Abstract
The nonmuscle myosin IIA heavy chain (Myh9) is strongly associated with adhesion structures of osteoclasts. In this study, we demonstrate that during osteoclastogenesis, myosin IIA heavy chain levels are temporarily suppressed, an event that stimulates the onset of cell fusion. This suppression is not mediated by changes in mRNA or translational levels but instead is due to a temporary increase in the rate of myosin IIA degradation. Intracellular activity of cathepsin B is significantly enhanced at the onset of osteoclast precursor fusion, and specific inhibition of its activity prevents myosin IIA degradation. Further, treatment of normal cells with cathepsin B inhibitors during the differentiation process reduces cell fusion and bone resorption capacity, whereas overexpression of cathepsin B enhances fusion. Ongoing suppression of the myosin IIA heavy chain via RNA interference results in formation of large osteoclasts with significantly increased numbers of nuclei, whereas overexpression of myosin IIA results in less osteoclast fusion. Increased multinucleation caused by myosin IIA suppression does not require RANKL. Further, knockdown of myosin IIA enhances cell spreading and lessens motility. These data taken together strongly suggest that base-line expression of nonmuscle myosin IIA inhibits osteoclast precursor fusion and that a temporary, cathepsin B-mediated decrease in myosin IIA levels triggers precursor fusion during osteoclastogenesis.
The final stages of osteoclastogenesis involve fusion of differentiated precursors from the monocyte/macrophage lineage (1). Although the membrane structural components regulating preosteoclast fusion are not well understood, in recent years a number of candidate cell surface molecules have been implicated, including receptors CD44 (2, 3), CD47 and its ligand macrophage fusion receptor (also known as signal regulatory protein α) (4–6), the purinergic receptor P2X7 (7), and the disintegrin and metalloproteinase ADAM8 (8). A recently identified receptor, the dendritic cell-specific transmembrane protein, is essential for osteoclast fusion both in vitro and in vivo (9, 10). More recently, the d2 subunit of proton-translocating vacuolar proton-translocating ATPases, a membrane subunit isoform expressed predominantly in osteoclasts, similarly was demonstrated to be required for fusion in vitro and in vivo (11). However, elucidation of the mechanisms by which these molecules may mediate cell fusion has proved to be difficult.
The mammalian class II myosin family consists of distinct isoforms expressed in skeletal, smooth, and cardiac muscle, as well as three nonmuscle forms designated IIA, IIB, and IIC (12–14). Although all class II molecules are composed of two heavy chains, two essential light chains, and two regulatory chains, their unique activities are a function of their particular heavy chain isoforms. Although the nonmuscle heavy chain isoforms share extensive structural homology, they have been shown to demonstrate distinct patterns of expression (15–18), enzyme kinetics and activation (12, 19–21), and cellular function (22–24). Knock-out of either myosin IIA or IIB results in embryonic lethality, although death derives from defects unique to each isoform (25, 26). In vitro, myosin IIA, a target of Rho kinase, has been shown to be involved in a wide variety of cellular functions, including cytokinesis, cell contractility, and adhesion and motility.
The actin cytoskeleton of osteoclasts possesses features unlike those of most mammalian cell types. First, osteoclasts do not possess stress fibers but instead form a meshwork of fine actin filaments throughout the cell (27–29). Osteoclasts express unusual attachment structures typified by the podosome, a form of adhesion structure most typically present in cells of the monocyte/macrophage lineage, dendritic cells, and smooth muscle cells. Podosomes are integrin-based cell-matrix contact structures that are notable for the presence of a short (0.5–1.0 μm) F-actin core surrounded by a ring of adaptor proteins, kinases, small GTPases, and regulators of endocytosis (30, 31). When cultured on glass, mature osteoclasts generate a belt of podosomes at the cell periphery. However, when cultured on bone, osteoclasts form a dense ring of podosome-like structures that is usually internal to the cell margins (32). This region, termed the sealing zone, surrounds a specialized membrane domain termed the ruffled border, from which protons and proteases are secreted to induce resorption of bone (1). We previously demonstrated that myosins IIA and IIB localize to distinct subcellular regions within osteoclasts, with MyoIIA2 strongly segregating to both podosomes and the actin ring of the sealing zone (28). Because of this distribution into osteoclast adhesion structures and findings in other cells showing MyoIIA to be associated with dynamic Rho-kinase-dependent functions, such as adhesion and locomotion, we hypothesized that MyoIIA may play a vital role in cell motility and the bone resorption function. In this study, we examined cellular expression of MyoIIA during osteoclastogenesis and, along with RNA interference-mediated suppression of the protein, have confirmed its role in cell spreading, motility, and sealing zone formation. However, this study also unexpectedly revealed a role for MyoIIA in regulating preosteoclast fusion during osteoclastogenesis.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture—Rabbit polyclonal antibodies against the MyoIIA heavy chain were previously generated (28) or were purchased from Sigma. Loading control antibodies to GAPDH and β-actin were purchased from Abcam. Protease inhibitors, excluding CA-074Me and CA-074, and an antibody to cathepsin B were purchased from EMD Biosciences. CA-074Me and CA-074 were obtained from Enzo Life Sciences/BioMol. Osteoclasts were generated either from RAW 264.7 macrophages (ATCC) or murine bone marrow, as previously described (28, 33, 34). Mature osteoclasts, as evidenced by a peripheral belt of podosomes (35), were present by days 5–7 of culture. Resorption and motility assays were performed as previously described (34, 36).
Immunocytochemistry and Microscopy—Osteoclasts were cultured either on glass coverslips or thinly cut ivory slices. Cells were fixed and permeabilized, as previously described (28, 33, 34, 36). Primary antibodies were added in a standard polyethylene glycol blocking buffer and were detected using Alexa-coupled secondary antibodies (Invitrogen). F-actin was labeled using Alexa-coupled phalloidin, also from Invitrogen. Nuclei were detected with bisbenzimide staining. Cells were visualized using either a Nikon 80i fluorescent microscope with SPOT camera software (Diagnostic Instruments) or a Zeiss 510 META laser-scanning confocal microscope (Campus Microscopy and Imaging Facility, The Ohio State University). Cell and sealing zone perimeters were measured using SigmaScan Pro 5.0 software (SPSS Science, Chicago, IL). Active cathepsin B was visualized using the Cathepsin B Magic Red real time detection kit (Immunochemistry Technologies, LLC, Bloomington, MN). The intensity of cathepsin B labeling was quantified using SigmaScan Pro 5.0 software.
Competitive RT-PCR of Myh9 mRNA—Murine myosin IIA heavy chain (Myh9) expression was determined by competitive RT-PCR. The corresponding sense primer was of the sequence 5′-AGACCGCTGATGCTATGAACCG-3′, whereas the antisense primer was of the sequence 5′-ATGGTCAGAGCAGTCTCAAGGC-3′. An internal standard cDNA template for the reaction was generated as previously described (37, 38). This template was transcribed in vitro using the MAXIscript system (Ambion), and 10 pg of the resulting RNA was added to 1 μg of osteoclast total cellular RNA prior to reverse transcription and PCR. These reactions were performed using the Superscript first strand synthesis system and Taq DNA polymerase, both from Invitrogen. The resulting RT-PCR products were run in a 2% gel and stained with ethidium bromide to visualize relative intensities of the bands, which were measured using Quantity One software (Bio-Rad).
