Abstract
The phytopathogenic fungus Fusarium graminearum secretes a very diverse pool of glycoside hydrolases (GHs) aimed at degrading plant cell walls. α-l-Arabinanases are essential GHs participating in the complete hydrolysis of hemicellulose, a natural resource for various industrial processes, such as bioethanol or pharmaceuticals production. Arb93A, the exo-1,5-α-l-arabinanase of F. graminearum encoded by the gene fg03054.1, belongs to the GH93 family, for which no structural data exists. The enzyme is highly active (1065 units/mg) and displays a strict substrate specificity for linear α-1,5-l-arabinan. Biochemical assays and NMR experiments demonstrated that the enzyme releases α-1,5-l-arabinobiose from the nonreducing end of the polysaccharide. We determined the crystal structure of the native enzyme and its complex with α-1,5-l-arabinobiose, a degradation product of α-Me-1,5-l-arabinotetraose, at 1.85 and 2.05Å resolution, respectively. Arb93A is a monomeric enzyme, which presents the six-bladed β-propeller fold characteristic of sialidases of clan GHE. The configuration of the bound arabinobiose is consistent with the retaining mechanism proposed for the GH93 family. Catalytic residues were proposed from the structural analysis, and site-directed mutagenesis was used to validate their role. They are significantly different from those observed for GHE sialidases.
The plant cell wall consists mainly of a complex aggregation of polysaccharides, such as cellulose, hemicellulose, and pectin. Hemicellulose is one of the most abundant renewable biopolymers on earth and constitutes an important source of energy for the biofuel industry. It represents 20–40% of plant biomass and is principally composed of pentoses, such as xylose and arabinose (1). Due to the high complexity and structural variability of this polysaccharide, many enzymes are necessary for its complete degradation (2). A number of microorganisms are able to break down hemicellulose, through the action of various glycoside hydrolases (GHs).2 The latter catalyze the cleavage of glycosidic bonds between sugars with either inversion or retention of the anomeric configuration (3). GHs have been classified into more than 114 different families based on their amino acid sequence similarity (CAZY (Carbohydrate Active Enzymes) server, available on the World Wide Web) (4, 5).
α-l-Arabinanases (EC 3.2.1.-) are accessory hemicellulases that hydrolyze α-l-arabinofuranosic linkages and act synergistically with other GHs to break down hemicellulose fully (6). These enzymes have become of interest in recent years because of their potential rate-limiting role in the degradation of lignocelluloses and their practical application in various industrial processes, such as the production of important medicinal compounds, the improvement of wine flavors, pulp treatment, juice clarification, the production of bioethanol, and the synthesis of oligosaccharides (7). According to the CAZY classification, α-l-arabinanases are present in six GH families (3, 43, 51, 54, 62, and 93) whose members display highly varying specificities. GH51 and GH54 are the most extensively characterized and contain mainly α-l-arabinofuranosidases, removing both α-1,2 and α-1,3 arabinofuranosyl moieties from arabinan and xylans (8–11). GH3 arabinofuranosidases are all bifunctional proteins acting on both β-1,4-xylose-xylose and β-1,3-xylose-arabinose bonds (12). GH62 enzymes have a specific arabinofuranose-debranching activity on xylan (13), whereas GH43 and GH93 proteins are known to hydrolyze α-1,5-linked arabinofuranose-oligosaccharides (14, 15). Nine three-dimensional structures ofα-l-arabinanases belonging to the GH43, -51, and -54 families have been solved to date (8–11, 14, 16–19). Barley glucanases, which are members of GH3, have been structurally characterized (20), and those of GH62 are predicted to display the same five-bladed β-propeller fold (clan GHF) as the well documented GH43 family (14).
GH93 is described as a family of exo-1,5-α-l-arabinanases in the CAZY data base. This family contains only few sequences obtained from bacterial and fungal genome sequencing, and the only characterized enzyme is Abnx, from Penicillium chrysogenum. Abxn cleaves arabinobiose from the nonreducing end of linear arabinan (15, 21). The products of its transglycosylation activity revealed net retention of the configuration of the anomeric carbon (22). GH93 is the only family of α-l-arabinanases for which neither structural nor detailed mechanistic information has been reported to date.
