Abstract
Sus1 is an evolutionary conserved protein that functions both in transcription and mRNA export and has been proposed to contribute to coupling these processes in yeast. Sus1 mediates its different roles as a component of both the histone H2B deubiquitinating module (Sus1-Sgf11-Ubp8-Sgf73) of the SAGA (Spt-Ada-Gcn5 acetyltransferase) transcriptional co-activator and the mRNA export complex, TREX-2 (Sus1-Sac3-Thp1-Cdc31). We have dissected the different functions of Sus1 with respect to its partitioning in transcription and export complexes using a mutational approach. Here we show that the sus1–10 (E18A, S19A, and G20A) and sus1–12 (V73A and D75A) alleles of Sus1 can be dissociated from TREX-2 while leaving its interaction with SAGA largely intact. Conversely, the binding to both TREX-2 and SAGA was impaired in the sus1–11 allele (G37A and W38A), in which two highly conserved residues were mutated. In vitro experiments demonstrated that dissociation of mutant Sus1 from its partners is caused by a reduced affinity toward the TREX-2 subunit, Sac3, and the SAGA factor, Sgf11, respectively. Consistent with the biochemical data, these sus1 mutant alleles showed differential genetic relationships with SAGA and mRNA export mutants. In vivo, all three sus1 mutants were impaired in targeting TREX-2 (i.e. Sac3) to the nuclear pore complexes and exhibited nuclear mRNA export defects. This study has implications for how Sus1, in combination with distinct interaction partners, can regulate diverse aspects of gene expression.
Gene expression machineries are functionally and physically coupled to ensure that transcription, RNA processing, RNA quality control, and nuclear mRNA export take place with high fidelity and efficiency (1–4). On a different but interdependent level, gene activity is regulated by the dynamic arrangement of chromosomes within the nucleus (5, 6). Studies in Saccharomyces cerevisiae have shown that nuclear pore complexes (NPCs)2 mediate tethering of activated genes to the nuclear periphery, thereby providing a platform for the integration of transcription and mRNA export (7–13). Sus1 was proposed to play a role in transcription-coupled mRNA export because of its presence in the SAGA transcriptional co-activator and the NPC-based TREX-2 mRNA exporter (14). Moreover, both complexes are involved in the repositioning of the activated GAL1 gene (and possibly other genes) to the NPCs (7, 8, 15).
SAGA (Spt-Ada-Gcn5 acetyltransferase) functions include histone acetylation and deubiquitination, nucleosome remodeling activity, and interactions with gene-specific activators and general transcription factors (16–18). Within SAGA, the 96-residue protein Sus1 has been shown to be an integral part of a tetrameric histone H2B-deubiquitinating (DUB) module (15, 19–24). The DUB module contains the protease Ubp8, Sus1, the small zinc finger protein Sgf11, and Sgf73. Sgf73 is the adaptor protein that anchors Sus1-Sgf11-Ubp8 at its N terminus and connects it to SAGA. Deletion of either SGF11 or SUS1 results in the dissociation of Ubp8 from Sgf73, implicating these proteins in the structural integrity of the DUB module (20, 21, 25). Importantly, Sgf73 together with Sus1 and Sgf11 are required for activation of Ubp8, which by itself is enzymatically inactive. The precise architecture of the DUB module has not been determined so far; however, Sus1 was shown to interact directly with the 99-residue protein Sgf11 both in vivo and in vitro (15). Overall, a regulated cycle of histone H2B ubiquitin addition (catalyzed by Rad6/Bre1) and removal (Ubp8) at the promoter and coding region of a gene triggers multiple steps of gene activation and influences both transcription initiation and elongation (26–30). Moreover, recent work has shown that SAGA-associated Sgf73 creates a link to TREX-2 by regulating TREX-2 assembly or stability, possibly through a chaperone activity (15). Specifically, Sgf73 is required for recruiting the TREX-2 factors Sac3 and Thp1 to SAGA and promotes the association of Sus1 with a distinct domain of Sac3. This assembly event is critical for TREX-2 function (see below).
The other Sus1-containing complex, TREX-2, functions in mRNA export as well as promoting transcription elongation and preventing DNA:RNA hybrid formation and genome instability (31–36). TREX-2 is composed of Sac3, Thp1, Cdc31, and Sus1. Recently, Sem1 was described as an additional TREX-2 subunit, but it is still unclear whether Sem1 is a stoichiometric component (37). Within TREX-2, Sus1 directly interacts with the Sac3 CID motif that also harbors a binding site for the calmodulin-like centrin Cdc31 (32). TREX-2 is mainly localized at the nuclear periphery and interacts physically and functionally with the general mRNA export receptor Mex67/Mtr2 (31). NPC tethering of TREX-2 depends on the nuclear basket protein, Nup1, and possibly other nucleoporins (31). Removal of the Sac3 CID strongly impairs TREX-2 targeting to the NPCs in vivo and triggers an mRNA export defect. Notably, the small Sus1 protein is important for NPC targeting of TREX-2, because SUS1 deletion causes TREX-2 dissociation from the NPCs (15).
In evolutionary terms, SAGA is well conserved in subunit composition and structural appearance and plays broad and important regulatory roles in transcription from yeast to flies and humans (18). Specifically, the Sus1-containing histone H2B DUB module of SAGA has human orthologues, which include ENY2 (Sus1), the protease USP22 (Ubp8), ATXN7L3 (Sgf11), and ATXN7 (Sgf73) (38–40). In analogy to yeast TREX-2, the Drosophila orthologue of Sus1, E(y)2, forms a complex with the Sac3 counterpart X-linked male sterile 2 (Xmas-2) and functions in mRNA export and gene-NPC anchorage (41). The Sac3 orthologue GANP was reported to suppress DNA recombination in mammalian cells, but whether GANP operates in mRNA export and gene positioning remains to be explored (42–44). Notably, a potential human orthologue of Cdc31, centrin 2, is associated with the NPCs and plays a role in the export of mRNA (45). The functional diversification of Sus1 as a component of both the SAGA histone DUB module and the TREX-2 mRNA export complex represents an intriguing example of molecular innovation during evolution. The small Sus1 protein can support both a sophisticated enzymatic mechanism and confer positional information for an NPC targeting event. Understanding Sus1 function therefore requires dissection of its separate SAGA- and TREX-2-related roles, integrated with an analysis of how SAGA and TREX-2 interact functionally.
