Abstract
Calmodulin (CaM) is a highly conserved intracellular calcium sensor. In plants, CaM also appears to be present in the apoplasm, and application of exogenous CaM has been shown to influence a number of physiological functions as a polypeptide signal; however, the existence and localization of its corresponding apoplasmic binding sites remain controversial. To identify the site(s) of action, a CaM-conjugated quantum dot (QD) system was employed for single molecule level detection at the surface of plant cells. Using this approach, we show that QD-CaM binds selectively to sites on the outer surface of the plasma membrane, which was further confirmed by high resolution transmission electron microscopy. Measurements of Ca2+ fluxes across the plasma membrane, using ion-selective microelectrodes, demonstrated that exogenous CaM induces a net influx into protoplasts. Consistent with these flux studies, calcium-green-dextran and FRET experiments confirmed that applied CaM/QD-CaM elicited an increase in cytoplasmic Ca2+ levels. These results support the hypothesis that apoplasmic CaM can act as a signaling agent. These findings are discussed in terms of CaM acting as an apoplasmic peptide ligand to mediate transmembrane signaling in the plant kingdom.
Calmodulin (CaM)2 is a conserved multifunctional calcium sensor that mediates intracellular Ca2+ signaling and regulates diverse cellular processes by interacting with calmodulin-binding proteins (1–3). Interestingly, in both animals and plants, CaM may also act as an extracellular agent to regulate physiological events (4). Consistent with this notion, extracellular CaM has been detected within the cell walls of a broad range of plant species (4, 5).
Functional studies have established that exogenously applied CaM can stimulate the proliferation of suspension-cultured plant cells (6) as well as affect intracellular activities of heterotrimeric G proteins and phospholipases in protoplasts (7, 8). Based on these findings, it has been proposed that, in plants, extracellular CaM may function as a signaling agent involved in the regulation of cell growth and development (4). However, as a 17-kDa hydrophilic protein, exogenously applied CaM could well be retrieved from the apoplasmic space and then exert its effects on components within the cytoplasm. Evidence against this hypothesis was provided by studies with Arabidopsis thaliana suspension-cultured cells in which it was shown that 24 h of incubation in exogenous CaM did not result in protein uptake or degradation (4).
To exert an effect from the apoplasm, it would seem logical to assume that a protein(s) within the plant plasma membrane would have a CaM-binding site exposed to the apoplasm. Although a number of studies have addressed the molecular mechanism(s) by which extracellular CaM might act as a signal (6, 9) and attempts have been made to identify extracellular CaM-binding proteins (4, 6), currently there is no direct evidence in support of the hypothesis that specific CaM-binding sites exist at the surface of plant cells.
To address this question, a CaM-conjugated quantum dot (QD) system was employed for single molecule level detection (10–13) at the surface of plant cells. These nanoparticles have several advantages over conventional fluorophores for light microscopic imaging, including their higher brightness and photostability (14, 15). In addition, because of their electron dense nature, QDs can be used for single labeling studies at the transmission electron microscope level (16, 17). Using this QD-CaM system, we demonstrate that QD-CaM binds selectively to sites on the outer surface of the plant plasma membrane. We also show by three independent methods that applied CaM can modulate Ca2+ fluxes across the plasma membrane, leading to alterations in cytoplasmic Ca2+ status. These findings support the hypothesis that, in plants, apoplasmic CaM can act as a signaling agent.
EXPERIMENTAL PROCEDURES
Cadmium Telluride QD Preparation—Briefly, sodium hydrogen telluride was prepared by mixing sodium borohydride and tellurium (molar ratio of 2:1) in water, and N2-saturated deionized water was then added to yield a final concentration of 47 mm sodium hydrogen telluride. Next, a precursor solution was prepared by dissolving CdCl2 (1 mm) and 3-mercaptopropionic acid (1.2 mm) in 50 ml of deionized water; the pH value was then adjusted to 9.0 by the stepwise addition of NaOH solution. An aliquot (100 μl) of oxygen-free solution containing fresh sodium hydrogen telluride, cooled to 0 °C, was then added to 10 ml of precursor solution and vigorously stirred. The resultant solution was put into a Teflon-lined stainless steel container and autoclaved at the reaction temperature for periods ranging from 5 to 180 min. Solutions containing the 3-mercaptopropionic acid-capped cadmium telluride QDs were then cooled to room temperature, and the fluorescence emission spectrum measured on a Fluorolog-3 spectrofluorometer (see supplemental Fig. S1 for a full description of the procedures employed).
Plant Material—Newly matured pollen grains were collected from tobacco (Nicotiana tabacum L.) and lily (Lilium longiflorum Thunb.) growing in a greenhouse at the Botanical Garden of the Institute of Botany, Chinese Academy of Sciences (Beijing, China). Tobacco BY-2 and A. thaliana suspension-cultured cells were propagated in the appropriate media (18). Transgenic ECFP-calmodulin-M13-EYFP A. thaliana plants were grown in a greenhouse (19).
Preparation of Protein-conjugated QD Probes—Purified plant CaM was prepared from A. thaliana according to Zielinski (20). The CaM tryptic peptides, CaM TR1C (N-terminal fragment containing residues 1–75) and CaM TR2C (C-terminal fragment containing residues 78–148) were kindly provided by Prof. Akke (Lund University). CmPP16, a 16-kDa RNA-binding protein from the pumpkin phloem sap, was expressed as a recombinant protein from Escherichia coli, as previously described (21).