Fusarium graminearum (teleomorph: Gibberella zeae) is a devastating pathogenic fungus that attacks various crop plants, such as maize, wheat, or barley. During the infection process, the fungus secretes a large and diverse panel of GHs that degrade the host cell wall. When F. graminearum grows in medium with plant cell wall material as the sole carbon source, as many as 11 CAZY families and 27 EC numbers are represented among the secreted enzymes (23). The diversity is particularly high for enzymes targeting hemicellulose, as recently shown by a transcriptional analysis revealing that 30 hemicellulase genes are transcribed in response to plant cell wall material (24). Two of these genes, fg03054.1 (in the F. graminearum genome data base (25); GeneID: 2785867) and fg03598.1 (GeneID: 2784780) are predicted to encode an exo-1,5-α-l-arabinanase of family GH93. Sequence alignment against protein structure sequences shows similarities with enzymes of clan GHE, which includes families GH33, -34, and -83. This clan is characterized by a six-bladed β-propeller fold and contains only sialidases/neuraminidases (EC 3.2.1.18) or trans-sialidases (EC 2.4.1-). Sialidases, or neuraminidases, catalyze the removal of terminal sialic acid residues from various glycoconjugates and are involved in pathogenesis of many human diseases, including influenza and cholera (26).
Here we report the production, biochemical characterization, and structural analysis of Arb93A, the exo-1,5-α-l-arabinanase from F. graminearum encoded by the gene fg03054.1. We describe the structure of the native enzyme and its complex with α-1,5-l-arabinobiose, which to our knowledge are the first crystal structures of a GH93 enzyme. Our data reveal that the enzyme binds linear α-1,5-l-arabinan and releases α-1,5-l-arabinobiose from the nonreducing end with net retention of the anomeric configuration. The catalytic residues identified through inspection of the active site were tested by site-directed mutagenesis. We propose a functional mechanism for α-1,5-l-arabinan hydrolysis and compare Arb93A structure with that of enzymes of the clan GHE.
EXPERIMENTAL PROCEDURES
Substrates, Strains, and Media—p-Nitrophenyl α-l-arabinofuranoside was purchased from Sigma. Wheat arabinoxylan, sugar beet arabinan, sugar beet debranched α-1,5-l-arabinan, and α-1,5-l-arabino-oligosaccharides with degrees of polymerization (DP) ranging from 1 to 6 were obtained from Megazyme.
F. graminearum (G. zeae) was originally isolated from diseased hops (Humulus lupulus) and identified by CABI Bioscience. F. graminearum was cultured at 23 °C on M3 medium (27) with hop cell wall as the sole carbon source at a concentration of 10 g/liter. The culture was performed as previously described (23).
Escherichia coli strains TOP10, XL1-Blue, and BL21 (DE3) (Invitrogen) were used for cloning procedures, site-directed mutagenesis, and protein expression, respectively. E. coli were grown in LB medium supplemented with 30 μg ml-1 kanamycin.
RNA Isolation, cDNA Synthesis, and Cloning of arb93A—Total RNA was extracted from F. graminearum with the RNaqueous-4PCR kit (Ambion) according to the manufacturer's instructions. cDNA was synthesized starting from the 3′-poly(A) of the mRNAs using the Smart cDNA synthesis kit (Clontech) and following the manufacturer's recommendations. The PCR products were directly used for amplification of arb93A with the primers 5′-ATCGGGTCTCCCATGAAGCCTGCCCAGAAGG-3′ and 5′-ATCGGGTCTCGAATTCTAAGACTTCATAACCTCAG-3′, incorporating BsaI restriction sites (underlined) for subsequent cloning. The amplification was carried out using iProof DNA polymerase (Bio-Rad). The resulting PCR product was purified on a Nucleospin device (Macherey-Nagel) and cloned into the pET30a+ plasmid (Novagen) previously digested with NcoI and EcoRI (Fermentas) using standard molecular biology procedures. The resulting pET30a-arb93A plasmid harbored the gene cloned in frame with the coding sequence of an N-terminal 6× histidine tag.
Site-directed Mutagenesis—Mutants of Arb93A were generated using a PCR-based QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions and using pET30a-arb93A as template. The following primer pairs were used to generate the mutated genes: 5′-CCTCACTCCTGTCTGGGCCCCTTTCCTGATGACCTAC-3′ and 5′-GTAGGTCATCAGGAAAGGGGCCCAGACAGGAGTGAGG-3′ for E170A; 5′-CAACGAGTACATCTACGTCTATGCATATGGTGGTGGCCCTAACCC-3′ and 5′-GGGTTAGGGCCACCACCATATGCATAGACGTAGATGTACTCGTTG-3′ for E242A; 5′-GACGTTCCTCAGCCAACTGCGGCCACCCGTTCTCTCCTCACTTTC-3′ and 5′-GAAAGTGAGGAGAGAACGGGTGGCCGCAGTTGGCTGAGGAACGTC-3′ for Y337A. The mutant DNA sequences were validated by sequencing.