Western Analysis—For Western analysis, osteoclast lysates were harvested with M-PER reagent (Pierce) and were run in precast polyacrylamide gels (Bio-Rad) and transferred to Hybond membrane (GE HealthCare). Primary antibodies were allowed to bind to the membranes using standard methodology and were detected using horseradish peroxidase-labeled secondary antibodies coupled with SuperSignal West Pico Chemiluminescent reagents (Pierce).
Pulse-Chase Analysis—Osteoclasts on days 0, 3, and 7 of differentiation were labeled for 2 h in methionine- and cysteine-free medium supplemented with 45 μCi/ml Tran35S-label (MP Biomedicals). The cells were then incubated in complete medium for various times before immunoprecipitation, performed as previously described (39). The precipitates were run in SDS-PAGE, and the gel was subsequently treated with Fluoro-Hance (Research Products International) and exposed to film. Quantity One software was used to quantify the intensity of the myosin IIA immunoprecipitate bands.
Knockdown and Overexpression of MyoIIA—siRNA oligonucleotides were designed and synthesized by Ambion. siRNA1, which was used for all relevant experiments, was of the sequence 5′-GGCUGAUUUCUGCAUUAUCtt-3′ (sense) and 5′-GAUAAUGCAGAAAUCAGCCtt-3′ (antisense). siRNA2, used to confirm many of the results, was of the sequence 5′-GGUGAACAAGGACGACAUCtt-3′ (sense) and 5′-GAUGUCGUCCUUGUUCACCtt-3′ (antisense). These siRNAs showed identity to no other myosin isoform or murine transcript. For all experiments, a nontargeting double-stranded RNA from Ambion was used as a negative control (C0), although siRNAs homologous to siRNA1 and siRNA2 but containing point mutations also were used for confirmation. The mutant siRNA1 (C1) was of the sequence 5′-GGCUGAUcagUGCAUUAUCtt-3′ (sense) and 5′-GAUAAUGCAcugAUCAGCCtt-3′ (antisense), whereas the mutant siRNA2 (C2) was of the sequence 5′-GGUGAACAcacACGACAUCtt-3′ (sense) and 5′-GAUGUCGUgugUGUUCACCtt-3′ (antisense). RAW264.7- and marrow-derived osteoclasts were transfected on day 4 of culture using 25–50 nm targeting or control siRNA. RAW264.7 cells were transfected with Lipofectamine 2000 (Invitrogen), and marrow-derived osteoclasts were transfected via electroporation, as previously described. Osteoclasts were transfected by these methods with >95% efficiency (34, 36). In other experiments, a human myosin IIA cDNA in the pEGFP-C3 vector (40) or the empty vector (Clontech) was transiently transfected into RAW264.7 cells on day 3 of RANKL treatment and assayed for cell perimeter and nuclear number on day 5. Transfection was performed by electroporation at 250 V/250 microfarads, and efficiency was >90%.
Overexpression of Cathepsin B—A murine cDNA clone of cathepsin B was obtained by RT-PCR from RAW264.7 cells and was subcloned into the pEF6/V5-His expression vector (Invitrogen). Following transfection of this construct into RAW264.7 macrophages, selection for stable transfectants was performed by selection with 3 μg/ml blasticidin. Two overexpressing clones were chosen for analysis.
Statistical Analysis—Pairwise comparisons of samples were performed using Student's t test. Linear regression was used to model the relationship between cell or sealing zone perimeter and the number of nuclei, allowing for different slope and intercept terms for each cell type, and was performed by the Center for Biostatistics (Ohio State University). In order to satisfy the assumptions of normality and constant variance, the data were modeled on the log scale. F-tests were used to determine the significance of type-specific slope and intercept parameters. Statistical comparison was deemed significant at p < 0.05.
RESULTS
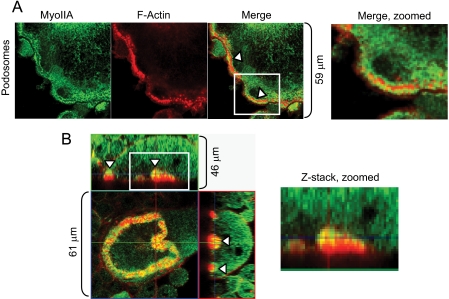
Myosin IIA Distribution in Osteoclasts—We previously showed that nonmuscle myosin IIA, but not IIB, was associated with podosomes and the actin ring of polarized osteoclasts (28). More detailed examination of these structures demonstrates that MyoIIA is distributed in the cloud of actin surrounding the podosome core but not the core itself. Fig. 1A illustrates immunocytochemistry of MyoIIA surrounding F-actin cores of podosomes (arrowheads). Additionally, detailed examination of the sealing zone of polarized osteoclasts showed that MyoIIA is distributed over the inner face of the actin ring (Fig. 1B, arrowheads). These results, demonstrating a strong presence of MyoIIA in adhesion structures, suggest a role in osteoclast motility and/or bone resorption.
FIGURE 1.
Myosin IIA is strongly localized to adhesion structures of osteoclasts. A, osteoclasts were cultured on glass coverslips and labeled for MyoIIA and F-actin. The arrowheads indicate MyoIIA surrounding the actin core of podosomes. B, osteoclasts were cultured on ivory and labeled for MyoIIA (green) and F-actin (red). Z-stack images demonstrate that MyoIIA is present at the inner face of the sealing zone (arrowheads). The boxed regions in A and B are magnified for closer inspection.
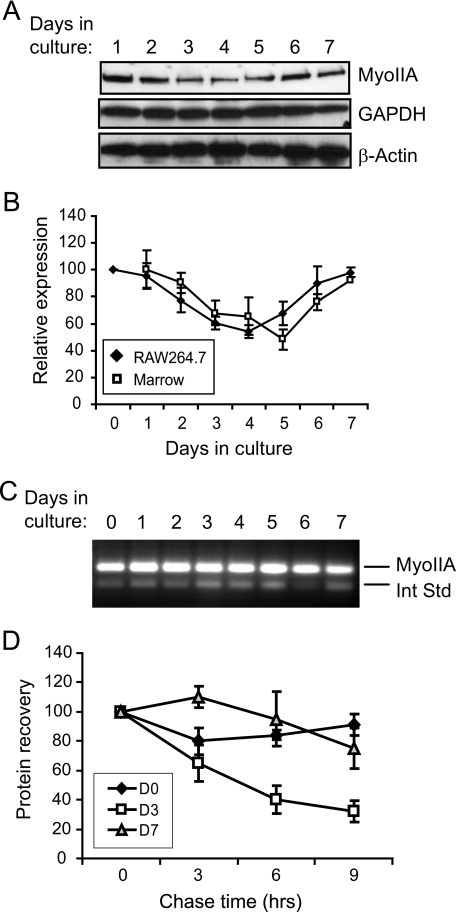
Myosin IIA Expression Is Transiently Decreased during Osteoclastogenesis—Two culture models of murine osteoclastogenesis are widely used to study this process. Mouse bone marrow precursors or the mouse macrophage cell line RAW264.7, when stimulated to differentiate with a soluble form of RANKL, can form mature osteoclasts over the course of 5–7 days in culture. Dividing precursors cease mitosis and initiate fusion after about 3 days, continuing to become more extensively multinucleated as long as fusion partners are in close proximity (41, 42). As a first step in examining the role of MyoIIA in osteoclast function, precursor cells were differentiated in the presence of RANKL over a 7-day culture period. Western blots demonstrated that differentiating osteoclasts temporarily decreased their cellular levels of the MyoIIA heavy chain during the middle of this differentiation period, corresponding to the time when precursor cells cease mitosis and begin to fuse (41, 42). Fig. 2A shows representative Western blots of this process in RAW264.7 cells, whereas the graph in Fig. 2B shows quantitation of three such experiments each for RAW264.7 cells and mouse marrow precursors. In both cases, MyoIIA levels decreased by about half by day 4 of culture and returned to baseline levels by day 7. In contrast, levels of β-actin and the housekeeping protein GAPDH remained constant (Fig. 2A). Additionally, expression levels of the MyoIIB isoform were unchanged over the same period (data not shown). To determine the mechanism behind this transient decrease, MyoIIA heavy chain mRNA levels were assayed by competitive RT-PCR. As shown in Fig. 2C, mRNA remained steady during the differentiation period, indicating that the temporary loss of MyoIIA resulted from a translational or post-translational mechanism. To further define this mechanism, pulse-chase analysis was performed on differentiating RAW 264.7 cells at days 0, 3, and 7 of culture to determine MyoIIA heavy chain half-life. Fig. 2D shows that although day 0 and day 7 cells exhibited only a 10–20% loss of MyoIIA after 9 h of chase, the day 3 cells lost ∼70% of their labeled MyoIIA over the same time period. These results demonstrate that the temporary decrease in MyoIIA expression during mid-osteoclastogenesis is due to increased degradation of the protein.