CaM, BSA, CmPP16, CaMTR1C, and CaMTR2C conjugation to QDs was performed as previously described (11, 22). The reaction volume was 0.2 ml of MES-NaOH buffer (pH 5.8) in which the concentration of QDs was 1 μm, and that of proteins was 100 μm. Concentrations of 1-ethyl-3-(3)-dimethylaminopropyl carbodiimide (EDC) and N-hydroxysuccinimide (NHS) were 1 and 0.3 mm, respectively. The solutions were incubated and vortexed at room temperature, generally for 4 h. QD-protein probes were separated from free protein by precipitation, through the addition of 0.6 ml of acetone, followed by centrifugation for 20 min. The resultant pellet was air-dried in the dark; prior to use, it was dispersed in phosphate-buffered saline (pH 7.2) containing 0.1% (v/v) β-mercaptoethanol.
The EDC-NHS-mediated coupling reaction was performed on the silylanized slides to confirm that the test proteins were being conjugated to the QD nanoparticles. EDC reacts with a carboxyl group to form an amine-reactive O-acylisourea intermediate, which can then react with an amine to yield a conjugate of the two molecules joined by a stable amide bond. The addition of sulfo-NHS stabilizes the amine-reactive intermediate by converting it to an amine-reactive sulfo-NHS ester, thus increasing the efficiency of the EDC-mediated coupling reaction (23). Either single QDs or QDs and test protein were added into this reaction system, and after 2 h, the slides were extensively washed with deionized water and observed with a Bio-Rad MRC 600 laser-scanning confocal microscope.
Protoplast Preparation—Isolation of pollen protoplasts was based on the method of Yu et al. (10), with some modifications. Purified mature tobacco and lily pollen was first germinated in a medium containing 1 mm KNO3, 1 mm Ca (NO3)2, 1 mm MgSO4, 1 mm H3BO3, and 12% sucrose (pH 5.8). When germinating pollen tubes were as long as the pollen grain diameter, they were transferred into enzymatic solution containing 1% cellulase (Onozuka R-10), 1% Macerozyme (R-10, Japan), 1 mm KCl, 0.8 mm MgSO4, 1 mm CaCl2, 10 mm MES, and 12% mannitol (pH 5.8). Intact pollen protoplasts were released after 1–3 h of enzymatic digestion at 30 °C in the dark. A cell wall-specific stain, Calcofluor White ST, was used to monitor the enzymatic removal of the wall. Pollen protoplast viability was confirmed using the vital dye, fluorescein diacetate, as previously described (10). After staining with 10 mg/ml fluorescein diacetate for 20 min, protoplasts were washed with fluorescein diacetate-free medium and observed with a Leica Dmire 2 fluorescence microscope.
Protoplasts from tobacco BY-2 and Arabidopsis suspension-cultured cells were isolated by adding 1% cellulase and 1% Macerozyme to the culture medium. Arabidopsis mesophyll protoplasts were isolated according to the procedure of Abel and Theologis (24). Released protoplasts were washed with their corresponding suspension media and W5 solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 5 mm glucose, and 1.5 mm MES-KOH, pH 5.6).
Cell Labeling and Analysis of QD-Protein Probes—To label cells with QD-protein probes, intact cells or their protoplasts were incubated in the appropriate germination or suspension culture medium containing QD-CaM, QD-BSA, QD-CmPP16, QD-CaMTR1C, QD-CaMTR2C, or QDs at 1 μm for 3 h at 25 °C. The cells were then carefully washed three to five times with the appropriate medium. The images were collected using a Bio-Rad MRC 600 laser-scanning confocal system attached to an Optiphot microscope (Nikon).
CaM Competitive Binding Assays—Equal quantities of lily pollen protoplasts were either incubated in medium containing 1 μm QD-CaM probe or were preincubated with 0, 1, 10, or 100 μm CaM, BSA, CmPP16, CaM TR1C, or CaM TR2C separately and then transferred to medium containing 1 μm QD-CaM probes. Protoplast-associated fluorescence was quantified by fluorimetry, using an F-4500 fluorospectrometer (Hitachi).
Detection of QD-CaM Probes by Transmission Electron Microscopy—Lily pollen and tobacco BY-2 protoplasts were incubated, for 3 h at 25 °C, in medium containing 1 μm QD-CaM, QD-CaM TR1C, QD-CaM TR2C, or QDs. After washing three to five times with their corresponding culture medium, the protoplasts were fixed, for 1 h at 4 °C in 1% glutaraldehyde in the appropriate medium. Protoplasts were then transferred for an additional 1 h at 4 °C to medium containing 2% glutaraldehyde followed by three washes with 100 mm phosphate buffer, pH 7.2, and then post-fixed for 3 h in the same buffer containing 1% OsO4. The protoplasts were then dehydrated in an ethanol series and finally embedded in Spurr's resin. No staining was performed to avoid contamination. Ultrathin sections were mounted on Formvar-coated grids and observed with a transmission electron microscope (JEOL 1210), operated at 80 kV.
Measurement of Ca2+ Fluxes—Net Ca2+ fluxes crossing the plasma membrane of lily pollen protoplasts were measured using the scanning ion-selective electrode technique (25, 26), using an Applicable Electronics, Inc. system (Beijing, China). Ca2+ ion-selective microelectrodes with an external tip diameter of ∼3 μm were manufactured and salinized with tributylchlorosilane and tips backfilled with commercially available ion-selective mixture (calcium ionophore I - mixture A, 21048; Fluka, Busch, Switzerland); only electrodes with Nernstian slopes of >25 mV were used.
Protoplasts were first adhered to glass coverslips treated with a poly-l-lysine (Sigma) solution. Gradients in Ca2+ activity next to individual protoplasts were measured by moving the Ca2+-selective microelectrode between two positions, in a preset excursion (5–30 μm), at a frequency of between 0.3 and 0.5 Hz. Three Ca2+-selective microelectrodes were equally spaced around each protoplast and were located within the equatorial plane of the protoplast. All of the cellular ion flux experiments were repeated at least three times, and a minimum of 10 protoplasts were used per treatment. Individual protoplasts were monitored in control bathing medium for 20 min, and only those displaying a stable pattern in which net uptake of Ca2+ was close to zero were used in subsequent experiments. Net Ca2+ fluxes were calculated as described (27).