Production of Recombinant Arb93A and Mutants—E. coli BL21 (DE3) cells transformed with pET30a+ containing the wild type (WT) or mutated arb93A gene were grown in 1 liter of LB-kanamycin, and protein expression was induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside at an A600 nm of 0.6–0.8. Protein was produced for 16–18 h at 16 °C. A selenomethionine-substituted protein was produced according to the method of Doublie (28).
Cells were harvested by centrifugation and sonicated in 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, and 25 mm imidazole. The lysate was subsequently centrifuged at 18,000 × g for 50 min and loaded onto a HisTrap HP 5-ml column (GE Healthcare) loaded with nickel sulfate and equilibrated in the aforementioned buffer. After washing with 100 ml of 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, and 50 mm imidazole, the protein was eluted with 15 ml of the same buffer supplemented with 500 mm imidazole. After a 3-fold dilution with 50 mm Tris-HCl, pH 8.0, the eluate was loaded onto a HiTrap Q-Sepharose Fast Flow 1-ml column (GE Healthcare) previously equilibrated with 50 mm Tris-HCl, pH 8.0. The flow-through containing the protein of interest was then directly loaded onto a HiTrap SP-Sepharose Fast Flow 1-ml column (GE Healthcare) previously equilibrated in the same buffer. Following a washing step with 30 ml of 50 mm Tris-HCl, pH 8.0, and 100 mm NaCl, the protein was eluted with 6 ml of the same buffer supplemented with 500 mm NaCl.
The N-terminal 6× histidine tag was cleaved overnight at room temperature with recombinant bovine enterokinase (Sigma) at 0.02 units/mg protein in 50 mm Tris-HCl, pH 8.0, 250 mm NaCl, 2 mm CaCl2, and 5% glycerol in a final volume of 13 ml. The reaction mix was diluted 2.5-fold with 50 mm Tris-HCl, pH 8.0, passed through a HisTrap HP 5-ml column to remove uncleaved protein, and loaded onto a HiTrap SP-Sepharose Fast Flow 1-ml column equilibrated with 50 mm Tris-HCl, pH 8.0, and 100 mm NaCl. After washing with 20 ml of 50 mm Tris-HCl, pH 8.0, and 100 mm NaCl, the protein was finally eluted with 50 mm Tris-HCl, pH 8.0, 500 mm NaCl, and 5% glycerol in highly concentrated fractions that were directly used for crystallization experiments. The average production yield was 30 mg/liter. The purity was checked by SDS-PAGE. The selenomethionine-substituted protein was purified following the same protocol, except that all buffers were supplemented with 5 mm β-mercaptoethanol. Final elution was performed with 50 mm Tris-HCl, pH 8.0, 500 mm NaCl, 5% glycerol, and 10 mm dithiothreitol.
Enzyme Activity Measurements—Specific activities were measured using 6.5 μm Arb93A and 0.5 mm substrate in 50 mm sodium acetate, pH 5, at 40 °C during 2 h. To determine the specificity of the enzyme, various substrates were tested. For p-nitrophenyl-α-l-arabinofuranoside, the rate of p-nitrophenol formed was determined by quantification at 410 nm using p-nitrophenol as the standard (extinction coefficient: 7 × 103 mm-1 cm-1) (29). For wheat arabinoxylan, sugar beet arabinan, and sugar beet debranched α-1,5-l-arabinan, the amount of reducing sugars released was determined with the 3,5-dinitrosalicylic acid method (30), using α-1,5-l-arabinobiose as a standard. One unit of enzyme activity was defined as the amount of enzyme releasing 1 μmol of α-1,5-l-arabinobiose equivalent/min.
For the determination of pH and temperature optima, the reactions were performed with sugar beet debranched α-1,5-l-arabinan as substrate, buffers ranging from pH 3 to 9 and temperatures ranging from 20 to 60 °C. The enzyme was incubated with the substrate in 50 mm sodium acetate for pH 3, 4, and 5; 50 mm sodium phosphate for pH 6, 7, and 8; and 50 mm Tris-HCl for pH 9.
Synthesis of Methyl Oligoarabinosides—The methyl-α-1,5-l-arabinotetraoside and methyl-α-1,5-l-arabinopentaoside used in NMR assays and crystallization trials were synthesized as described in the supplemental material, via protocols similar to those used for the preparation of other arabinofuranosides (31, 32).