FIGURE 2.
Myosin IIA is transiently down-regulated during osteoclastogenesis. A, osteoclasts were cultured to maturity over a 7-day period. Cell lysates probed on immunoblots for MyoIIA, GAPDH, and β-actin show a transient decrease in MyoIIA during mid-osteoclastogenesis. B, multiple experiments like that in A, for both RAW264.7- and marrow-derived cells, were quantified. Results shown are the mean of three experiments ± S.D. C, MyoIIA mRNA levels during osteoclastogenesis were examined by competitive RT-PCR. The upper band represents the signal from MyoIIA mRNA, whereas the lower band represents an internal standard. D, pulse-chase analysis was performed to determine the half-life of MyoIIA protein on day 0, 3, or 7 of RAW264.7 culture. Each culture demonstrated similar efficiencies of incorporation of 35S-labeled amino acids, and immunoprecipitation was performed from equivalent cpm. Results shown are the average of three experiments ± S.D.
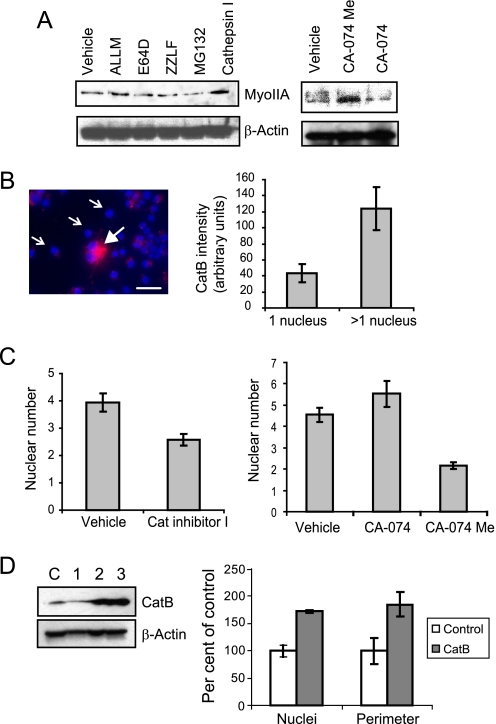
Cathepsin B Regulates Myosin IIA Levels and Osteoclast Fusion—Differentiating osteoclasts were tested with a panel of specific protease inhibitors to identify the class of enzyme responsible for the transient decrease in MyoIIA expression. RAW264.7 cells on day 3 of culture were treated with a panel of these compounds for 6 h. The proteasomal inhibitors MG132 and ZLLF did not suppress MyoIIA degradation; nor did the cysteine protease inhibitor E64 (Fig. 3A) or the specific calpain inhibitor calpeptin (data not shown). A single nonspecific inhibitor of cysteine proteases, ALLM, produced a slight suppression of degradation. However, because ALLM is established as having a degree of reactivity with cathepsins B and L, cathepsin inhibitor I (which inhibits B and, to a lesser extent, L) also was tested on differentiating osteoclasts. Cells treated with cathepsin inhibitor I expressed MyoIIA at levels greater than that of control cells or cells undergoing any other treatment. These data indicate a role for cathepsin B (catB) in promoting MyoIIA degradation. To explore this finding further, differentiating osteoclasts were treated with either CA-074 or CA-074Me, specific inhibitors of cathepsin B. Although CA-074 is mostly impermeant to cell membranes, CA-074Me is modified with a cleavable methyl ester that allows cell entry. As shown in Fig. 3A, treatment with CA-074Me, but not CA-074, strongly increased MyoIIA levels, demonstrating further that MyoIIA degradation is promoted by intracellular cathepsin B.
FIGURE 3.
Cathepsin B levels during osteoclastogenesis correlate with the extent of cell fusion. A, a panel of protease inhibitors was added to RAW264.7 osteoclasts on day 3 of culture. Only cell-permeant cathepsin B inhibitors (cathepsin inhibitor I and CA-074Me) were able to prevent degradation of MyoIIA. B, differentiating osteoclast cultures were labeled for active cathepsin B on day 3. A multinucleated cell (closed arrow) demonstrates higher levels of labeling than mononuclear cells (open arrows). Scale bar, 50 μm. At right, the labeling intensities of day three cells were quantified and expressed as mean ± S.D.; p < 0.00001. C, the addition of cell permeant cathepsin B inhibitors to cultures on days 3–4 results in suppression of osteoclast multinucleation. Results shown are mean ± S.D. For cathepsin inhibitor I versus vehicle, p < 0.01; for CA-074Me versus vehicle, p < 0.0001; for CA-074 versus vehicle, p > 0.1. D (left), Western analysis shows catB expression in clonal RAW264.7 macrophages stably transfected with either empty vector (lane C) or catB-expressing plasmid (lanes 1–3). Only clones 2 and 3 demonstrated overexpression of catB and were chosen for analysis. Right, average relative nuclear number and cell perimeter are shown for mature osteoclasts derived from clone 2 or the empty vector control. Clone 3 showed similar results. Data are expressed as mean ± S.D.; for both nuclei and perimeter, p < 0.05.
Previous studies (43) have demonstrated increasing levels of both catB protein and activity in lysates from differentiating human osteoclast precursors. We confirmed and extended these studies by taking advantage of a commercially available cell-permeant cathepsin B substrate that fluoresces upon cleavage, allowing visual identification of active catB within live cells. Examination of osteoclasts at varying stages of maturation demonstrated a distinct rise in active catB levels when osteoclast precursors shifted from a mononucleated to multinucleated stage. Fig. 3B, left, shows an example of day 3 osteoclast cultures containing a mixture of mononuclear (open arrows) and multinuclear (closed arrowhead) cells. Multinucleated cells demonstrated intense active catB labeling. Quantification of staining intensity in day 3 cultures revealed a clear rise in active enzyme levels as preosteoclasts underwent multinucleation (Fig. 3B, right). This rise in activity temporally correlates with the loss of MyoIIA expression demonstrated in Fig. 2A.