Measurements of Cytosolic Ca2+ Levels—Pollen protoplasts were first microinjected with the Ca2+-sensitive dye, Calcium Green-1 linked to a 10-kDa dextran (Invitrogen) and then given a 1-h recovery period at 25 °C. Subsequently, 1 μm CaM, QD-CaM, CaM TR1C, CaM TR2C, or single QDs were added to the bathing medium containing 1 mm KCl, 0.8 mm MgSO4, 1 mm CaCl2, 10 mm MES, and 12% mannitol (pH 5.8). Sequential fluorescence images were then acquired by using a Leica TCS 4D laser-scanning confocal microscope: excitation wavelength, 488 nm; emission spectra collected between 515 and 550 nm.
Cytoplasmic Ca2+ levels were also investigated by means of FRET, according to Adachi and Tsubata (28). Mesophyll protoplasts, derived from transgenic A. thaliana plants expressing the ECFP-calmodulin-M13-EYFP cameleon, were first analyzed under a laser-scanning confocal microscope, and images were collected using LSM 510 software (Carl Zeiss, Jena, Germany). For ECFP analysis, we used an excitation wavelength of 458 nm, and emission spectra were collected between 475 and 490 nm; for EYFP, we used an excitation wavelength of 514 nm, and emission spectra were collected between 530 to 600 nm. The images were again collected in a time course following the introduction to the bathing medium of 1 μm CaM, QD-CaM, CaM TR1C, CaM TR2C, or single QDs. FRET ratios for these various treatments were calculated, using Image J software, as the ratio of EYFP to ECFP intensity, according to the following formula: ratio = 1000·EYFP/ECFP.
RESULTS
General Nonspecific Binding of QD-Protein Probes to Cell Walls—To investigate whether CaM-binding sites exist on the surface of intact plant cells, we first prepared water-soluble and photostable QD (supplemental Fig. S1). These freshly prepared QDs were then used to conjugate CaM and various control proteins to yield luminescent test probes (supplemental Fig. S2). As controls for our binding studies, we prepared QD-BSA and QD-CmPP16; the latter is a hydrophilic 16-kDa protein equivalent in size to CaM (21, 29). Two CaM tryptic fragments were also prepared; QD-CaM TR1C and QD-CaM TR2C, containing residues 1–75 and residues 78–148, respectively (30, 31).
As a control, we first determined whether nonconjugated QDs could permeate and/or adhere to the surface of plant cells. For this purpose, N. tabacum (tobacco) and L. longiflorum (lily) pollen grains, as well as tobacco BY-2 and A. thaliana suspension-cultured cells, were incubated for 3 h in medium + 1 μm QDs. After a thorough rinse with incubation medium, the cells were examined under a fluorescence microscope. Both control and QD-treated pollen grains (supplemental Fig. S3) and suspension-cultured cells (supplemental Fig. S4) were devoid of fluorescent signals, thus indicating that nonconjugated QDs do not adhere to, or become bound by, these plant cell walls.
Equivalent experiments were next performed in which cells were incubated for 3 h in 1 μm QD-CaM probe. Here, strong green fluorescent signals were detected around the cell walls of tobacco and lily pollen grains (supplemental Fig. S3) as well as BY-2 and Arabidopsis suspension-cultured cells (supplemental Fig. S4). To test for the specificity of this CaM binding, the cells were incubated with 1 μm QD-CmPP16 or QD-BSA probes. Both probes also yielded strong green fluorescent signals with tobacco and lily pollen grains (supplemental Fig. S3) as well as BY-2 and Arabidopsis suspension-cultured cells (supplemental Fig. S4). Taken together, these experiments indicated that the observed CaM binding to plant cell walls is likely caused by steric hindrance rather than by CaM interacting with a specific protein(s).
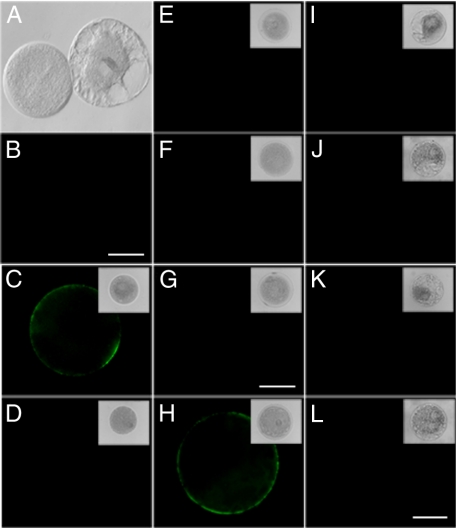
CaM Binds to the Protoplast Plasma Membrane—To test for the presence of CaM-binding sites on the external surface of the plasma membrane, a series of experiments was next performed with protoplasts isolated from both pollen grains (supplemental Fig. S5) and suspension-cultured cells. Control experiments performed with free QDs established that they did not bind to the surface of tobacco pollen protoplasts (Fig. 1, A and B). In contrast, when incubated with 1 μm QD-CaM probe, tobacco and lily pollen protoplasts exhibited clear fluorescent signals on the outer surface of the plasma membrane (Fig. 1, C and H, respectively). Importantly, no fluorescent signals were observed after protoplasts were incubated with the control probes QD-CmPP16 (Fig. 1, D and I) or QD-BSA (Fig. 1, E and J). These results provided support for the hypothesis that the QD-CaM probe can interact, specifically, with a protein(s) located at the outer surface of the plasma membrane. Interestingly, neither QD-CaM TR1C (Fig. 1, F and K) nor QD-CaM TR2C probe (Fig. 1, G and L) bound to the protoplast surface. This finding suggested that only native CaM can bind to the putative plasma membrane-located binding site(s).
FIGURE 1.