Kinetic Parameter Determination—Kinetic measurements were performed with α-1,5-l-arabinan and α-1,5-l-arabino-oligosaccharides with DPs ranging from 1 to 6 as substrate. Typical reaction conditions were 6.5 μm enzyme in a final volume of 22 μl of sodium acetate, pH 5, with substrate concentrations ranging from 0.2 to 35 mm. After 4 min at 40 °C, the reactions were stopped by heating to 95 °C. Twenty μl were then used for derivatization with 8-aminonaphthalene-1,3,6-trisulfonic acid (Molecular Probes) and analysis by polysaccharide analysis using carbohydrate gel electrophoresis (PACE), as described previously (33). After electrophoresis, the intensities of bands were quantified with the QuantityOne software (Bio-Rad). Measurements were performed in duplicate. Michaelis-Menten kinetic parameters were derived using nonlinear regression formulas of the Prism software (GraphPad).
NMR Experiments—Substrate and buffers were exchanged by lyophilization at least twice from D2O. Exchange into deuterated buffer was performed for the enzyme by 10× dilutions and concentration on a Vivaspin 500 10 K (VivaSciences) three times.
1H NMR spectra were acquired with a Bruker Avance 400 spectrometer operating at a frequency of 400.13 MHz in 5-mm NMR tubes. Spectra were recorded with 4006 Hz spectral width, 32,768 data points, 4.089 s acquisition time, 0.1 s relaxation delay, and up to 16 scans. A substrate reference spectrum was acquired before adding the enzyme, and two experiments were performed. 30 μl of 500 mm sodium acetate, pH 4.5, and 2.5 m NaCl and 70 μl of protein concentrated to 9 mg ml-1 in 50 mm sodium acetate, pH 4.5, and 250 mm NaCl were added to 500 μl of methyl-α-1,5-l-arabinotetraoside to give a final concentration of 25.6 μm for the enzyme and 13.4 mm for the substrate. A spectrum was recorded after 5 min and then every 15 min for 3 h, and a final spectrum was recorded after 18 h at 25 °C. In the second experiment, 35 μl of enzyme concentrated to 7.8 mg ml-1 in 50 mm Tris-DCl, pH 7.5, 250 mm NaCl were added to 560 μl of methyl-α-1,5-l-arabinopentoside in the same buffer to give a final concentration of 11 μm for the enzyme and 11 mm for the substrate. A spectrum was recorded after 5 min and then every 5 min for 3 h and then every hour for 20 h at 20 °C. Integration was performed using the solvent peak (D2O) as reference, and an integral of 1 was used for one proton.
Crystallization, Data Collection, Structure Solution, and Refinement—Crystals were obtained by the hanging drop vapor diffusion method using 2-μl drops containing a 50:50 (v/v) mix of reservoir solution combined with protein solution at 20 °C. Native crystals were obtained with a solution containing 10% polyethylene glycol 20000, 100 mm Tris, pH 8.5, and 100 mm MgCl2 and the protein at 7.8 mg ml-1. Selenomethionine crystals were obtained with a solution containing 25% polyethylene glycol 550MME, 100 mm Hepes, pH 7.5, and the protein at 11.6 mg ml-1. The complex was obtained after soaking selenomethionine-substituted crystals, obtained in 22% polyethylene glycol 4000, 100 mm Hepes, pH 7.5, and 50 mm ammonium sulfate, for 2 h at room temperature with 1 mm α-Me-1,5-arabinotetraose. 20% glycerol was added as cryoprotectant when necessary prior to freezing in liquid nitrogen at 100 K.
Native, selenomethionine-substituted, and complex data were collected on a single crystal at the European Synchrotron Radiation Facility (Grenoble, France) on ID14-1, ID29, and ID14-2 beamlines, respectively, using ADSC detectors. Data were processed using XDS (34) before being scaled and converted to structure factors using SCALA in the CCP4 suite program (35). All further computing was performed using the CCP4 suite unless otherwise stated. Statistics for data collections are shown in Table 1.
TABLE 1.