In this study we report a comprehensive mutational analysis of Sus1 aimed at defining the molecular requirements for its association with either SAGA or TREX-2. Our data show that mutational uncoupling of Sus1-ligand interactions results in selective functional impairments in transcription-coupled mRNA export.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Microbiological Techniques—The S. cerevisiae strains used in this study are listed in supplemental Table 1. Deletion disruption and C-terminal TAP tagging at the genomic locus were performed as described previously (46–48). A two-step allele replacement method was devised to generate strains expressing nontagged and C-terminally TAP- or FLAG-tagged Sus1 wild-type or mutant variants (see below).
Plasmids used in this study are listed in supplemental Table 2. The site-directed sus1 mutants were generated by fusion PCR, and the correctness of the cloned DNA fragments was verified by sequencing. All recombinant DNA techniques were done according to standard procedures using Escherichia coli DH5α for cloning and plasmid propagation.
Preparation of media, yeast transformation, and genetic manipulations were performed according to established methods. For selection of yeast transformants on nourseothricin (clonNAT)-containing plates, YPD plates were supplemented with 100 μg/ml nourseothricin (Werner BioAgents). Tetrad dissection was performed using a Singer MSM micromanipulator.
Genomic SUS1 Gene Replacement—To replace genomic wild-type SUS1 with nontagged and TAP- or FLAG-tagged sus1 alleles, we devised a novel two-step allele replacement strategy. In a first step, haploid sus1::klURA3 (URA3 gene from Kluyveromyces lactis) deletion disruption strains were obtained by tetrad dissection of a heterozygous diploid SUS1/sus1::klURA3 strain that had been created by transformation of a klURA3 PCR cassette, bearing short flanking homology regions of the SUS1 promoter and terminator sequence into the diploid W303 strain background. The haploid sus1::klURA3 null mutant strains were then co-transformed with DNA fragments (1 μg) containing wild-type and mutant SUS1, SUS1-TAP, or SUS1-FLAG alleles, excised by XhoI/BamHI digestion from pRS315-SUS1/sus1, pRS315-SUS1/sus1-TAP, or pRS316-SUS1/sus1-FLAG, and empty pRS315 vector (100 ng) for selection of transformants on SDC-Leu plates. To select for clones that had lost the klURA3 marker because of a site-specific recombination event, transformants were replica-plated onto 5-FOA-containing plates. To verify the correctness of the allele replacement, clones that grew on 5-FOA-containing medium were analyzed by colony PCR and sequencing.
Affinity Purifications—TAP-tagged proteins were affinity-purified according to published methods (49). Proteins were detected by SDS-PAGE on NuPAGE 4–12% polyacrylamide gels (Invitrogen) with subsequent colloidal Coomassie Brilliant Blue G (Sigma) staining or by Western blot analysis. Mass spectrometric identification of the proteins contained in Coomassie-stained bands was performed as described (50). The following primary antibodies were used for Western analysis: anti-Arc1 (Hurt laboratory), anti-CBP (BioCat), anti-Cdc31 (from E. Schiebel, ZMBH, Heidelberg University, Germany), anti-FLAG (Sigma), and anti-Sac3 (from R. Kölling, Hohenheim University, Germany).
In Vitro Binding Assays—Recombinant proteins were expressed in LB medium in E. coli BL21 codon plus RIL cells (Stratagene). Expression was induced by addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 23 °C for 3 h. Hexahistidine-tagged Sus1 was purified by metal affinity chromatography and imidazole elution. The GST-Sac3-(573–805)-Cdc31 heterodimer was created by co-expression of GST-Sac3-(573–805) and Cdc31. GST-tagged proteins were purified on GSH beads and eluted with GSH. Recombinant Sus1 proteins were then mixed with GST-Sgf11 or with GST-Sac3-(573–805)-Cdc31 at a 2:1 molar ratio, respectively, in a buffer containing 100 mm NaCl, 0.5 mm dithiothreitol, 50 mm HEPES, pH 7.5. Proteins were incubated with GSH beads for 30 min at 16 °C, washed in the same buffer (4 °C), and eluted with GSH. After trichloroacetic acid precipitation, the samples were separated by SDS-PAGE (12% gel, MES buffer) and visualized by Coomassie staining.
Live Cell Imaging and Fluorescence Microscopy—Prior to live imaging, cells were grown to mid-log phase in YPD (integration strains) or SDC-Leu (plasmid-based) liquid medium. Fluorescence microscopy was performed using an Imager Z1 (Carl Zeiss) microscope equipped with a 100×/63× NA 1.4 Plan-Apo-Chromat oil immersion lens (Carl Zeiss) and using DICIII, HE-EGFP, or 4,6-diamidino-2-phenylindole filters. Images were acquired with an AxioCam MRm camera and AxioVision 4.3 software (Carl Zeiss).
In situ hybridization of poly(A)+ RNA was performed according to Ref. 51. Prior to fixation, cells were grown to an A600 of 0.3 at 30 °C and then shifted to 37 °C for 2 h.
Measurement of Global H2B Ubiquitin Levels—Immunoprecipitation of FLAG-tagged histone H2B was performed as described previously (52).