Detection of CaM-binding sites on the plasma membrane of protoplasts prepared from tobacco and lily pollen grains. A, brightfield image of tobacco pollen protoplasts. B, fluorescence micrograph of same protoplasts in A incubated with 1 μm QD; note the absence of any fluorescent signal at the protoplast surface. C, tobacco pollen protoplast incubated with 1 μm QD-CaM probe. Note the presence of a clear fluorescent signal along the plasma membrane. D–G, tobacco pollen protoplasts incubated with 1 μm QD-CmPP16, QD-BSA, QD-CaMTR1C, and QD-CaMTR2C probes, respectively. Note the absence of any fluorescent signal at the protoplast surface. H, lily pollen protoplast incubated with 1 μm QD-CaM probe. Note the presence of a clear fluorescent signal along the plasma membrane. I–L, lily pollen protoplasts incubated with 1 μm QD-CmPP16, QD-BSA, QD-CaMTR1C, and QD-CaMTR2C probes, respectively. Note the absence of any fluorescent signal at the protoplast surface. The magnification of A is the same as for B, C–F are the same as G, and I–K are the same as L. All scale bars, 5 μm.
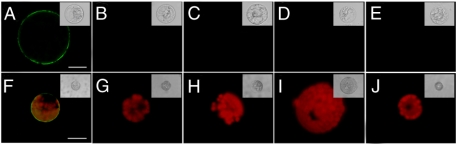
Equivalent experiments were next performed on protoplasts prepared from BY-2 and Arabidopsis suspension-cultured cells. Here, incubation with the QD-CaM probe gave a clear fluorescent signal associated with the plasma membrane (Fig. 2, A and F). Again, no signal was detected in the presence of QD-CmPP16 (Fig. 2, B and G), QD-BSA (Fig. 2, C and H), QD-CaM TR1C (Fig. 2, D and I), or QD-CaM TR2C (Fig. 2, E and J) probe. These results suggested that the plasma membrane-localized CaM-binding site(s) may be present on a wide range of plant cells.
FIGURE 2.
Detection of CaM-binding sites on the plasma membrane of protoplasts prepared from tobacco BY-2 and A. thaliana suspension-cultured cells. A–E, fluorescence micrographs of BY-2 cell protoplasts incubated with protein-conjugated QD. A, protoplast incubated with 1 μm QD-CaM probe; note the presence of a clear fluorescent signal along the plasma membrane. B–E, protoplasts incubated with QD-CmPP16, QD-BSA, QD-CaMTR1C, and QD-CaMTR2C probes, respectively. Note the absence of any fluorescent signal at the protoplast surface. F–J, fluorescence micrographs of A. thaliana protoplasts incubated with protein-conjugated QD. F, protoplast incubated with 1 μm QD-CaM probe; note the presence of a clear fluorescent signal along the plasma membrane. G–J, protoplasts incubated with QD-CmPP16, QD-BSA, QD-CaMTR1C, and QD-CaMTR2C probes, respectively. Note the absence of any fluorescent signal at the protoplast surface. The red signal represents chlorophyll autofluorescence. The insets are the corresponding brightfield images of the fluorescence micrographs. The magnifications of A–E are uniform and F–J are uniform; all scale bars, 2.5 μm.
Binding of QD-CaM Probe Is Specific—To further test the binding specificity of the QD-CaM probe to the plasma membrane-localized site(s), we next performed a series of competition assays. Lily protoplasts were first incubated for 1 h in medium containing various concentrations of CaM, BSA, CmPP16, CaM TR1C, or CaM TR2C, followed by transfer to medium containing 1 μm QD-CaM probe. After a 2-h incubation period, protoplasts were washed thoroughly, and fluorescence intensity was quantified using fluorimetry. The fluorescence level associated with the bound QD-CaM probe was found to decrease as a function of increasing CaM concentration in the pretreatment medium (supplemental Fig. S6). In marked contrast, the pretreatment with BSA, CmPP16, CaM TR1C, or CaM TR2C had no effect on the level of QD-CaM binding to lily protoplasts. These results indicate that only native CaM can compete with the QD-CaM probe for binding to a specific target protein(s), presumably located on the outer surface of the lily pollen protoplast plasma membrane.
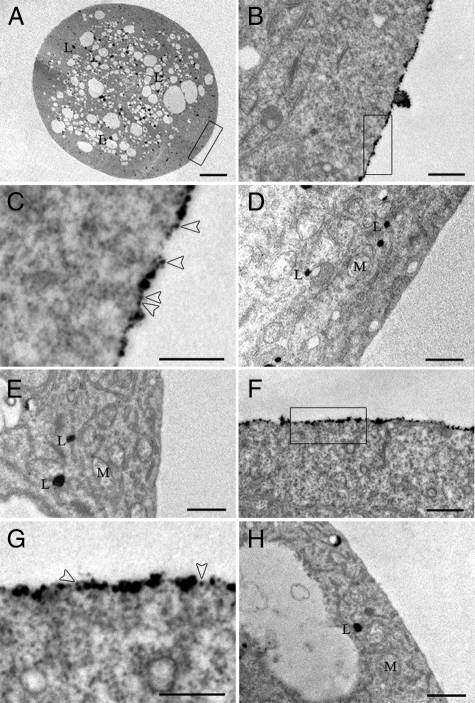
High Resolution Imaging Locates QD-CaM Probe to the Plasma Membrane—An aliquot of the lily protoplasts used in the above QD-CaM binding studies was processed for chemical fixation and embedment. Sectioned material was examined by high resolution transmission electron microscopy to test for the presence of QD-CaM probes bound to the outer surface of the plasma membrane. Electron dense particles, associated with the QD-CaM probe, were routinely observed on the surface of lily protoplast plasma membranes (Fig. 3, A–C). In contrast, few if any particles were found along the plasma membrane of lily protoplasts incubated with single QDs (Fig. 3D). Specificity of QD-CaM binding was further confirmed by the absence of electron dense particles along the plasma membrane of lily protoplasts incubated with QD-CaM TR1C probe (Fig. 3E).