To determine whether inhibition of cathepsin B activity had any effect on osteoclast formation, cathepsin B inhibitors were added to day 3 preosteoclasts, and the resulting cells were examined the next day. Fig. 3C, left, illustrates that the addition of cathepsin inhibitor I produced a significant decrease in multinucleation, relative to vehicle-only controls. Similar results were obtained when CA-074Me, but not CA-074, was added to cultures (Fig. 3C, right). These results demonstrate that inhibitors of intracellular cathepsin B suppress osteoclast fusion. Further, to determine the effects of catB gain of function, a murine cDNA clone was isolated by RT-PCR and overexpressed in RAW264.7 macrophages (Fig. 3D, left). Upon differentiation into mature osteoclasts, catB-overexpressing cells demonstrated an 84% increase in nuclear number, with a similar accompanying increase in cell perimeter (Fig. 3D, right). These results suggest that cathepsin B activity has an effect on osteoclast fusion by promoting proteolysis of MyoIIA.
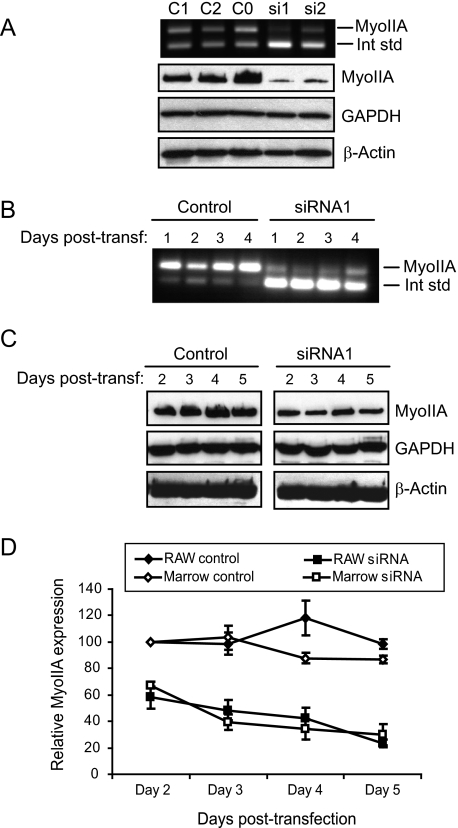
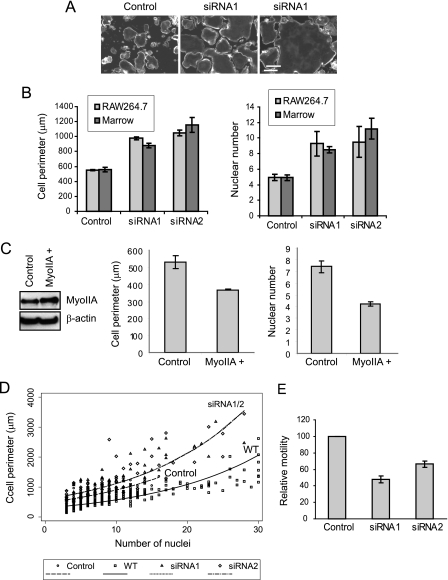
RNA Interference-mediated Suppression of Myosin IIA—The results above suggest that modulation of MyoIIA levels plays a key role in osteoclastogenesis. To test the role of MyoIIA more directly, RNA interference was used to knock down MyoIIA expression in the latter half (day 4 onward) of the osteoclast differentiation process, thus preventing recovery of its expression levels after the normal transient decrease. Two siRNAs specific to the MyoIIA heavy chain were designed and tested for efficacy in both RAW264.7 and mouse marrow cells, as were three control double-stranded oligonucleotides. Cells were transfected on day 4 of culture and assayed for MyoIIA mRNA and protein levels 2 and 3 days later, respectively. Fig. 4A (top) demonstrates that both targeting siRNAs diminished levels of MyoIIA mRNA, whereas the three control double-stranded RNA oligonucleotides (C0–C2) had no effect. The lower panels of Fig. 4A confirm a significant loss of MyoIIA protein after siRNA treatment, whereas cellular GAPDH and β-actin levels remained constant. The remaining panels of Fig. 4 illustrate the time course of diminished MyoIIA expression achieved with siRNA1. Notable decreases in MyoIIA mRNA were evident as early as 1 day post-transfection (Fig. 4B), with suppressed protein levels evident 1 day later (Fig. 4C). GAPDH and β-actin levels were unaffected over the length of the time course. Quantitation of multiple experiments with siRNA1 showed MyoIIA protein to drop to ∼60% of control levels by 2 days post-transfection in both RAW264.7 and marrow cells and to diminish further to about 30% of controls by 5 days post-transfection (Fig. 4D). In a single time course experiment, similar results were obtained with siRNA2 in RAW264.7 cells (data not shown). These results demonstrate that MyoIIA levels can be efficiently and specifically suppressed in the latter half of the osteoclast differentiation process.
FIGURE 4.
Myosin IIA levels are efficiently suppressed via RNA interference. A (top), competitive RT-PCR shows specific knockdown of MyoIIA mRNA with two siRNAs but not with three control oligonucleotides (C0–C2) 2 days post-transfection. Bottom, Western blot analysis for MyoIIA, GAPDH, and β-actin at 3 days post-transfection. B, a time course shows the effects of siRNA1 on MyoIIA mRNA for 4 days post-transfection. C, transfection of siRNA1 produces a sustained suppression of MyoIIA protein through 5 days post-transfection. D, multiple experiments like those in C were quantified and graphed. Results shown are the means of at least three experiments ± S.D.
Alteration of Myosin IIA Levels Modulates Osteoclast Fusion, Spreading, and Motility—An immediately obvious consequence of MyoIIA suppression was the generation of osteoclasts with larger surface areas than normal osteoclasts or control-transfected cells. Generation of podosomal actin cores appeared unaffected. Fig. 5A shows photomicrographs of phalloidin-labeled control- and siRNA-treated cells, demonstrating the large size of cells subjected to RNA interference. The left and center panels illustrate typical fields of control- and siRNA-treated cells, whereas the right panel shows an example of the enormous size to which the siRNA-treated cells could develop (all photographs are at the same scale). These differences were quantified in Fig. 5B (left), which shows that for both siRNA1- and siRNA2-treated cells, average cell perimeter roughly doubled when compared with controls. To determine whether the large siRNA-treated cells were the product of increased preosteoclast fusion, control- or siRNA-treated cells (at 4 days following transfection) were labeled with bisbenzimide, and their nuclei were enumerated. Fig. 5B, right, illustrates that nuclear number approximately doubled in either RAW264.7 or marrow cultures when cells were treated either with siRNA1 or siRNA2. To determine whether siRNA-treated cells might undergo greater fusion than controls due to enhanced cell survival, nuclear number and apoptosis (via a terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling assay) were measured in control and siRNA-treated samples. No significant differences were seen (data not shown).
FIGURE 5.