RESULTS
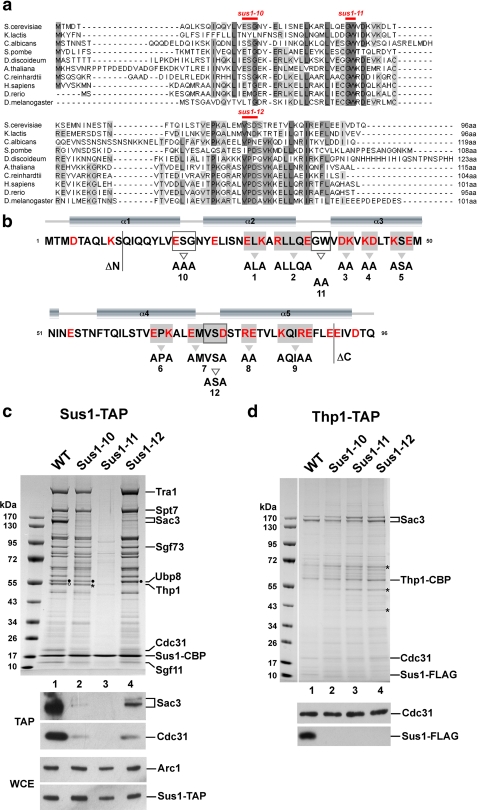
Sus1 is an evolutionary conserved protein in eukaryotes (Fig. 1a) with no homologues in viral, archaeal, and eubacterial genomes. Several Sus1 core residues are strongly conserved, whereas the N and C termini of the protein exhibit variable lengths and little sequence conservation. Secondary structure predictions indicate a primarily α-helical topology for Sus1 with five putative α-helices connected by short loop regions (Fig. 1b). We sought to generate a battery of sus1 mutants that could establish whether Sus1 employs distinct or overlapping interaction surfaces to bind to its two known ligands as follows: the CID (Cdc31 interaction domain) within the C terminus of Sac3 (32) or Sgf11, a subunit of the SAGA histone H2B DUB module (15, 25). A “clustered charged-to-alanine mutagenesis” strategy (53) was initially used to probe for potential interaction surfaces on Sus1 (Fig. 1b). This approach exploits the fact that charged residues are generally exposed at the protein surface rather than being buried in the hydrophobic core of the molecule. Sus1-TAP purifications were then performed to biochemically check whether the interaction with either SAGA or TREX-2 or both was perturbed. Normally, Sus1 affinity-purified by the TAP method efficiently co-enriches both SAGA (including the SAGA-like SLIK(SALSA) complex) and TREX-2 (14). Surprisingly, none of the nine mutants (sus1-1 to 9) significantly altered Sus1 binding to SAGA or TREX-2 (supplemental Fig. S1A). We found that deleting the less conserved N- and C-terminal parts of the protein (sus1ΔN1–10 and sus1ΔC91–96; see Fig. 1b) also did not impair the interaction of Sus1 with either SAGA or TREX-2 (data not shown). The remarkable tolerance of Sus1 toward this systematic replacement of charged residues may suggest that the interaction of Sus1 with its partners is based on other types of interaction (e.g. hydrophobic) or requires more extensive mutations to be disrupted.
FIGURE 1.
Conserved residues differentially regulate association of Sus1 with SAGA or TREX-2. a, multiple sequence alignment of Sus1 from representative species including fungi, protozoa, plants and metazoa. The alignment was generated with ClustalW2, and conserved residues were shaded using JalView. b, secondary structure prediction for Sus1 was calculated from PsiPred. For schematic depiction of Sus1 mutational strategies, mutant alleles are numbered (sus1ΔN, sus1–1 to sus1–12, sus1ΔC). All Sus1 charged residues are indicated in red. Residues mutated according to the clustered charge to alanine algorithm are shaded in gray, and gray arrowheads denote sequence changes. Open boxes/open arrowheads depict (partially) conserved Sus1 residues/sequence changes with a predicted localization within protein loops. N- and C-terminal truncations are indicated. Positions of sus1–10, sus1–11 and sus1–12 mutations are also indicated in a. c, affinity purification of genomically expressed Sus1-TAP from wild-type (WT), sus1–10, sus1–11, and sus1–12 strains. Eluates were analyzed by SDS-PAGE (4–12% gradient gels, MOPS buffer) and Coomassie staining. Indicated bands were assigned according to their molecular mass (15). Filled circles designate Ubp8 and open circles designate Thp1. Asterisk indicates a contaminant, eEF-1α, which runs slightly below Thp1. Ubp8, Thp1, and eEF-1α were determined by mass spectrometry. Eluates (TAP) were analyzed by Western blotting using anti-Sac3 and anti-Cdc31 antibodies. Sus1-TAP expression levels (WCE) were determined by anti-ProtA detection (ProtA is part of the TAP tag) and normalization with Arc1 (a yeast cytosolic marker protein) levels. d, Thp1-TAP affinity purification from the indicated sus1 mutant allele backgrounds. Indicated Coomassie-stained bands were determined by mass spectrometry. Asterisks label contaminants of the purification (from top to bottom: keratin, Eno2, and Tdh3). FLAG-tagged Sus1 and Cdc31 were immunodetected by anti-FLAG and anti-Cdc31 antibodies, respectively.
Next, we engineered point-specific mutations in putative loop regions in Sus1 that lie between the α-helices predicted by sequence analysis (Fig. 1b; termed sus1–10, sus1–11, and sus1–12). Each of these amino acid changes involved residues, such as Gly and Asp, that are characteristically present in flexible surface loops in proteins. Additionally, some of the selected residues (e.g. the Gly37Trp38 pair) are highly conserved in evolution (Fig. 1a). Notably, affinity purification of TAP-tagged Sus1–10 and Sus1–12 mutant proteins from yeast showed a pronounced loss of TREX-2 factors (i.e. Sac3, Thp1, and Cdc31), but typical SAGA factors (e.g. Tra1 and Spt7), including the DUB module components Sgf73, Sgf11, and Ubp8, were still co-enriched (Fig. 1c). Western blot analysis revealed that the loss of TREX-2 factors was more severe in sus1–10 than sus1–12 (Fig. 1c). These data indicate that whereas Sus1–10 and Sus1–12 are impaired in their interaction with TREX-2, the structural integrity of the SAGA DUB module (Sus1-Sgf11-Ubp8-Sgf73) remains largely unaffected in these sus1 mutants. In contrast, the purification of TAP-tagged Sus1–11 displayed a striking loss of both TREX-2 and SAGA subunits (Fig. 1c). Because Sgf11 was also absent from this purification, it is conceivable that a reduced affinity between Sus1 and its direct interaction partner Sgf11 may have dissociated Sus1 from the DUB module and hence from the entire SAGA complex (see below). The decrease or lack of a biochemical interaction observed for the different Sus1 mutant proteins is not because of major alterations in Sus1 protein levels. Wild-type and mutant Sus1 proteins exhibited similar expression levels in yeast, although we noticed a slight reduction in the total amount of Sus1–11 (Fig. 1c).