FIGURE 3.
Localization of QD-CaM probe to the outer surface of the plasma membrane of both lily pollen and tobacco BY-2 protoplasts. A–E, transmission electron micrographs of lily pollen protoplasts incubated with QD-CaM probe or unbound QDs. A, low magnification image of a protoplast incubated with QD-CaM probe. B, higher magnification of area highlighted in A. Note that the QD-CaM probe was confined to the outer surface of the protoplast. C, higher magnification of the area highlighted in B. Arrowheads identify single QD-CaM probes bound to the outer surface of the plasma membrane. D and E, lily protoplast incubated with unbound QDs and QD-CaM TR1C probe, respectively. No particles were detected bound to the plasma membrane. F, presence of QD-CaM probe bound to the plasma membrane of a BY-2 protoplast. G, higher magnification of the area highlighted in F. Arrowheads identify single QD-CaM probes bound to the outer surface of the plasma membrane. H, BY-2 protoplast incubated with QD-CaM TR1C probe; no particles were bound to the plasma membrane. Scale bars, A, 10 μm; B–H, 0.5 μm. M, mitochondria; L, lipid.
Parallel studies were also performed with protoplasts prepared from BY-2 suspension-cultured cells. Again, incubation of these protoplasts with QD-CaM probe resulted in the presence of particles bound to the outer plasma membrane surface (Fig. 3, F and G). However, protoplasts incubated with the QD-CaM TR1C probe (Fig. 3H) or single QDs (data not shown) were found to be devoid of bound particles. Taken together, these results provide further support for the hypothesis that the plant plasma membrane contains a protein(s) that can bind to extracellular CaM.
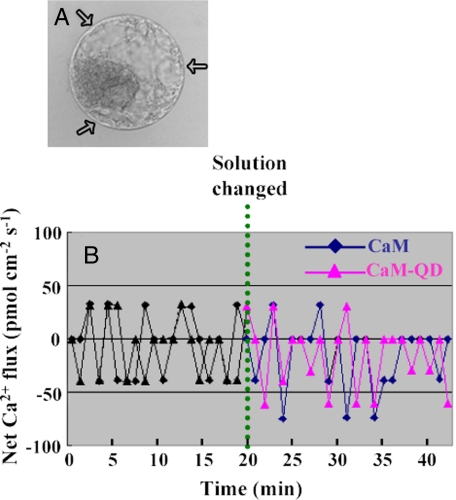
CaM and QD-CaM Probe Stimulate Ca2+ Influx across the Protoplast Plasma Membrane—To investigate the possible biological function, in lily pollen protoplasts, of extracellular CaM and the QD-CaM probe on intracellular Ca2+, we next used the scanning ion-selective electrode technique to investigate net Ca2+ flux (27). Three scanning ion-selective electrodes were used to simultaneously record the Ca2+ flux crossing the plasma membrane of individual protoplasts (Fig. 4, A and B). Monitoring of these fluxes in normal bathing medium for 20 min established that the net uptake of Ca2+ was close to zero. The addition of either 1 μm CaM or QD-CaM probe resulted in a change of this condition to a net Ca2+ influx. This alteration away from flux equilibrium was generally apparent within 10 min of adding CaM/QD-CaM (Fig. 4B). Importantly, the addition of QDs alone, or QD-CaM TR1C probe to the bathing medium did not significantly alter the Ca2+ flux status of the protoplast (data not shown).
FIGURE 4.
Exogenously applied CaM alters the Ca2+ flux across the plasma membrane of protoplasts isolated from lily pollen grains. A, lily protoplast illustrating the location of the three scanning ion-selective electrodes (arrows) employed to simultaneously record the plasma membrane Ca2+ fluxes. B, affect of CaM or QD-CaM probe on net Ca2+ flux. Note that both CaM and QD-CaM caused a net influx of Ca2+ into lily protoplasts. The data represent the average values obtained for the three scanning ion-selective electrodes. The positive values represent Ca2+ efflux, whereas the negative values represent influx.
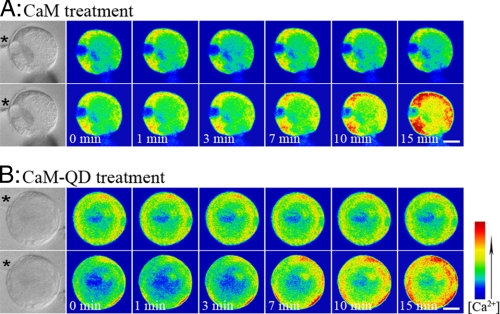
Parallel experiments were also performed with calcium-green-dextran (32) to monitor the affects of CaM and the QD-CaM probe on cytoplasmic Ca2+ levels in lily pollen protoplasts. Following microinjection-based loading of calcium-green-dextran into the cytoplasm of lily protoplasts, the resting intracellular Ca2+ level was monitored for 15 min (Fig. 5). Introduction of CaM or QD-CaM probe was found to elicit a clear increase in the cytoplasmic Ca2+ level within 7–15 min (Fig. 5, A and B, respectively). In contrast, no change in the resting intracellular Ca2+ levels were detected in equivalent experiments performed in the presence of single QDs or the QD-CaM TR1C probe (data not shown). Taken together, our scanning ion-selective electrode experiments and calcium-green-dextran studies provide support for the hypothesis that addition of extracellular CaM or QD-CaM probe can induce a consistent change in Ca2+ flux across the protoplast plasma membrane.
FIGURE 5.