Alteration of myosin IIA levels during osteoclastogenesis alters cell fusion and spreading. A, control- or siRNA1-transfected cultures were labeled with fluorescent phalloidin to show the peripheral podosome belt. Scale bar, 200 μm. B, RAW264.7- or marrow-derived osteoclasts were treated with control oligonucleotides, siRNA1, or siRNA2. Left, perimeters of siRNA-treated marrow cells were statistically different from controls (p < 0.001), as were the perimeters of siRNA-treated RAW264.7 cells (p < 0.00001). Right, nuclei were enumerated in RAW264.7- or marrow-derived osteoclasts treated with control or siRNA oligonucleotides. siRNA-treated RAW264.7 cells demonstrated greater nuclear number than controls (p < 0.05 for both siRNA1 and -2), as did marrow cells (p < 0.01 for both siRNA1 and -2). C, myosin IIA-GFP was transiently expressed in differentiating RAW264.7 osteoclasts, as demonstrated by Western analysis (left). The shift in molecular weight caused by the GFP fusion was not clearly visible due to the high molecular mass of MyoIIA. The pEGFP-C3 vector was transfected as a control, and with both constructs, transfection efficiency was >90%. In MyoIIA-overexpressing cells, both cell perimeter and nuclear number were decreased (graphs); p < 0.005. D, nuclear number versus cell perimeter was plotted for wild type (WT) RAW264.7 osteoclasts as well as for control- or siRNA-transfected cells. For the perimeter outcome, the slopes of the log-transformed data were not significantly different (p = 0.253), and thus a common slope was used in the final model. Pairwise comparisons between each siRNA and control cells showed significant differences at the intercept and at 10 nuclei (all p values < 0.001). E, siRNA-treated RAW264.7 osteoclasts showed diminished motility in migration assays relative to controls; p < 0.05.
In a separate experiment, RAW264.7 macrophages were transfected with control or siRNA1 nucleotides and cultured in the absence of RANKL to determine whether knockdown of MyoIIA stimulated fusion in the absence of preosteoclast differentiation. After 7 days, control-treated macrophages possessed an average of 1.01 ± 0.01 nuclei, whereas siRNA-treated cells possessed 3.78 ± 0.71 nuclei (n > 100 for each treatment). These results clearly indicate that suppression of MyoIIA levels alone is sufficient to induce macrophage multinucleation without a requirement for RANKL-mediated differentiation. The resulting multinucleated cells possessed none of the hallmark features of osteoclasts, since they were negative for tartrate-resistant acid phosphatase and did not demonstrate increased expression of osteoclast differentiation markers, such as the vacuolar proton-translocating ATPase a3 subunit (44, 45) or β3 integrin (46) (data not shown). Thus, MyoIIA-regulated fusion occurs independently of RANKL-induced differentiation. However, RANKL treatment appears to enhance fusion, since MyoIIA knockdown in the presence of RANKL produces greater multinucleation than MyoIIA knockdown alone. This may be due in part to RANKL-mediated up-regulation of cathepsin B expression via NF-κB-responsive elements in its promoter (47).
As an additional indicator of the direct role of MyoIIA in regulating osteoclast fusion, a MyoIIA-GFP fusion protein was transiently expressed in differentiating RAW264.7 osteoclasts, resulting in a ∼2-fold overexpression of MyoIIA, and cell perimeter and nuclear number were assessed. Cells were transfected on day 3 following RANKL treatment, when endogenous MyoIIA levels drop, and assayed on day 5. Fig. 5C demonstrates that increased MyoIIA levels resulted in decreased cell perimeter and nuclear number, consistent with a role in its inhibition of osteoclast fusion.
Although these data demonstrate a role for MyoIIA in mediating osteoclast fusion, numerous studies in other cell types have suggested a role for this protein in cell spreading and motility (22, 48–50). Therefore, we sought to determine whether the larger surface area of siRNA-treated cells was a result solely of increased preosteoclast fusion or whether cell spreading also was affected. We first investigated whether we could quantify a relationship between nuclear number and cell perimeter in normal osteoclasts generated from RAW264.7 cells. A survey of ∼300 cells at different stages of maturation indicated that cell perimeter increases proportionately to nuclear number (Fig. 5D, wild type (WT) cells). In further experiments, control- and siRNA-treated cells also were examined for the relationship between nuclear number and cell perimeter. These cells were transfected on day 4 of differentiation and assayed 4 days later, using the same protocol as that described in the legend to Fig. 4. As shown in Fig. 5D, control-treated cells, which possessed up to 16 nuclei, had a slightly greater perimeter than wild type cells of the same nuclear number. This increased spreading may be attributable to the lipid-based transfection methods used in this study. In contrast, siRNA-treated cells, which possessed up to 28 nuclei, demonstrated perimeters that were similar to control cells at low nuclear number but increased rapidly as nuclear number increased. These results, which were very similar for both siRNA1 and siRNA2, show that loss of MyoIIA increases not only cell fusion but also cell spreading in highly multinucleated cells, resulting in formation of extremely large osteoclasts, such as that shown in Fig. 5A. Nearly identical results were obtained from marrow-derived osteoclasts (data not shown).
Because myosin IIA plays a role in cell adhesion, we assessed the effects of its suppression on cell motility. Most motility assays are dependent on cell size, relying either on the ability of cells to migrate through filters of a particular pore size or on their ability to migrate and generate a path over a given surface area. Because normal MyoIIA-suppressed cells are much larger than their control counterparts, we needed to produce culture conditions in which the siRNA-treated cells produced were of similar size as the controls. To achieve this, differentiating RAW264.7 osteoclast cultures were washed extensively just prior to transfection with either anti-MyoIIA or control siRNAs. This treatment resulted in removal of mononuclear cells and inhibited further cell fusion. The resulting cultures produced cells very similar in nuclear number and perimeter (control, 4.4 ± 2.8 nuclei, 501.4 ± 206.0 μm, n = 47; siRNA1, 4.8 ± 2.5 nuclei, 508.1 ± 216 μm, n = 53). When tested for osteopontin-directed migration (Fig. 5E), the siRNA-treated cells showed significantly diminished motility (inhibition of ∼35–50%), indicating a role for MyoIIA in osteoclast migration.
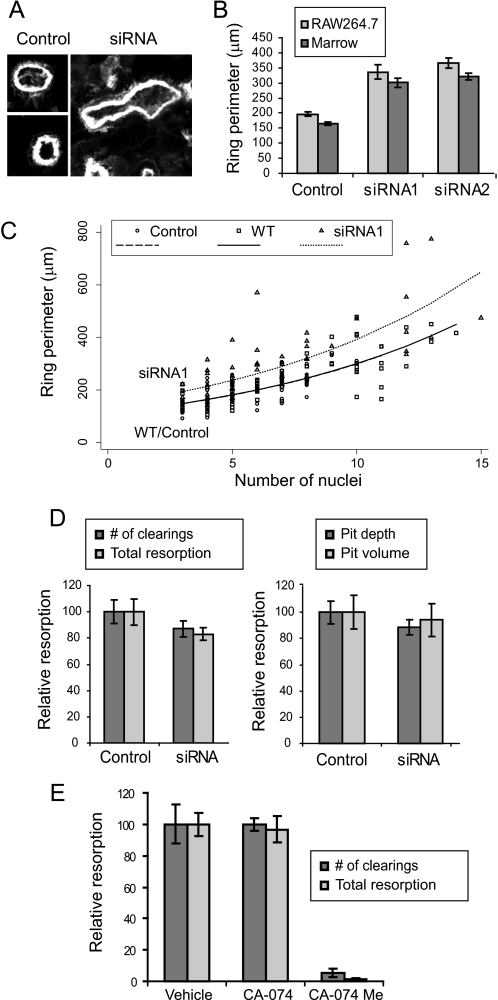
Effects of MyoIIA and Cathepsin B on Sealing Zone Formation and Bone Resorption—Because of the presence of MyoIIA in the sealing zone of polarized osteoclasts, we determined whether actin ring formation was affected by suppression of its expression. When plated on bone, siRNA-treated cells demonstrated obviously larger actin rings than control cells (Fig. 6A). The increased size of actin rings is demonstrated graphically in Fig. 6B. To determine whether the enlarged actin rings were a consequence of the enhanced overall size of the cells or a direct effect of MyoIIA suppression on ring formation, we first examined the relationship between cell size and actin ring circumference in normal polarized RAW264.7-derived osteoclasts on bone. As demonstrated in Fig. 6C (wild type (WT) cells), it was determined that there exists a relationship between nuclear number and ring circumference (i.e. larger cells tend to generate larger rings). When control- and siRNA-transfected cells were assayed for nuclear number and actin ring size, the control cells behaved similarly to wild type. In contrast, siRNA-treated cells generated actin rings ∼25% larger than expected, indicating a modest but significant effect of myosin IIA suppression on actin ring size. Both siRNAs produced similar results; however, only siRNA1 is shown for figure clarity. Additionally, siRNA1 was tested on marrow-derived osteoclasts. This treatment produced results similar to those in RAW264.7 cells; the actin rings generated by the siRNA were ∼38% greater in perimeter than control transfectants (not shown).