To characterize the effects of the different sus1 alleles on TREX-2 subunit composition, we purified TREX-2 via TAP-tagged Thp1 and used Western blotting to determine the amount of bound mutant Sus1 proteins. Consistent with the results of the Sus1-TAP purifications, FLAG-tagged Sus1–10, Sus1–11, and Sus1–12 were specifically absent from the TREX-2 complex, whereas the other TREX-2 subunits Sac3 and Cdc31 were efficiently co-enriched with Thp1 (Fig. 1d). Taken together, the biochemical data indicate that sus1–10 and sus1–12 uncouple Sus1 from TREX-2, whereas the sus1–11 mutation effectively impairs the Sus1 interaction with both TREX-2 and SAGA.
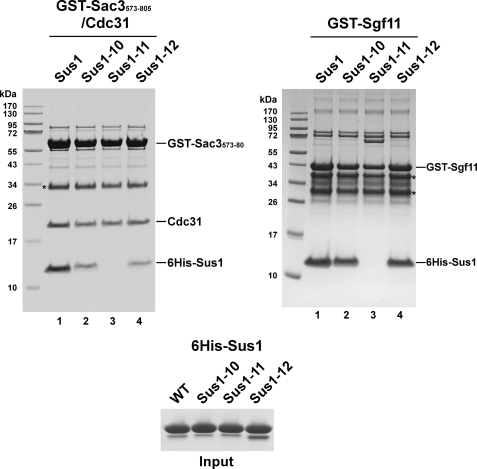
To confirm that the dissociation of Sus1 from TREX-2 or SAGA is caused primarily by a reduced affinity between Sus1-Sac3 and Sus1-Sgf11, we reconstituted these Sus1-ligand interactions in vitro (Fig. 2). Recombinant wild-type and mutant Sus1 proteins were first assayed for their ability to bind to a Sac3-(573–805)-Cdc31 heterodimer. This C-terminal fragment of Sac3 harbors the binding sites for Sus1 and Cdc31. Wild-type Sus1 bound to Sac3-(573–805)-Cdc31 very efficiently, whereas the binding was reduced with Sus1–10 and Sus1–12 and largely abolished for the Sus1–11 mutant. On the other hand, binding of Sus1–10 and Sus1–12 to Sgf11 was largely unaffected, whereas Sus1–11 failed to interact with Sgf11 (Fig. 2). We consider it unlikely that the perturbed Sus1-ligand interactions were caused by severe misfolding of the Sus1 mutants. CD spectra for the mutant Sus1 proteins were determined and found to be virtually indistinguishable from those of the wild-type protein (all showed prominent negative ellipticities at 220 nm consistent with the presence of an α-helical conformation; data not shown). In summary, the in vitro experiments largely recapitulate the Sus1-TAP affinity purifications, with Sus1–11 showing a global binding defect, whereas Sus1–10/-12 display a selective Sac3 interaction defect. We observed that the extent of Sus1–10/-12 dissociation from Sac3 in vitro is not as severe as expected from the yeast affinity purifications. We believe this was caused mainly by the excess of proteins present in the in vitro assay, although an unprotected Sus1-binding site on Sac3 may render TREX-2 susceptible to proteolytic attack in vivo, thus aggravating Sus1 dissociation from Sac3. This possibility is also consistent with the reduced yield of Thp1-TAP purifications in all sus1 mutants (data not shown).
FIGURE 2.
Sus1 mutant proteins bind to Sac3 and Sgf11 with different affinities in vitro. Recombinant GST-Sac3-(573–805)-Cdc31 complex or GST-Sgf11 was immobilized on GSH beads and incubated with the indicated recombinant 6xHis-Sus1 wild-type (WT) or mutant proteins (input). Proteins bound to the beads were eluted with GSH and analyzed by SDS-PAGE (12% gel, MES buffer) and Coomassie staining. Asterisks indicate Sac3 and Sgf11 degradation products, as determined by mass spectrometry. Note that GST-Sac3-(573–805) harbors the entire CID, which binds to both Cdc31 and Sus1.