Time course of CaM/QD-CaM probe-induced changes in intracellular Ca2+ levels in lily pollen protoplasts. A, upper panels, pseudocolor images of the resting intracellular Ca2+ concentration in a lily protoplast loaded with calcium green-dextran (control). Lower panels, affect on the intracellular Ca2+ level upon adding 1 μm CaM to the bathing solution. B, upper panels, pseudocolor images of the resting intracellular Ca2+ concentration in a lily protoplast loaded with calcium green-dextran (control). Lower panels, affect on the intracellular Ca2+ level upon adding 1 μm QD-CaM to the bathing solution. Scale bars, 2.5 μm. The bar on the right shows the relationship between Ca2+ concentration and cell pseudocolor. The asterisks indicate the locations of micropipettes used to introduce calcium green-dextran.
FRET-based Ca2+ Measurement by Laser Scanning Confocal Microscopy—To measure the influence of extracellular CaM/QD-CaM on intracellular Ca2+ levels by a noninvasive method, we next isolated mesophyll protoplasts from transgenic Arabidopsis plants expressing a fluorescent indicator cameleon (28). These cells contained the ECFP-calmodulin-M13-EYFP Ca2+ reporter system for which confocal microscopy and FRET technology can be used, at the single-cell level, to measure the dynamics of cytoplasmic Ca2+ in response to applied treatments.
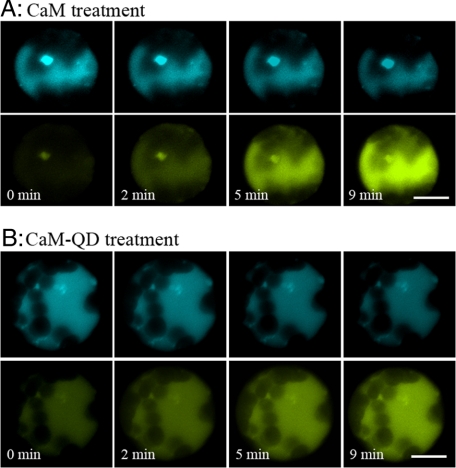
FRET analysis performed on these Arabidopsis protoplasts revealed a rise in cytoplasmic Ca2+ shortly after CaM or the QD-CaM probe was introduced into the bathing medium. A time course of FRET measurements showed that marked FRET occurred within 2 min after protoplasts were given a 1 μm CaM (Fig. 6A) or QD-CaM probe (Fig. 6B) treatment. No change was elicited in response to single QDs, CaMTR1C, or CaMTR2C (data not shown). Each treatment was examined on 20 protoplasts, and a consistent response was observed in all but one or two protoplasts.
FIGURE 6.
FRET-based measurement of CaM/CaM-QD-induced changes in Ca2+ levels in mesophyll protoplasts isolated from transgenic ECFP-calmodulin-M13-EYFP Arabidopsis plants. The images were collected by laser scanning confocal microscopy; ECFP was excited at 458 nm. FRET was detected when protoplasts were incubated with 1 μm CaM (A) and 1 μm CaM-QD (B). Scale bars, 2.5 μm.
Ratio increases resulted from a rise in the fluorescence of the acceptor EYFP. However, no perceptible or very small decreases in the fluorescence of the donor ECFP were observed (Fig. 6). This was due in large part to the high levels of chloroplast autofluorescence present in these Arabidopsis protoplasts, which caused a high background against which ECFP changes were measured. In any event, the mean for the ratio of EYFP/ECFP fluorescence showed a significant increase in protoplasts after treatment with CaM or the QD-CaM probe (supplemental Fig. S7).
DISCUSSION
Like animals, plant cells may make use of peptide signals (33). Although some 10 polypeptide hormones or putative polypeptide signals have been reported in plants, only a few have been well characterized at the genetic and biochemical level (34, 35). It has also been proposed that extracellular CaM may serve as a polypeptide signal in plants (5, 6, 8, 9). Experimental confirmation of the existence and localization of such CaM-binding sites on the cell surface was the subject of the present study. Here, we used CaM-labeled QDs to detect CaM-binding sites on the cell surface at both the light and electron microscopic levels in intact plants and their protoplasts.
Synthesized QD-Protein Probes are Highly Specific and Photostable—The preparation of biocompatible water-soluble QDs is a key step in the use of semiconductor QDs for live cell imaging. We first synthesized mercaptopropionic acid capped cadmium telluride QDs with good luminescence properties using the protocol of Chen and Zhong (36). Carbodiimide was used to mediate the formation of amide linkages between a carboxylate and an amine group to synthesize luminescent probes of CaM-QDs and other protein-QDs. To detect whether CaM and other proteins were successfully bioconjugated to the QD nanoparticles, the EDC-NHS-mediated coupling reaction on the silylanized slides was performed. These experiments indicated that bioconjugation is specific, and these proteins are indeed covalently bound to the nanocrystal surface.
Presence of the Membrane-localized Specific CaM-binding Sites Was Universal in Living Plant Cells, Whereas CaM Binding to the Cell Wall Was Most Likely Caused by Steric Hindrance—Our fluorescent labeling results with protoplasts isolated from a range of cell types and different plant species revealed that CaM-QDs can bind to the plasma membrane via an interaction with a specific binding site(s). In contrast, our studies on intact plant cells demonstrated that not only QD-CaM but also QD-BSA and QD-CmPP16 appeared to bind to the cell wall. Hence, the observed cell wall-associated CaM-binding most likely is due to steric hindrance rather than CaM interacting with specific binding sites within the matrix of the wall.
Specificity in the interaction between the QD-CaM probe and binding site(s) located on the membrane surface is supported by two lines of evidence. First, neither QD-BSA, QD-CmPP16, QD-CaM TR1C, nor QD-CaM TR2C became bound to the protoplast membrane. Second, increasing levels of single QDs, QD-BSA, or QD-CmPP16 had no effect on the extent to which QD-CaM bound to protoplasts. The fact that CaM, but not QD-CaM TR1C or QD-CaM TR2C, competed with QD-CaM binding also provided further support for the notion that the interaction with the target protein is dependent on CaM being in its native conformation. Taken together, these findings lead us to conclude that specific CaM-binding proteins are membrane-localized instead of cell wall-associated.
