric cancers and increased risk of metabolic syndrome (43, 44). Finally, this alternative repair pathway through new unexpected results would pave the road for further investigations to establish its structural base and biological relevance, therefore challenging the existing paradigm.
Supplementary Material
Acknowledgments
We thank J. Laval, D. Averbeck, J.-M. Saucier, and A. Kuzminov for helpful discussion. We are grateful to Hiroshi Ide (Hiroshima University, Japan), Grigory Dianov (MRC, Harwell, Oxfordshire, UK), and Dmitry Zharkov (ICBFM, Novosibirsk, Russia) for the recombinant proteins.
Author's Choice—Final version full access.
This work was supported by grants from the Fondation pour la Recherche Médicale “EQUIPES FRM 2007,” FP6 Euroatom Grant RISC-RAD FI6R-CT-2003-508842, CNRS France-Pologne GRDE 182, Institut National du Cancer (INCa), Electricité de France (EDF), and National Center for Biotechnology, Kazakhstan (to M. K. S.) and from La Ligue Contre le Cancer “Equipe Labellisée 2006,” EDF, and INCa (to F. R.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental figures S1–S4.
Footnotes
The abbreviations used are: ICL, DNA interstrand cross-link; 8-MOP, 8-methoxypsoralen; XL, duplex and/or three-stranded oligonucleotide containing single unhooked ICL; MA, psoralen-thymine monoadduct; MAp, pyron side MA; MAf, furan side MA; HMT, 4′-hydroxy-methyl-4,5′,8-trimethylpsoralen; Ang, angelicin; 3CP, 3-carbethoxypsoralen; MePy, 7 methylpyrido(3,4-c)psoralen; BER, base excision repair; NER, nuclear excision repair; XPF/ERCC1, Xeroderma Pigmentosum group F/Excision Repair Cross-Complementing 1 endonuclease complex; NEIL1, human Nei-like oxidative DNA glycosylase 1; AP, apurinic/apyrimidinic; APE1, major human AP endonuclease 1; TLS, translesion synthesis; hAGT, human O6-alkylguanine-DNA alkyltransferase; UVA, ultraviolet light A.
References
- 1.Dronkert, M. L., and Kanaar, R. (2001) Mutat. Res. 486 217–247 [DOI] [PubMed] [Google Scholar]
- 2.McHugh, P. J., Spanswick, V. J., and Hartley, J. A. (2001) Lancet Oncol. 2 483–490 [DOI] [PubMed] [Google Scholar]
- 3.Cimino, G. D., Gamper, H. B., Isaacs, S. T., and Hearst, J. E. (1985) Annu. Rev. Biochem. 54 1151–1193 [DOI] [PubMed] [Google Scholar]
- 4.Cole, R. S. (1971) Biochim. Biophys. Acta 254 30–39 [DOI] [PubMed] [Google Scholar]
- 5.Johnston, B. H., and Hearst, J. E. (1981) Biochemistry 20 739–745 [DOI] [PubMed] [Google Scholar]
- 6.Eichman, B. F., Mooers, B. H., Alberti, M., Hearst, J. E., and Ho, P. S. (2001) J. Mol. Biol. 308 15–26 [DOI] [PubMed] [Google Scholar]
- 7.Hwang, G. S., Kim, J. K., and Choi, B. S. (1996) Biochem. Biophys. Res. Commun. 219 191–197 [DOI] [PubMed] [Google Scholar]
- 8.Spielmann, H. P., Dwyer, T. J., Sastry, S. S., Hearst, J. E., and Wemmer, D. E. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 2345–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raschle, M., Knipsheer, P., Enoiu, M., Angelov, T., Sun, J., Griffith, J. D., Ellenberger, T. E., Scharer, O. D., and Walter, J. C. (2008) Cell 134 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanada, K., Budzowska, M., Modesti, M., Maas, A., Wyman, C., Essers, J., and Kanaar, R. (2006) EMBO J. 25 4921–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niedernhofer, L. J., Odijk, H., Budzowska, M., van Drunen, E., Maas, A., Theil, A. F., de Wit, J., Jaspers, N. G., Beverloo, H. B., Hoeijmakers, J. H., and Kanaar, R. (2004) Mol. Cell. Biol. 24 5776–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuraoka, I., Kobertz, W. R., Ariza, R. R., Biggerstaff, M., Essigmann, J. M., and Wood, R. D. (2000) J. Biol. Chem. 275 26632–26636 [DOI] [PubMed] [Google Scholar]
- 13.Fisher, L. A., Bessho, M., and Bessho, T. (2008) J. Biol. Chem. 283 1275–1281 [DOI] [PubMed] [Google Scholar]
- 14.Minko, I. G., Harbut, M. B., Kozekov, I. D., Kozekova, A., Jakobs, P. M., Olson, S. B., Moses, R. E., Harris, T. M., Rizzo, C. J., and Lloyd, R. S. (2008) J. Biol. Chem. 283 17075–17082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nojima, K., Hochegger, H., Saberi, A., Fukushima, T., Kikuchi, K., Yoshimura, M., Orelli, B. J., Bishop, D. K., Hirano, S., Ohzeki, M., Ishiai, M., Yamamoto, K., Takata, M., Arakawa, H., Buerstedde, J. M., Yamazoe, M., Kawamoto, T., Araki, K., Takahashi, J. A., Hashimoto, N., Takeda, S., and Sonoda, E. (2005) Cancer Res. 65 11704–11711 [DOI] [PubMed] [Google Scholar]