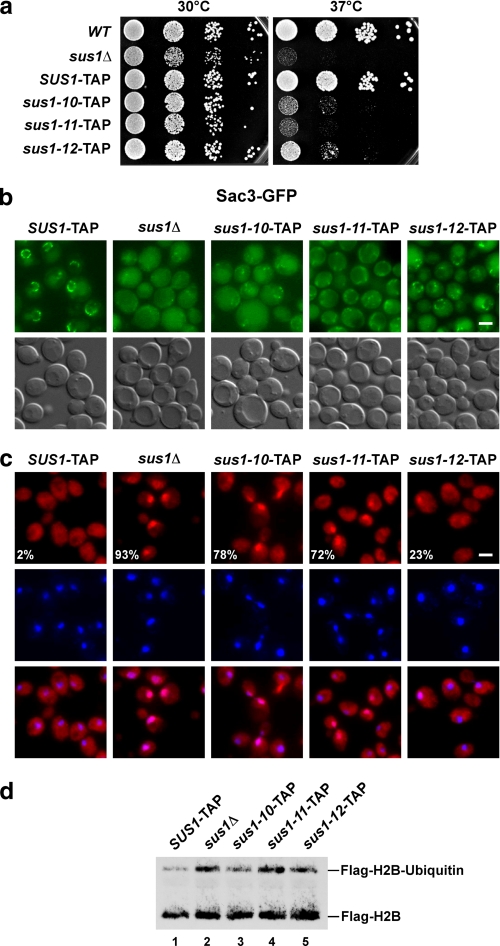
Next, we determined the functional consequences of the selective dissociation of Sus1 from TREX-2 by examining the mutant growth phenotypes. Although sus1Δ cells have a mild growth defect at 30 °C on glucose-containing media (YPD), these cells are strongly retarded in growth at 37 °C (Fig. 3a). Cells expressing TAP-tagged SUS1 grow like untagged wild-type cells at both temperatures making it unlikely that the epitope tag introduces major adverse effects on Sus1 function. The sus-10, sus1-11, and sus1-12 mutants all showed robust temperature-sensitive phenotypes at 37 °C with sus1-11 cells having the most severe growth defect comparable with sus1Δ cells. We observed a graded loss of SUS1 function in vivo, as sus1-12 was less severely growth-retarded as compared with sus1–10 and sus1–11.
FIGURE 3.
Uncoupling of Sus1 from Sac3 causes TREX-2 mislocalization and mRNA export defects in vivo. a, growth analysis of SUS1-TAP wild-type (WT) and mutant cells on rich medium (YPD) at 30 and 37 °C. Cell density was normalized, and 10-fold serial dilutions were prepared and then plated. Plates were incubated for 2 days. b, subcellular localizations of Sac3-GFP in the indicated wild-type and mutant sus1 strains are shown. Fluorescence microscopic and Nomarski photographs of representative cells grown at 30 °C are shown (scale bar, 2 μm). c, analysis of nuclear mRNA export in the indicated wild-type and sus1 mutant strains. Exponentially growing cells were shifted to 37 °C and grown for 2 more hours before detection of poly(A)+ RNA by fluorescent in situ hybridization with Cy3-labeled oligo(dT) probes. DNA was stained with 4,6-diamidino-2-phenylindole. Numbers indicate the percentage of cells (∼200 cells counted in each case) that exhibit an apparent mRNA export defect. Scale bar, 2 μm. d, analysis of global histone H2B ubiquitin levels. Anti-FLAG-H2B immunoprecipitates derived from SUS1-TAP cells, sus1Δ cells, and the indicated mutants. Recovered proteins were analyzed by SDS-PAGE and Western blotting using an anti-FLAG antibody to detect unmodified FLAG-H2B and ubiquitinated FLAG-H2B.
Previously, we showed that Sus1 is required for the efficient targeting of TREX-2 to the NPCs (15). TREX-2 association with the NPCs is necessary for mRNA export and presumably mediates the dynamic tethering of activated genes (e.g. GAL1) to the nuclear periphery (32). The largest TREX-2 component, Sac3, displays a distinct nuclear rim staining in wild-type cells; however, in sus1Δ cells Sac3-GFP is strongly mislocalized to the cytoplasm and nucleoplasm (Fig. 3b) (15). Consistent with a requirement of Sus1 binding to Sac3 for proper TREX-2 localization, sus1–10, sus1–11, and sus1–12 cells all showed an altered pattern of Sac3-GFP staining. The sus1–11 allele, which is globally affected in its interaction with SAGA and TREX-2, showed the strongest Sac3 mislocalization, similar in severity to the deletion of SUS1. However, defective TREX-2 targeting cannot be attributed to the function of Sus1 within SAGA, because deletion of UBP8 (with a concomitant loss of Sus1 from SAGA) does not affect TREX-2 localization (15). Therefore, the pronounced Sac3 mislocalization seen with the sus1–11 allele is specifically related to the role of Sus1 within TREX-2. Accordingly, the TREX-2-defective sus1–10 and sus-12 alleles also caused Sac3 mislocalization, but to a somewhat lesser extent with sus1–12 having the mildest phenotype. This observation is consistent with the finding that TAP-purified Sus1–12 exhibits a stronger residual interaction with Sac3 as compared with Sus1–10 (Fig. 1c). Taken together, the disruption of the Sus1-Sac3CID interaction causes graded TREX-2 mislocalization phenotypes and underscores the importance of the small Sus1 protein in targeting TREX-2 to NPCs.
Next, we characterized the sus1 mutant alleles with respect to nuclear mRNA export at the restrictive temperature (37 °C). When assayed by fluorescent in situ hybridization, nuclear export of poly(A)+ RNA is impaired in all mutants, although to different degrees. The sus1–10 and sus1–11 alleles exhibited robust mRNA export defects, whereas sus1–12 had a milder impairment with fewer cells affected (Fig. 3c). These results correlate with the cell biological and growth properties of sus1–12 (see above). Because sus1–10 and sus1-12 specifically fail to incorporate into TREX-2, the bulk mRNA export block can be attributed to the function of Sus1 in TREX-2 rather than SAGA.
Although the DUB module (Sus1-Sgf11-Ubp8-Sgf73) appears to be assembled correctly in the sus1–10 and sus-12 mutants (see Fig. 1c), it is still possible that Ubp8 enzymatic activity is dysregulated. Therefore, we measured in vivo histone H2B ubiquitin levels as a readout for Ubp8 function. Previously, we reported that deletion of SUS1 increases global H2B ubiquitin levels as a consequence of Ubp8 dissociation from SAGA (25). Ubp8 when uncoupled from its DUB module cofactors is enzymatically inactive (15, 21). Consistent with a complete dissociation of Sus1–11 from the DUB module, the global level of ubiquitin at H2B Lys123 was increased in the sus1–11 mutant to a similar magnitude as in a sus1Δ strain (Fig. 3d). The sus1–10 and sus1–12 mutants exhibited a slight increase in H2B ubiquitin levels when compared with wild type, which may hint to some reduction of Ubp8 activity even though Sus1–10/-12 association appears to be largely intact on a structural level.