Membrane Localization of Specific CaM-binding Sites Confirmed by Transmission Electron Microscope—The tightly packed atoms in the core and shell of the QD nanocrystal make them electron-dense moieties, and thus, they can be visualized by electron microscopy (16). In the present study, localization of CaM-binding sites on the plant cell surface, as determined by light microscopy, was further confirmed using high resolution electron microscopy. These experiments clearly revealed that the QD-CaM probe was bound to the outer surface of the plasma membrane. Again, specificity of this binding to a putative receptor was supported by the absence of signal detected with QDs alone, QD-BSA, QD-CmPP16, QD-CaM TR1C, or QD-CaM TR2C.
CaM-binding sites Function in Ca2+ Signaling—Intracellular signal transduction pathways, especially those involving secondary messengers, provided another avenue to test the hypothesis that extracellular CaM can act as an apoplasmic peptide signal (37). In support of this notion, our ion-selective microelectrode studies provided direct evidence that application of either CaM or QD-CaM induced a change in the steady state condition of the protoplast from no net Ca2+ uptake to a net influx of Ca2+. Importantly, the time course for this change in net flux matched the response in cytoplasmic Ca2+ measured by calcium-green-dextran and FRET-based experiments using the ECFP-calmodulin-M13-EYFP cameleon. Again, the inability of QDs alone, QD-CaM TR1C, or QD-CaM TR2C to induce changes in cytoplasmic Ca2+ levels supports our conclusion that this response is specific and elicited by the applied CaM.
Although these collective findings are consistent with QD-CaM eliciting an increase in intracellular Ca2+ concentration through a transmembrane mechanism activated by QD-CaM binding to an external site on a plasma membrane receptor, our studies do not preclude the involvement of other ligands. Here, it is important to note that the Arabidopsis genome contains some 50 members of the EF-hand family of small, acidic Ca2+-binding proteins in addition to CaM (38). Future studies will be performed to test whether additional CaM-like proteins can similarly activate this response pathway.
The biological role for this CaM-mediated change in cytosolic Ca2+ level remains to be elucidated. If extracellular CaM is indeed the specific physiological ligand that activates this response pathway, then it becomes important to question how CaM might gain entry into the apoplasm. Under normal conditions, CaM is generally localized to the cytosol (39). Furthermore, CaM lacks any identifiable signal sequence(s) that would potentially target it for secretion into the apoplasm. Hence, its entry into the cell wall may occur as a direct result of cellular damage. CaM is an abundant protein and small in size; thus, its release upon local damage could well allow it to permeate through the walls of neighboring cells. Activation of the transmembrane signaling pathway by CaM may serve to inform these surrounding cells of local tissue damage.
In summary, our results clearly demonstrated that the apoplasmic CaM-binding sites were widely present on surface in plant cells using the synthesized CaM-QD luminescent probes. It was revealed via labeling and competition assays that CaM-binding proteins were membrane-localized instead of cell wall-associated as reported previously. Transmission electron microscopy showed the precise localization of the extracellular CaM-binding sites on the membrane surface, providing the most direct proof for the existence of apoplasmic CaM-binding sites. In addition, we confirmed that both CaM and CaM-QDs could mediate intracellular secondary messenger Ca2+ concentrations when binding to the plant plasma membrane. Taken together, our results suggested that QD-CaM, in addition to its traditional role in mediating the intracellular Ca2+ signal pathway, is able to function as an apoplasmic signal in the regulation of plant growth and development after interaction with its extracellular binding sites.
Supplementary Material
Acknowledgments
We thank Prof. Mikael Akke (Lund University, Sweden) for providing CaMTR1C and CaMTR2C.
This work was supported by National Key Basic Research Program Grant 2006CB910600 from MOST, Key Program Grant 30730009, and General Program Grant 30600030 from the National Science Fund of China.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1–S7.
Footnotes
The abbreviations used are: CaM, calmodulin; BSA, bovine serum albumin; FRET, fluorescence resonance energy transfer; QD, quantum dot; MES, 4-morpholineethanesulfonic acid; EDC, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide; NHS, N-hydroxysuccinimide; ECFP, enhanced cyan fluorescent protein; EYFP, enhanced yellow fluorescent protein.