FIGURE 6.
Effects of myosin IIA and cathepsin B on sealing zone formation and resorption. A, examples of sealing zones from control- or siRNA-treated cells on ivory are shown. Cells were labeled with fluorescent phalloidin and photographed at the same scale. B, RAW264.7- or marrow-derived osteoclasts were treated with control oligonucleotides, siRNA1, or siRNA2. The sealing zone perimeters of siRNA-treated marrow cells were statistically different from controls, as were the perimeters of siRNA-treated RAW264.7 cells (all p values < 0.000001). C, nuclear number versus actin ring perimeter was plotted for wild type (WT) RAW264.7 osteoclasts as well as for control- or siRNA-transfected cells. As in the cell perimeter data above, the slopes of the log-transformed data were not significantly different (p = 0.925), leading to the use of a common slope model. Comparison of wild type and control cells showed no significant difference, whereas the actin rings of siRNA1 cells were significantly different from controls at 10 nuclei and at the intercept (p < 0.001). D, control- or siRNA1-treated osteoclasts were assayed for resorptive capacity on synthetic bone substrate (left) or ivory slices (right); no significant differences were noted between controls and siRNA treatment. E, treatment of osteoclasts (days 3–6 of culture) with CA-074Me, but not CA-074, dramatically reduced bone resorptive capacity. For both number of clearings and total resorption, p < 0.00005.
Bone resorption in MyoIIA-suppressed cells was tested in two types of assays. The resorptive capacity of osteoclasts was assayed on synthetic bone substrate (Fig. 6D, left) or on ivory slices (Fig. 6D, right). In neither case was resorption affected by the siRNA1-mediated loss of MyoIIA expression, despite the diminished motility of these cells. We expect that maintenance of resorptive capacity may be a function of the siRNA-treated cells possessing more or larger sealing zones, which could counteract the effects of lowered motility. Although suppressing MyoIIA levels in mature osteoclasts does not affect resorptive capacity, it was previously discovered that treatment of mature rat osteoclasts with CA-074Me, but not CA-074, reduced the ability of these cells to resorb bone. Further, CA-074Me, but not CA-074, reduced bone resorption in vivo when administered subcutaneously (51). To test the effects of cathepsin B inhibition on bone resorption in our system, RAW264.7-derived osteoclasts were treated with 50 μm CA-074 or CA-074Me on days 3–6 following RANKL treatment. As shown in Fig. 6E, CA-074Me but not CA-074 produced a dramatic inhibition of resorptive capacity, consistent with previous reports using other osteoclast models. Thus, our results indicate that cathepsin B plays a critical role in osteoclast formation and function by regulating myosin IIA levels and osteoclast fusion.
DISCUSSION
The roles of myosin IIA in cell function have been studied extensively in model systems from lower eukaryotes to mammalian cells. However, this study is the first detailed examination of myosin IIA in osteoclasts. Although modulation of osteoclast spreading and motility was an expected effect of MyoIIA suppression, we also found, perhaps more importantly, that this myosin plays a crucial role in mediating cell fusion.
Alteration of the actin cytoskeleton is a critical process in cell-cell fusion. Cytoskeletal rearrangements have been implicated in modulating fusion-related events ranging from migration to cell adhesion to the fusion process itself (reviewed in Ref. 52). The mechanism by which myosin IIA suppresses cell fusion in osteoclasts is unclear. One possibility is that MyoIIA, which is distributed fairly homogeneously in monocytic/macrophage osteoclast precursors, may provide intracellular tension or even a physical barrier that prevents fusion from occurring. Alternately, MyoIIA may play a role in limiting cell-cell contact in these precursors through interaction with cell surface proteins. For example, it is possible that myosin IIA may play a role in fusion by interacting indirectly with membrane proteins such as d2, dendritic cell-specific transmembrane protein, CD44, and ADAM8, thus regulating their functions.
Our data clearly demonstrate that regulated proteolysis of myosin IIA during osteoclastogenesis stimulates precursor fusion. Although regulated proteolysis of myosin IIA might appear to be an unusual mechanism for changing its expression, previous studies in smooth muscle cells showed that nonmuscle myosin levels during cell division are regulated by changes in protein half-life (53). Our data additionally show that cathepsin B appears to promote the proteolysis seen during osteoclastogenesis. Previous studies have indicated negative effects of intracellular cathepsin B inhibitors on osteoclast activity and, more recently, formation, although the mechanisms by which cathepsin B promotes bone resorption have remained a mystery (51, 54). In this study, we have identified a role for cathepsin B in osteoclast function through its modulation of myosin IIA levels, and subsequently, osteoclast formation.
At this point, it is unclear how cathepsin B might promote myosin IIA degradation. It does not appear that cathepsin B activates other general proteases, which would then cleave myosin IIA, based on our studies of protease inhibitor panels. It is possible that cathepsin B directly degrades myosin IIA, although the mechanism by which these proteins might associate is unknown. However, although cathepsin B is known primarily as a lysosomal enzyme, it retains endopeptidase activity at neutral pH and can function in extracellular compartments and the cytosol. Nonlysosomal cathepsin B has been implicated in both pathological and physiological processes. In cells of the monocyte/macrophage lineage, cytosolic cathepsin B was suggested to regulate secretion of tumor necrosis factor-α induced by inflammatory stimuli (55). Further, multiple reports have implicated nonlysosomal cathepsin B in promoting myoblast fusion. First, cathepsin B activity increases during fusion of postmitotic myoblasts into myotubes (56, 57). Diminution of cathepsin B levels by either retroviral gene trapping or cell-permeant inhibitors negatively affects myoblast fusion (56, 58). Most recently, cathepsin B has been shown to localize to the cytoplasmic face of the plasma membrane of differentiating myoblasts at caveolae, suggesting a role in mediating cytoskeletal rearrangements required for cell fusion (59). Moreover, a panel of protease inhibitors very similar to ours was used to examine the enzymatic basis for proteolytic breakdown of actin-rich dendritic spines in N-methyl-d-aspartate-stimulated neurons. Only inhibitors of cathepsin B were capable of preventing this degradation, and the cytosolic protein myristoylated alanine-rich C kinase substrate was shown to be a target of the enzyme (60). Thus, cathepsin B-mediated degradation of cytoskeletal proteins may be a universal mechanism by which extensive changes in cell shape are mediated. More studies are required to determine how cathepsin B functionally interacts with the cytoskeleton.