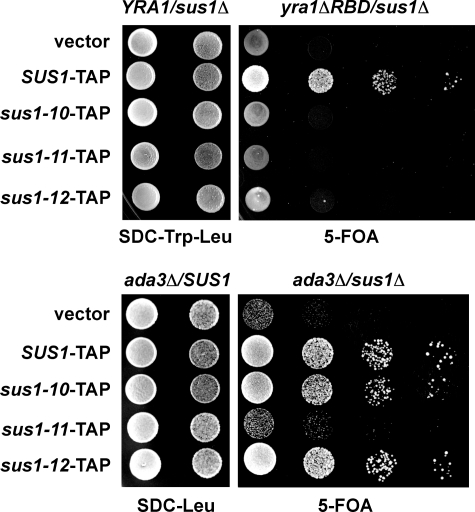
In vivo, SUS1 is embedded in a network of functional interactions with key factors operating in transcription-coupled mRNA export (14). To analyze how weakening of Sus1-ligand interactions would perturb the SUS1 genetic network, we assayed growth phenotypes of the generated sus1 alleles in combination with a mutant allele of a typical mRNA export factor (i.e. Yra1) or a bona fide SAGA subunit (i.e. Ada3). Previously, we demonstrated that sus1Δ is synthetically lethal when combined with the yra1ΔRBD mutant (14) and synthetically enhanced with the ada3Δ allele.3 Consistent with the biochemical data, sus1–11 was genetically linked to both yra1ΔRBD and ada3Δ (Fig. 4). In contrast, sus1–10 and sus1–12 were not genetically linked to ada3Δ. However, sus1–10 and sus1–12 exhibit a synthetic lethal interaction with yra1ΔRBD consistent with their pronounced biochemical defect toward TREX-2 (Fig. 1c). We also surveyed the initial set of mutants, sus1–1 to sus1–9 (Fig. 1b), for genetic interactions with yra1ΔRBD and temperature-sensitive phenotypes, however, without detecting any functional impairments (supplemental Fig. 1, b and c). Taken together, the genetic data suggest that SUS1 function critically depends on intact interactions between Sus1 and its partners. Furthermore, the rather selective biochemical defects of the mutant sus1 alleles are matched by differential genetic defects that perturb either the mRNA export pathway alone (i.e. YRA1) or in combination with the transcription/chromatin remodeling pathway (i.e. ADA3).
FIGURE 4.
sus1 mutant alleles differentially perturb the genetic interactions of SUS1. The yra1Δsus1Δ double mutant strain expressing both the wild-type YRA1 allele (URA3 plasmid) and yra1ΔRBD allele (TRP1 plasmid) were transformed with the wild-type SUS1 and the indicated sus1 mutant alleles on LEU2 plasmids. Growth was followed on SDC-Trp-Leu plates and on 5-FOA plates after shuffling out the URA3 cover plasmid. The ada3Δsus1Δ strain was transformed both with a URA3 plasmid containing wild-type SUS1 and the indicated sus1 mutant alleles on LEU2 plasmids. Growth was followed on SDC-Leu plates and on 5-FOA plates after shuffling out the URA3 cover plasmid. Cell density was normalized, and 10-fold serial dilutions were prepared and then plated. Plates were incubated for 2–3 days at 30 °C.
DISCUSSION
In this study we have employed a spectrum of methods to dissect structurally and functionally the role of Sus1 within the TREX-2 and SAGA components of the gene expression machinery. Taken together, our mutational analysis showed that Sus1, via the sus1–10 or sus1–12 alleles, can be uncoupled from Sac3 (i.e. TREX-2), although its interaction with Sgf11 (i.e. SAGA) remains largely unaffected. These mutations were clustered in putative loops between the α-helices predicted by sequence analysis. On the other hand, via the sus1–11 allele, in which two highly conserved amino acids (Gly37Trp38) were mutated, Sus1 was concomitantly impaired in its interaction with both TREX-2 and SAGA. The Gly37Trp38 pair could be involved either in a direct molecular contact to Sac3 and Sgf11 or be important in stabilizing the putative α-helical conformation of Sus1 needed for binding these partners. Independent of which scenario is true, Sus1 clearly contains residues that are important for both types of interaction. We have not been able to selectively eliminate the Sus1-Sgf11 interaction without also impairing the Sus1-Sac3 interaction. However, within SAGA Sus1 is part of a biochemically stable DUB module, which may involve additional contacts between the Sus1-Sgf11 heterodimer and Ubp8 or Sgf73. Taken together, our findings suggest that there are substantial differences in the way Sus1 is bound to its ligands, although certain requirements are common to both interaction partners.
How could structural differences in the binding sites for Sgf11 and Sac3 impact on Sus1 function? Several lines of biochemical and genetic evidence suggest that SAGA and TREX-2 are physically and functionally linked; moreover, both machineries function in GAL1 gene gating. A major question is whether Sus1 plays entirely separate roles in histone deubiquitination and TREX-2 NPC targeting. Alternatively, Sus1 is involved in some way in inter-complex communication. It is intriguing that SAGA-associated Sgf73 plays a crucial role in physically recruiting Sac3 and Thp1 to SAGA. In contrast, deletion of Ubp8 with a concomitant loss of Sus1 from SAGA did not have a major effect on Sac3-Thp1 binding (15). This observation argues against SAGA-bound Sus1 being the predominant mediator of the interaction with TREX-2. Nevertheless, it is possible that Sus1 might be dynamically exchanged between its TREX-2- and SAGA-binding sites, when a SAGA-TREX-2 intermediate is formed. For example TREX-2 might be subject to dynamic NPC binding and release (generated by Sus1-Sac3 association/dissociation) in synchronization with transcriptional events. Support for such a model is provided by the observation that activated GAL1 does not become statically attached to a single coordinate of the nuclear periphery but exhibits a constrained, peripheral sliding motion when visualized by dynamic imaging studies (7). It is interesting to speculate that de novo formation of the SAGA histone deubiquitination module could involve competitive capture of TREX-2-bound Sus1 molecules, which may in turn cause transient release of TREX-2 from the NPCs. Accordingly, the reverse reaction might disassemble the DUB module and inhibit histone deubiquitination while supporting TREX-2 tethering at the pore. Such dynamic shuttling of Sus1 may specifically affect the SAGA-TREX-2 machinery involved in gene gating and could be subject to many other types of regulation, such as post-translational modification. Although further work is required to approach these questions, the Sus1 mutants generated in this study provide a powerful basis for exploring the spectrum of interactions involved in the integration of transcription and nuclear mRNA export.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Tables 1 and 2.