Data collection and refinement statistics
| Data set | Selenomethionine | Native | Selenomethionine/Arabinobiose |
|---|---|---|---|
| Space group | P212121 | P212121 | P212121 |
| a, b, c (Å) | 48.84, 92.83, 99.43 | 48.34, 90.49, 92.50 | 48.62, 86.88, 108.17 |
| Resolution (outer shell) (Å)a | 48.85-2.45 (2.58-2.45) | 46.27-1.85 (1.95-1.85) | 48.62-2.05 (2.16-2.05) |
| Measured/Unique reflections | 219,533/17,270 | 122,420/35,097 | 109,746/29,532 |
| Multiplicity | 12.7 (13) | 3.5 (3.2) | 3.7 (3.8) |
| Completeness (%) | 100 (100) | 99.2 (96.3) | 99.9 (99.8) |
| Rmergeb | 0.116 (0.3972) | 0.083 (0.394) | 0.128 (0.427) |
| Mean I/σI | 21.3 (7.7) | 12.3 (2.9) | 8.3 (2.8) |
| Wilson plot B | 30 | 13.4 | 16.2 |
| Refinement | |||
| Rcryst/Rfreec | 15/17.8 | 16.8/21.6 | |
| Root mean square deviation bonds (Å/degrees) | 0.014/1.49 | 0.015/1.58 | |
| Protein atoms/B factor (Å2) | 2916/12.25 | 2862/13.18 | |
| Ligand atoms/B factor (Å2) | 19/17.9 | ||
| Heteroatoms/B factor (Å2) | 20/23.41 | 32/33.6 | |
| Water molecules/B factor (Å2) | 415/23.87 | 318/25.3 | |
| Protein Data Bank code | 2W5N | 2W5O |
Values in parenthesis refer to the highest resolution shell
Rmerge = Σ||I – 〈I〉|/|Σ〈I〉|
Rcryst = (Σ||Fo – Fc||)/(Σ||Fo||)
Initial phases and a first model for Arb93A were obtained by the single wavelength anomalous dispersion (SAD) method using the selenomethionine-substituted protein and autoSHARP (36). Five selenium positions were found and refined with a phasing power ANO of 0.877. Solvent flattening with an estimate of 48.2% solvent resulted in excellent and easily interpretable maps in which ARP/warp could build a first model (37). This model was used to solve the structure of the native protein by molecular replacement using AMORE (38). The model was then updated with ARP/wARP. The complex structure was solved by molecular replacement using PHASER and the native coordinates as a search model (39).
The structures were refined using REFMAC (40) iterated with manual rebuilding in COOT (41). The incorporation of the ligand was performed after inspection of the mFo - DFc weighted maps. Water molecules were introduced automatically using Coot and inspected manually. The stereochemical quality of the model was assessed with the program Procheck (42) prior to deposition in the Protein Data bank under the codes 2W5N and 2W5O for the native and complex structures, respectively. Details of the model quality are given in Table 1. All figures were drawn with the PyMOL Molecular Graphics System (DeLano Scientific, Palo Alto, CA) unless stated otherwise.
RESULTS
Cloning and Expression—The arb93A coding sequence isolated from RNA of F. graminearum was 1,146 bp in length and corresponded to 381 amino acids. The protein was predicted to harbor an N-terminal signal peptide with a cleavage site between Ala16 and Lys17. Therefore, only the DNA encoding the mature protein of 366 amino acids was cloned in pET30a+ for production in E. coli BL21 (DE3) cells. The apparent molecular mass after purification was ∼40,000 Da, consistent with the theoretical value of 41,077 Da. A Blast search against sequences in the Protein Data Bank revealed that the closest structurally characterized protein to Arb93A was the sialidase from Micromonospora viridifaciens (Protein Data Bank code 1EUS) with 20.9% sequence identity (43).
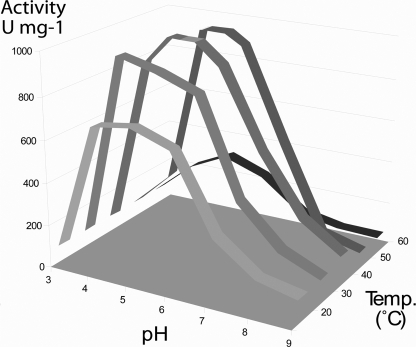
Activity Assay and Substrate Specificity—Enzymatic analysis at different pH and temperatures revealed an optimum activity at pH 5 and 40 °C. Fig. 1 summarizes the pH/temperature dependences of Arb93A. The protein is highly efficient in a large range of pH and temperature values; more than 80% of maximal activity is conserved at pH 4–6 and 30–50 °C.
FIGURE 1.
Effect of pH and temperature on the activity of Arb93A from F. graminearum. One unit of enzyme activity (U) was defined as the amount of enzyme releasing 1 μmol of α-1,5-l-arabinobiose equivalent/min.