In addition to our findings showing a role for MyoIIA in osteoclast precursor fusion, we also showed it to regulate cell spreading and formation of the sealing zone. Knockdown of MyoIIA increased both cell perimeter and sealing zone perimeter. Further, in migration assays, MyoIIA-suppressed cells demonstrated somewhat lower osteopontin-directed motility than control cells. These results are consistent with studies in foreskin fibroblasts showing that MyoIIA-suppressed human foreskin fibroblasts demonstrated decreased directional migration (61). Although overall resorptive capacity was not markedly affected by knockdown of MyoIIA, inhibition of intracellular cathepsin B, which results in heightened levels of MyoIIA, dramatically diminished resorption. We attribute this finding at least in part to high levels of MyoIIA preventing osteoclast formation, although it is possible that other cathepsin B-dependent mechanisms may be involved.
In summary, this study revealed a not unexpected role for myosin IIA in mediating osteoclast spreading, motility, and sealing zone formation but also indicated a surprising function in regulating cell fusion during late osteoclastogenesis. These data also indicate a role for cathepsin B as the upstream regulator of MyoIIA-mediated multinucleation in osteoclasts. These results add to our knowledge of a process that is poorly understood in osteoclasts and indeed in other cell types, such as myoblasts, that undergo regulated fusion. Because differentiation of preosteoclasts without subsequent fusion results in poor resorptive capacity and potentially, effects on osteoblast function (11), defining the steps involved in this process is important to our discernment of the cellular events that regulate skeletal health. Future studies to elucidate how cytoskeletal rearrangements may affect membrane structure, including membrane protein distribution, should provide useful insights into the process of osteoclast fusion.
Acknowledgments
We thank the Campus Microscopy and Imaging Facility (The Ohio State University) for continued technical advice and support. We also thank Greg Young at the Center for Biostatistics (The Ohio State University Medical Center) for statistical analyses.
This work was supported, in whole or in part, by National Institutes of Health Grants AR051515 and DK052131 (to B. S. L.) and HL-45788 and P20-RR16440 (to R. B. W.).
Footnotes
The abbreviations used are: MyoIIA, myosin nonmuscle IIA heavy chain; RANKL, receptor activator of NF-κB ligand; RT-PCR, reverse transcription followed by PCR; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; catB, cathepsin B; siRNA, small interfering RNA.
References
- 1.Teitelbaum, S. L. (2000) Science 289 1504-1508 [DOI] [PubMed] [Google Scholar]
- 2.Cui, W., Ke, J. Z., Zhang, Q., Ke, H. Z., Chalouni, C., and Vignery, A. (2006) Blood 107 796-805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sterling, H., Saginario, C., and Vignery, A. (1998) J. Cell Biol. 143 837-847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han, X., Sterling, H., Chen, Y., Saginario, C., Brown, E. J., Frazier, W. A., Lindberg, F. P., and Vignery, A. (2000) J. Biol. Chem. 275 37984-37992 [DOI] [PubMed] [Google Scholar]
- 5.Lundberg, P., Koskinen, C., Baldock, P. A., Lothgren, H., Stenberg, A., Lerner, U. H., and Oldenborg, P. A. (2007) Biochem. Biophys. Res. Commun. 352 444-448 [DOI] [PubMed] [Google Scholar]
- 6.Saginario, C., Sterling, H., Beckers, C., Kobayashi, R., Solimena, M., Ullu, E., and Vignery, A. (1998) Mol. Cell Biol. 18 6213-6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinberg, T. H., and Hiken, J. F. (2007) Purinergic Signal. 3 53-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, S. J., Han, J. H., and Roodman, G. D. (2001) J. Bone Miner. Res. 16 814-822 [DOI] [PubMed] [Google Scholar]
- 9.Kukita, T., Wada, N., Kukita, A., Kakimoto, T., Sandra, F., Toh, K., Nagata, K., Iijima, T., Horiuchi, M., Matsusaki, H., Hieshima, K., Yoshie, O., and Nomiyama, H. (2004) J. Exp. Med. 200 941-946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yagi, M., Miyamoto, T., Sawatani, Y., Iwamoto, K., Hosogane, N., Fujita, N., Morita, K., Ninomiya, K., Suzuki, T., Miyamoto, K., Oike, Y., Takeya, M., Toyama, Y., and Suda, T. (2005) J. Exp. Med. 202 345-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, S. H., Rho, J., Jeong, D., Sul, J. Y., Kim, T., Kim, N., Kang, J. S., Miyamoto, T., Suda, T., Lee, S. K., Pignolo, R. J., Koczon-Jaremko, B., Lorenzo, J., and Choi, Y. (2006) Nat. Med. 12 1403-1409 [DOI] [PubMed] [Google Scholar]
- 12.Golomb, E., Ma, X., Jana, S. S., Preston, Y. A., Kawamoto, S., Shoham, N. G., Goldin, E., Conti, M. A., Sellers, J. R., and Adelstein, R. S. (2004) J. Biol. Chem. 279 2800-2808 [DOI] [PubMed] [Google Scholar]
- 13.Saez, C. G., Myers, J. C., Shows, T. B., and Leinwand, L. A. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 1164-1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simons, M., Wang, M., McBride, O. W., Kawamoto, S., Yamakawa, K., Gdula, D., Adelstein, R. S., and Weir, L. (1991) Circ. Res. 69 530-539 [DOI] [PubMed] [Google Scholar]
- 15.Kolega, J. (1998) J. Cell Sci. 111 2085-2095 [DOI] [PubMed] [Google Scholar]
- 16.Maupin, P., Phillips, C. L., Adelstein, R. S., and Pollard, T. D. (1994) J. Cell Sci. 107 3077-3090 [DOI] [PubMed] [Google Scholar]
- 17.Conrad, A. H., Jaffredo, T., and Conrad, G. W. (1995) Cell Motil. Cytoskeleton 31 93-112 [DOI] [PubMed] [Google Scholar]
- 18.Rochlin, M. W., Itoh, K., Adelstein, R. S., and Bridgman, P. C. (1995) J. Cell Sci. 108 3661-3670 [DOI] [PubMed] [Google Scholar]
- 19.Kovacs, M., Wang, F., Hu, A., Zhang, Y., and Sellers, J. R. (2003) J. Biol. Chem. 278 38132-38140 [DOI] [PubMed] [Google Scholar]
- 20.Rosenfeld, S. S., Xing, J., Chen, L. Q., and Sweeney, H. L. (2003) J. Biol. Chem. 278 27449-27455 [DOI] [PubMed] [Google Scholar]