Footnotes
The abbreviations used are: NPC, nuclear pore complex; SAGA, Spt-Ada-Gcn5 acetyltransferase; DUB, deubiquitinating; GST, glutathione S-transferase; MES, 4-morpholineethanesulfonic acid; 5-FOA, 5-fluoroorotic acid; MOPS, 4-morpholinepropanesulfonic acid; TAP, tandem affinity purification; CID, Cdc31 interaction domain.
S. Lutz and E. Hurt, unpublished data.
References
- 1.Maniatis, T., and Reed, R. (2002) Nature 416 499-506 [DOI] [PubMed] [Google Scholar]
- 2.Köhler, A., and Hurt, E. (2007) Nat. Rev. Mol. Cell Biol. 8 761-773 [DOI] [PubMed] [Google Scholar]
- 3.Luna, R., Gaillard, H., Gonzalez-Aguilera, C., and Aguilera, A. (2008) Chromosoma 117 319-331 [DOI] [PubMed] [Google Scholar]
- 4.Iglesias, N., and Stutz, F. (2008) FEBS Lett. 582 1987-1996 [DOI] [PubMed] [Google Scholar]
- 5.Akhtar, A., and Gasser, S. M. (2007) Nat. Rev. Genet. 8 507-517 [DOI] [PubMed] [Google Scholar]
- 6.Brown, C. R., and Silver, P. A. (2007) Curr. Opin. Genet. Dev. 17 100-106 [DOI] [PubMed] [Google Scholar]
- 7.Cabal, G. G., Genovesio, A., Rodriguez-Navarro, S., Zimmer, C., Gadal, O., Lesne, A., Buc, H., Feuerbach-Fournier, F., Olivo-Marin, J. C., Hurt, E. C., and Nehrbass, U. (2006) Nature 441 770-773 [DOI] [PubMed] [Google Scholar]
- 8.Chekanova, J. A., Abruzzi, K. C., Rosbash, M., and Belostotsky, D. A. (2008) RNA (N. Y.) 14 66-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taddei, A., Van Houwe, G., Hediger, F., Kalck, V., Cubizolles, F., Schober, H., and Gasser, S. M. (2006) Nature 441 774-778 [DOI] [PubMed] [Google Scholar]
- 10.Dieppois, G., Iglesias, N., and Stutz, F. (2006) Mol. Cell. Biol. 26 7858-7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brickner, J. H., and Walter, P. (2004) PLoS Biol. 2 e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abruzzi, K. C., Belostotsky, D. A., Chekanova, J. A., Dower, K., and Rosbash, M. (2006) EMBO J. 25 4253-4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casolari, J. M., Brown, C. R., Komili, S., West, J., Hieronymus, H., and Silver, P. A. (2004) Cell 117 427-439 [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Navarro, S., Fischer, T., Luo, M. J., Antunez, O., Brettschneider, S., Lechner, J., Perez-Ortin, J. E., Reed, R., and Hurt, E. (2004) Cell 116 75-86 [DOI] [PubMed] [Google Scholar]
- 15.Köhler, A., Schneider, M., Cabal, G. G., Nehrbass, U., and Hurt, E. (2008) Nat. Cell Biol. 10 707-715 [DOI] [PubMed] [Google Scholar]
- 16.Grant, P. A., Duggan, L., Cote, J., Roberts, S. M., Brownell, J. E., Candau, R., Ohba, R., Owen-Hughes, T., Allis, C. D., Winston, F., Berger, S. L., and Workman, J. L. (1997) Genes Dev. 11 1640-1650 [DOI] [PubMed] [Google Scholar]
- 17.Larschan, E., and Winston, F. (2001) Genes Dev. 15 1946-1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniel, J. A., and Grant, P. A. (2007) Mutat. Res. 618 135-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla, A., Stanojevic, N., Duan, Z., Sen, P., and Bhaumik, S. R. (2006) Mol. Cell. Biol. 26 3339-3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingvarsdottir, K., Krogan, N. J., Emre, N. C., Wyce, A., Thompson, N. J., Emili, A., Hughes, T. R., Greenblatt, J. F., and Berger, S. L. (2005) Mol. Cell. Biol. 25 1162-1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, K. K., Florens, L., Swanson, S. K., Washburn, M. P., and Workman, J. L. (2005) Mol. Cell. Biol. 25 1173-1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henry, K. W., Wyce, A., Lo, W. S., Duggan, L. J., Emre, N. C., Kao, C. F., Pillus, L., Shilatifard, A., Osley, M. A., and Berger, S. L. (2003) Genes Dev. 17 2648-2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMahon, S. J., Pray-Grant, M. G., Schieltz, D., Yates, J. R., III, and Grant, P. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8478-8482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daniel, J. A., Torok, M. S., Sun, Z. W., Schieltz, D., Allis, C. D., Yates, J. R., III, and Grant, P. A. (2004) J. Biol. Chem. 279 1867-1871 [DOI] [PubMed] [Google Scholar]
- 25.Köhler, A., Pascual-Garcia, P., Llopis, A., Zapater, M., Posas, F., Hurt, E., and Rodriguez-Navarro, S. (2006) Mol. Biol. Cell 17 4228-4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weake, V. M., and Workman, J. L. (2008) Mol. Cell 29 653-663 [DOI] [PubMed] [Google Scholar]
- 27.Wyce, A., Xiao, T., Whelan, K. A., Kosman, C., Walter, W., Eick, D., Hughes, T. R., Krogan, N. J., Strahl, B. D., and Berger, S. L. (2007) Mol. Cell 27 275-288 [DOI] [PubMed] [Google Scholar]