References
- 1.Ikura, M., and Ames, J. B. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouche, N., Yellin, A., Snedden, W. A., and Fromm, H. (2005) Annu. Rev. Plant Biol. 56 435–466 [DOI] [PubMed] [Google Scholar]
- 3.Reddy, A. S. N. (2001) Plant Sci 15 59–64 [Google Scholar]
- 4.Cui, S. J., Guo, X. Q., Chang, F., Cui, Y. W., Ma, L. G., Sun, Y., and Sun, D. Y. (2005) J. Biol. Chem. 280 31420–31427 [DOI] [PubMed] [Google Scholar]
- 5.Biro, R. L., Sun, D. Y., Serlin, B. S., Terry, M. E., Datta, N., Sopory, S. K., and Roux, S. J. (1984) Plant Physiol. 75 382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao, G. H., Hou, L. X., Ding, C. B., Cui, S. J., and Sun, D. Y. (2005) Planta 222 428–437 [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y. L., Huang, R. F., Xiao, Y. M., Lu, P., Chen, J., and Wang, X. C. (2004) Plant Physiol. 136 4096–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma, L. G., Xu, X. D., Cui, S. J., and Sun, D.Y. (1999) Plant Cell 11 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang, Z. L., Ma, L. G., Wang, X. C., and Sun, D. Y. (2003) Prog. Nat. Sci. 13 678–682 [Google Scholar]
- 10.Yu, G. H., Liang, J. G., He, Z. K., and Sun, M. X. (2006) Chem. Biol. 13 723–731 [DOI] [PubMed] [Google Scholar]
- 11.Ravindran, S., Kim, S., Martin, R., Lord, E. M., and Ozkan, C. S. (2005) Nanotechnology 16 1–4 [Google Scholar]
- 12.Mansson, A., Balaz, M., Bunk, R., Nicholls, I. A., Omling, P., Tagerud, S., and Montelius, L. (2004) Biochem. Biophys. Res. Commun. 314 529–534 [DOI] [PubMed] [Google Scholar]
- 13.Dahan, M., Lévi, S., Luccardini, C., Rostaing, P., Riveau, B., and Triller, A. (2003) Science 302 442–445 [DOI] [PubMed] [Google Scholar]
- 14.Chalmers, N. I., Palmer, R. J., Du-Thumm, L., Sullivan, R., Shi, W., and Kolenbrander, P. E. (2007) Appl. Environ. Microbiol. 73 630–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grecco, H. E., Lidke, K. A., Heintzmann, R., Lidke, D. S., Spagnuolo, C., Martinez, O. E., Jares-Erijman, E. A., and Jovin, T. M. (2005) Microsc. Res. Tech. 65 169–179 [DOI] [PubMed] [Google Scholar]
- 16.Giepmans, B. N. G., Deerinck, T. J., Smarr, B. L., Jones, Y. Z., and Ellisman, M. H. (2005) Nat. Meth. 2 743–749 [DOI] [PubMed] [Google Scholar]
- 17.Nisman, R., Dellaire, G., Ren, Y., Li, R., and Bazett-Jones, D. P. (2004) J. Histochem. Cytochem. 52 13–18 [DOI] [PubMed] [Google Scholar]
- 18.Miao, Y., and Jiang, L. (2007) Nat. Protocols 2 2348–2353 [DOI] [PubMed] [Google Scholar]
- 19.Allen, G. J., Kwak, J. M., Chu, S. P., Liopis, J., Tsien, R. Y., Harper, J. F., and Schroeder, J. I. (1999) Plant J. 19 735–747 [DOI] [PubMed] [Google Scholar]
- 20.Zielinski, R. E. (2001) Methods in Molecular Biology: Production of Recombinant Plant Calmodulin Isoforms in Bacteri. Calcium-Binding Proteins (Vogel, H. J., ed) Vol. 172, p. 143, Humana Press Inc., Totowa, NJ [DOI] [PubMed] [Google Scholar]
- 21.Xoconostle-Cázares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H. L., Monzer, J., Yoo, B. C., McFarland, K. C., Franceschi, V. R., and Lucas, W. J. (1999) Science 283 94–98 [DOI] [PubMed] [Google Scholar]
- 22.Chan, W. C. W., and Nie, S. M. (1998) Science 281 2016–2018 [DOI] [PubMed] [Google Scholar]
- 23.DeSilva, N. S. (2003) Am. J. Respir. Cell Mol. Biol. 29 757–770 [DOI] [PubMed] [Google Scholar]
- 24.Abel, S., and Theologis, A. (1994) Plant J. 5 421–427 [DOI] [PubMed] [Google Scholar]
- 25.Kochian, L. V., Schaff, J. E., Kuhtreiber, W., Jaffe, L. F., and Lucas, W. J. (1992) Planta 188 601–610 [DOI] [PubMed] [Google Scholar]
- 26.Wu, X. Q., Chen, T., Zheng, M. Z., Chen, Y. M., Teng, N. J., Šamaj, J., Baluška, F., and Lin, J. X. (2008) J. Proteome. Res. 7 4299–4312 [DOI] [PubMed] [Google Scholar]
- 27.Vincent, P., Chua, M., Nogue, F., Fairbrother, A., Mekeel, H., Xu, Y., Allen, N., Bibikova, T. N., Gilroy, S., and Bankaitis, V. A. (2005) J. Cell Biol. 168 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adachi, T., and Tsubata, T. (2008) Biochem. Biophys. Res. Commun. 283 5972–5972 [DOI] [PubMed] [Google Scholar]
- 29.Duval, F. D., Renard, M., Jaquinod, M., Biou, V., Francçoise Montrichard, F., and Machere, D. (2002) Plant J. 32 481–493 [DOI] [PubMed] [Google Scholar]
- 30.Lundström, P., and Akke, M. (2004) J. Am. Chem. Soc. 126 928–935 [DOI] [PubMed] [Google Scholar]
- 31.Waltersson, Y., Linse, S., Brodin, P., and Grundström, T. (1993) Biochemistry 32 7866–7871 [DOI] [PubMed] [Google Scholar]
- 32.Goddard, H., Manison, N. F. H., Tomos, D., and Brownlee, C. (2000) Proc. Natl. Acad. Sci. U. S. A. 15 1932–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marx, J. (1996) Science 237 1338–1339 [DOI] [PubMed] [Google Scholar]
- 34.Matsubayashi, Y. (2003) J. Cell Sci. 116 3863–3870 [DOI] [PubMed] [Google Scholar]
- 35.Bahyrycz, A., and Konopinska, D. (2007) J. Peptide Sci. 13 787–797 [DOI] [PubMed] [Google Scholar]
- 36.Chen, B., and Zhong, P. (2005) Anal. Bioanal. Chem. 381 98–992 [Google Scholar]
- 37.Shang, Z. L., Ma, L. G., Zhang, H. L., He, R. H., Wang, X. C., Cui, S. J., and Sun, D. Y. (2005) Plant Cell Physiol. 46 598–608 [DOI] [PubMed] [Google Scholar]
- 38.McCormack, E., Tsai, Y. C., and Braam J. (2005) Trends Plant Sci. 10 383–389 [DOI] [PubMed] [Google Scholar]
- 39.Vos, J. W., and Hepler, P. K. (1998) Protoplasma 201 158–171 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.