- 16.Shen, X., Jun, S., O'Neal, L. E., Sonoda, E., Bemark, M., Sale, J. E., and Li, L. (2006) J. Biol. Chem. 281 13869–13872 [DOI] [PubMed] [Google Scholar]
- 17.Cipak, L., Watanabe, N., and Bessho, T. (2006) Nat. Struct. Mol. Biol. 13 729–733 [DOI] [PubMed] [Google Scholar]
- 18.Hoy, C. A., Thompson, L. H., Mooney, C. L., and Salazar, E. P. (1985) Cancer Res. 45 1737–1743 [PubMed] [Google Scholar]
- 19.De Silva, I. U., McHugh, P. J., Clingen, P. H., and Hartley, J. A. (2000) Mol. Cell. Biol. 20 7980–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hitomi, K., Iwai, S., and Tainer, J. A. (2007) DNA Repair (Amst.) 6 410–428 [DOI] [PubMed] [Google Scholar]
- 21.Hazra, T. K., Izumi, T., Boldogh, I., Imhoff, B., Kow, Y. W., Jaruga, P., Dizdaroglu, M., and Mitra, S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 3523–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandaru, V., Sunkara, S., Wallace, S. S., and Bond, J. P. (2002) DNA Repair (Amst.) 1 517–529 [DOI] [PubMed] [Google Scholar]
- 23.Couve-Privat, S., Mace, G., Rosselli, F., and Saparbaev, M. K. (2007) Nucleic Acids Res. 35 5672–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, X., Peterson, C. A., Zheng, H., Nairn, R. S., Legerski, R. J., and Li, L. (2001) Mol. Cell. Biol. 21 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumaresan, K. R., Ramaswamy, M., and Yeung, A. T. (1992) Biochemistry 31 6774–6783 [DOI] [PubMed] [Google Scholar]
- 26.Sladek, F. M., Munn, M. M., Rupp, W. D., and Howard-Flanders, P. (1989) J. Biol. Chem. 264 6755–6765 [PubMed] [Google Scholar]
- 27.Cheng, S., Sancar, A., and Hearst, J. E. (1991) Nucleic Acids Res. 19 657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Houten, B., Gamper, H., Hearst, J. E., and Sancar, A. (1986) J. Biol. Chem. 261 14135–14141 [PubMed] [Google Scholar]
- 29.Cheng, S., Van Houten, B., Gamper, H. B., Sancar, A., and Hearst, J. E. (1988) J. Biol. Chem. 263 15110–15117 [PubMed] [Google Scholar]
- 30.Kanne, D., Rapoport, H., and Hearst, J. E. (1984) J. Med. Chem. 27 531–534 [DOI] [PubMed] [Google Scholar]
- 31.Dou, H., Theriot, C. A., Das, A., Hegde, M. L., Matsumoto, Y., Boldogh, I., Hazra, T. K., Bhakat, K. K., and Mitra, S. (2008) J. Biol. Chem. 283 3130–3140 [DOI] [PubMed] [Google Scholar]
- 32.Hegde, M. L., Theriot, C. A., Das, A., Hegde, P. M., Guo, Z., Gary, R. K., Hazra, T. K., Shen, B., and Mitra, S. (2008) J. Biol. Chem. 283 27028–27037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsons, J. L., Zharkov, D. O., and Dianov, G. L. (2005) Nucleic Acids Res. 33 4849–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsons, J. L., Kavli, B., Slupphaug, G., and Dianov, G. L. (2007) Biochemistry 46 4158–4163 [DOI] [PubMed] [Google Scholar]
- 35.Zharkov, D. O., Golan, G., Gilboa, R., Fernandes, A. S., Gerchman, S. E., Kycia, J. H., Rieger, R. A., Grollman, A. P., and Shoham, G. (2002) EMBO J. 21 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doublie, S., Bandaru, V., Bond, J. P., and Wallace, S. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10284–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia, L., Shafirovich, V., Geacintov, N. E., and Broyde, S. (2007) Biochemistry 46 5305–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boiteux, S., Bichara, M., Fuchs, R. P., and Laval, J. (1989) Carcinogenesis 10 1905–1909 [DOI] [PubMed] [Google Scholar]
- 39.Vassylyev, D. G., Kashiwagi, T., Mikami, Y., Ariyoshi, M., Iwai, S., Ohtsuka, E., and Morikawa, K. (1995) Cell 83 773–782 [DOI] [PubMed] [Google Scholar]
- 40.Fang, Q., Noronha, A. M., Murphy, S. P., Wilds, C. J., Tubbs, J. L., Tainer, J. A., Chowdhury, G., Guengerich, F. P., and Pegg, A. E. (2008) Biochemistry 47 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coste, F., Ober, M., Le Bihan, Y. V., Izquierdo, M. A., Hervouet, N., Mueller, H., Carell, T., and Castaing, B. (2008) Chem. Biol. 15 706–717 [DOI] [PubMed] [Google Scholar]
- 42.Svoboda, D. L., Taylor, J. S., Hearst, J. E., and Sancar, A. (1993) J. Biol. Chem. 268 1931–1936 [PubMed] [Google Scholar]
- 43.Shinmura, K., Tao, H., Goto, M., Igarashi, H., Taniguchi, T., Maekawa, M., Takezaki, T., and Sugimura, H. (2004) Carcinogenesis 25 2311–2317 [DOI] [PubMed] [Google Scholar]
- 44.Vartanian, V., Lowell, B., Minko, I. G., Wood, T. G., Ceci, J. D., George, S., Ballinger, S. W., Corless, C. L., McCullough, A. K., and Lloyd, R. S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 1864–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.