- 28.Fleming, A. B., Kao, C. F., Hillyer, C., Pikaart, M., and Osley, M. A. (2008) Mol. Cell 31 57-66 [DOI] [PubMed] [Google Scholar]
- 29.Pavri, R., Zhu, B., Li, G., Trojer, P., Mandal, S., Shilatifard, A., and Reinberg, D. (2006) Cell 125 703-717 [DOI] [PubMed] [Google Scholar]
- 30.Pascual-Garcia, P., Govind, C. K., Queralt, E., Cuenca-Bono, B., Llopis, A., Chavez, S., Hinnebusch, A. G., and Rodriguez-Navarro, S. (2008) Genes Dev. 22 2811-2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer, T., Strässer, K., Racz, A., Rodriguez-Navarro, S., Oppizzi, M., Ihrig, P., Lechner, J., and Hurt, E. (2002) EMBO J. 21 5843-5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer, T., Rodriguez-Navarro, S., Pereira, G., Racz, A., Schiebel, E., and Hurt, E. (2004) Nat. Cell Biol. 6 840-848 [DOI] [PubMed] [Google Scholar]
- 33.Jones, A. L., Quimby, B. B., Hood, J. K., Ferrigno, P., Keshava, P. H., Silver, P. A., and Corbett, A. H. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 3224-3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei, P., Stern, C. A., Fahrenkrog, B., Krebber, H., Moy, T. I., Aebi, U., and Silver, P. A. (2003) Mol. Biol. Cell 14 836-847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez-Aguilera, C., Tous, C., Gomez-Gonzalez, B., Huertas, P., Luna, R., and Aguilera, A. (2008) Mol. Biol. Cell 19 4310-4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauer, A., and Kölling, R. (1996) J. Cell Sci. 109 1575-1583 [DOI] [PubMed] [Google Scholar]
- 37.Wilmes, G. M., Bergkessel, M., Bandyopadhyay, S., Shales, M., Braberg, H., Cagney, G., Collins, S. R., Whitworth, G. B., Kress, T. L., Weissman, J. S., Ideker, T., Guthrie, C., and Krogan, N. J. (2008) Mol. Cell 32 735-746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, Y., Lang, G., Ito, S., Bonnet, J., Metzger, E., Sawatsubashi, S., Suzuki, E., Le Guezennec, X., Stunnenberg, H. G., Krasnov, A., Georgieva, S. G., Schule, R., Takeyama, K., Kato, S., Tora, L., and Devys, D. (2008) Mol. Cell 29 92-101 [DOI] [PubMed] [Google Scholar]
- 39.Zhang, X. Y., Varthi, M., Sykes, S. M., Phillips, C., Warzecha, C., Zhu, W., Wyce, A., Thorne, A. W., Berger, S. L., and McMahon, S. B. (2008) Mol. Cell 29 102-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weake, V. M., Lee, K. K., Guelman, S., Lin, C. H., Seidel, C., Abmayr, S. M., and Workman, J. L. (2008) EMBO J. 27 394-405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurshakova, M. M., Krasnov, A. N., Kopytova, D. V., Shidlovskii, Y. V., Nikolenko, J. V., Nabirochkina, E. N., Spehner, D., Schultz, P., Tora, L., and Georgieva, S. G. (2007) EMBO J. 26 4956-4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida, M., Kuwahara, K., Shimasaki, T., Nakagata, N., Matsuoka, M., and Sakaguchi, N. (2007) Genes Cells 12 1205-1213 [DOI] [PubMed] [Google Scholar]
- 43.Kuwahara, K., Tomiyasu, S., Fujimura, S., Nomura, K., Xing, Y., Nishiyama, N., Ogawa, M., Imajoh-Ohmi, S., Izuta, S., and Sakaguchi, N. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10279-10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takei, Y., Swietlik, M., Tanoue, A., Tsujimoto, G., Kouzarides, T., and Laskey, R. (2001) EMBO Rep. 2 119-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Resendes, K. K., Rasala, B. A., and Forbes, D. J. (2008) Mol. Cell. Biol. 28 1755-1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longtine, M. S., McKenzie, A., Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998) Yeast 10 953-961 [DOI] [PubMed] [Google Scholar]
- 47.Janke, C., Magiera, M. M., Rathfelder, N., Taxis, C., Reber, S., Maekawa, H., Moreno-Borchart, A., Doenges, G., Schwob, E., Schiebel, E., and Knop, M. (2004) Yeast 21 947-962 [DOI] [PubMed] [Google Scholar]
- 48.Kressler, D., Roser, D., Pertschy, B., and Hurt, E. (2008) J. Cell Biol. 181 935-944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Séraphin, B. (1999) Nat. Biotechnol. 17 1030-1032 [DOI] [PubMed] [Google Scholar]
- 50.Nissan, T. A., Bassler, J., Petfalski, E., Tollervey, D., and Hurt, E. C. (2002) EMBO J. 21 5539-5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segref, A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Lührmann, R., and Hurt, E. C. (1997) EMBO J. 16 3256-3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kao, C. F., and Osley, M. A. (2003) Methods (San Diego) 31 59-66 [DOI] [PubMed] [Google Scholar]
- 53.Wertman, K. F., Drubin, D. G., and Botstein, D. (1992) Genetics 132 337-350 [DOI] [PMC free article] [PubMed] [Google Scholar]