- 21.Wang, F., Kovacs, M., Hu, A., Limouze, J., Harvey, E. V., and Sellers, J. R. (2003) J. Biol. Chem. 278 27439-27448 [DOI] [PubMed] [Google Scholar]
- 22.Cai, Y., Biais, N., Giannone, G., Tanase, M., Jiang, G., Hofman, J. M., Wiggins, C. H., Silberzan, P., Buguin, A., Ladoux, B., and Sheetz, M. P. (2006) Biophys. J. 91 3907-3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandquist, J. C., Swenson, K. I., Demali, K. A., Burridge, K., and Means, A. R. (2006) J. Biol. Chem. 281 35873-35883 [DOI] [PubMed] [Google Scholar]
- 24.Swailes, N. T., Colegrave, M., Knight, P. J., and Peckham, M. (2006) J. Cell Sci. 119 3561-3570 [DOI] [PubMed] [Google Scholar]
- 25.Conti, M. A., Even-Ram, S., Liu, C., Yamada, K. M., and Adelstein, R. S. (2004) J. Biol. Chem. 279 41263-41266 [DOI] [PubMed] [Google Scholar]
- 26.Tullio, A. N., Accili, D., Ferrans, V. J., Yu, Z. X., Takeda, K., Grinberg, A., Westphal, H., Preston, Y. A., and Adelstein, R. S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 12407-12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akisaka, T., Yoshida, H., Inoue, S., and Shimizu, K. (2001) J. Bone Miner. Res. 16 1248-1255 [DOI] [PubMed] [Google Scholar]
- 28.Krits, I., Wysolmerski, R. B., Holliday, L. S., and Lee, B. S. (2002) Calcif. Tissue Int. 71 530-538 [DOI] [PubMed] [Google Scholar]
- 29.Turksen, K., Kanehisa, J., Opas, M., Heersche, J. N., and Aubin, J. E. (1988) J. Bone Miner. Res. 3 389-400 [DOI] [PubMed] [Google Scholar]
- 30.Linder, S., and Kopp, P. (2005) J. Cell Sci. 118 2079-2082 [DOI] [PubMed] [Google Scholar]
- 31.Jurdic, P., Saltel, F., Chabadel, A., and Destaing, O. (2006) Eur. J. Cell Biol. 85 195-202 [DOI] [PubMed] [Google Scholar]
- 32.Luxenburg, C., Geblinger, D., Klein, E., Anderson, K., Hanein, D., Geiger, B., and Addadi, L. (2007) PLoS ONE 2 e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMichael, B. K., Kotadiya, P., Singh, T., Holliday, L. S., and Lee, B. S. (2006) Bone 39 694-705 [DOI] [PubMed] [Google Scholar]
- 34.McMichael, B. K., and Lee, B. S. (2008) Exp. Cell Res. 314 564-573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Destaing, O., Saltel, F., Geminard, J. C., Jurdic, P., and Bard, F. (2003) Mol. Biol. Cell 14 407-416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotadiya, P., McMichael, B. K., and Lee, B. S. (2008) Bone 43 951-960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeyaraj, S., Dakhlallah, D., Hill, S. R., and Lee, B. S. (2005) J. Biol. Chem. 280 37957-37964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, B. S., Holliday, L. S., Krits, I., and Gluck, S. L. (1999) J. Bone Miner. Res. 14 2127-2136 [DOI] [PubMed] [Google Scholar]
- 39.Lee, B. S., Gluck, S. L., and Holliday, L. S. (1999) J. Biol. Chem. 274 29164-29171 [DOI] [PubMed] [Google Scholar]
- 40.Wei, Q., and Adelstein, R. S. (2000) Mol. Biol. Cell 11 3617-3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi, N., Udagawa, N., Tanaka, S., Murakami, H., Owan, I., Tamura, T., and Suda, T. (1994) Dev. Biol. 163 212-221 [DOI] [PubMed] [Google Scholar]
- 42.Tanaka, S., Takahashi, N., Udagawa, N., Tamura, T., Akatsu, T., Stanley, E. R., Kurokawa, T., and Suda, T. (1993) J. Clin. Invest. 91 257-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blair, H. C., Sidonio, R. F., Friedberg, R. C., Khan, N. N., and Dong, S. S. (2000) J. Cell Biochem. 78 627-637 [DOI] [PubMed] [Google Scholar]
- 44.Toyomura, T., Oka, T., Yamaguchi, C., Wada, Y., and Futai, M. (2000) J. Biol. Chem. 275 8760-8765 [DOI] [PubMed] [Google Scholar]
- 45.Kornak, U., Schulz, A., Friedrich, W., Uhlhaas, S., Kremens, B., Voit, T., Hasan, C., Bode, U., Jentsch, T. J., and Kubisch, C. (2000) Hum. Mol. Gen. 9 2059-2063 [DOI] [PubMed] [Google Scholar]
- 46.McHugh, K. P., Kitazawa, S., Teitelbaum, S. L., and Ross, F. P. (2001) J. Cell Biochem. 81 320-332 [DOI] [PubMed] [Google Scholar]
- 47.Bien, S., Ritter, C. A., Gratz, M., Sperker, B., Sonnemann, J., Beck, J. F., and Kroemer, H. K. (2004) Mol. Pharmacol. 65 1092-1102 [DOI] [PubMed] [Google Scholar]
- 48.Takizawa, N., Ikebe, R., Ikebe, M., and Luna, E. J. (2007) J. Cell Sci. 120 3792-3803 [DOI] [PubMed] [Google Scholar]
- 49.Betapudi, V., Licate, L. S., and Egelhoff, T. T. (2006) Cancer Res. 66 4725-4733 [DOI] [PubMed] [Google Scholar]
- 50.Wylie, S. R., and Chantler, P. D. (2003) Mol. Biol. Cell 14 4654-4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hill, P. A., Buttle, D. J., Jones, S. J., Boyde, A., Murata, M., Reynolds, J. J., and Meikle, M. C. (1994) J. Cell Biochem. 56 118-130 [DOI] [PubMed] [Google Scholar]
- 52.Chen, E. H., Grote, E., Mohler, W., and Vignery, A. (2007) FEBS Lett. 581 2181-2193 [DOI] [PubMed] [Google Scholar]
- 53.Grainger, D. J., Hesketh, T. R., Metcalfe, J. C., and Weissberg, P. L. (1991) Biochem. J. 277 145-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim, M. H., Kim, B. T., Min, Y. K., and Kim, S. H. (2008) Amino Acids 34 497-506 [DOI] [PubMed] [Google Scholar]
- 55.Ha, S. D., Martins, A., Khazaie, K., Han, J., Chan, B. M., and Kim, S. O. (2008) J. Immunol. 181 690-697 [DOI] [PubMed] [Google Scholar]
- 56.Jane, D. T., DaSilva, L., Koblinski, J., Horwitz, M., Sloane, B. F., and Dufresne, M. J. (2002) J. Cell Biochem. 84 520-531 [PubMed] [Google Scholar]
- 57.Jane, D. T., and Dufresne, M. J. (1994) Biochem. Cell Biol. 72 267-274 [DOI] [PubMed] [Google Scholar]
- 58.Jane, D. T., Morvay, L. C., Allen, F., Sloane, B. F., and Dufresne, M. J. (2002) Biochem. Cell Biol. 80 457-465 [DOI] [PubMed] [Google Scholar]
- 59.Jane, D. T., Morvay, L., Dasilva, L., Cavallo-Medved, D., Sloane, B. F., and Dufresne, M. J. (2006) Biol. Chem. 387 223-234 [DOI] [PubMed] [Google Scholar]
- 60.Graber, S., Maiti, S., and Halpain, S. (2004) Neuropharmacology 47 706-713 [DOI] [PubMed] [Google Scholar]
- 61.Even-Ram, S., Doyle, A. D., Conti, M. A., Matsumoto, K., Adelstein, R. S., and Yamada, K. M. (2007) Nat. Cell Biol. 9 299-309 [DOI] [PubMed] [Google Scholar]