To determine the substrate specificity, the activity of Arb93A was measured on four different substrates (Table 2). The protein is not active on p-nitrophenyl α-l-arabinofuranoside, indicating that more than one arabinofuranose moiety is necessary for cleavage. This was confirmed by PACE analysis of the hydrolysis products from oligoarabinofuranoside substrates with DPs 2–6, showing that the lowest DP of the released sugars was 2 (data not shown). Arb93A is probably not able to cleave arabinose or arabino-oligosaccharide substitutions of a xylan backbone, because no activity was detected on wheat arabinoxylan. Release of reducing sugars was observed with arabinan and debranched α-1,5-l-arabinan, revealing activities of 113 ± 45 and 1065 ± 60 units/mg, respectively. Contrary to debranched α-1,5-l-arabinan, the α-1,5-linked backbone of l-arabinan is substituted by α-1,3-linked (and possibly some α-1,2-linked) l-arabinofuranosyl residues. The difference in activity may therefore be explained by the fact that only α-1,5-linkages are hydrolyzed by Arb93A. When digesting arabinan, only arabinobiose and arabinose are produced (data not shown), indicating that the enzyme degrades the substrate in an exo-type mode of action. Thus, Arb93A can be defined as an exo-1,5-α-l-arabinanase releasing α-1,5-l-arabinobiose (arabinose being a side product of arabinotriose degradation) similarly to its homologue Abxn from P. chrysogenum 31B (22).
TABLE 2.
Substrate specificity of Arb93A measured at pH 5 and 40 °C One unit of enzyme activity was defined as the amount of enzyme releasing 1 μmol of α-1,5-l-arabinobiose equivalent min.
| Substrate | Specific activity |
|---|---|
| units/mg | |
| p-Nitrophenyl α-l-arabinofuranoside | 0 |
| Wheat arabinoxylan | 0 |
| Sugar beet arabinan | 113 ± 45 |
| Sugar beet debranched α-1,5-l-arabinan | 1,065 ± 60 |
Enzyme Kinetics—The kinetic parameters of Arb93A toward arabino-oligosaccharides with DPs 2–6 and debranched arabinan (DP > 100) were examined at pH 5 and 40 °C (Table 3). The kcat values are very similar for all of the substrates, varying between 0.81 × 105 and 1.13 × 105 s-1. Consequently, the observed differences of catalytic efficiencies (kcat/Km) are only due to the differences in affinity (Km). Except for DP4, the affinity of the enzyme for the substrates tends to increase with the length of the arabinofuranose-oligosaccharide chain. α-1,5-l-Arabinotetraose (DP4), for which the enzyme displays a Km of 9.26 ± 0.44 mm, is more resistant to hydrolysis than the other oligomers. There is no significant difference between the Km of Arb93A for DP3 and DP5. Comparison of the kcat/Km of the native Abnx purified from P. chrysogenum cultures with that of Arb93A shows that Arb93A activity is about 1000-fold stronger than Abnx activity on DP4/5/6 (44).
TABLE 3.
Michaelis-Menten kinetic parameters of Arb93A measured on arabino-oligosaccharides with various DPs DP2, DP3, DP4, DP5, DP6, and DP > 100 correspond respectively to α-1,5-l-arabinobiose, α-1,5-l-arabinotriose, α-1,5-l-arabinotetraose, α-1,5-l-arabinopentaose, α-1,5-l-arabinohexaose, and debranched α-1,5-l-arabinan from sugar beet. Hydrolysis reactions were performed at pH 5 and 40 °C. ND, not determined.
| Substrate | kcat | kcat/Km | Km |
|---|---|---|---|
| s–1 | s–1m–1 | m | |
| DP2 | 0 | 0 | 0 |
| DP3 | (1.13 ± 0.16) × 105 | (3.17 ± 0.45) × 107 | (3.56 ± 0.39) × 10–3 |
| DP4 | (1.05 ± 0.03) × 105 | (1.13 ± 0.03) × 107 | (9.26 ± 0.44) × 10–3 |
| DP5 | (0.81 ± 0.02) × 105 | (2.05 ± 0.06) × 107 | (3.96 ± 0.51) × 10–3 |
| DP6 | (0.94 ± 0.03) × 105 | (4.66 ± 0.14) × 107 | (2.03 ± 0.65) × 10–3 |
| DP >100 | ND | ND | (0.75 ± 0.05) × 10–3 |
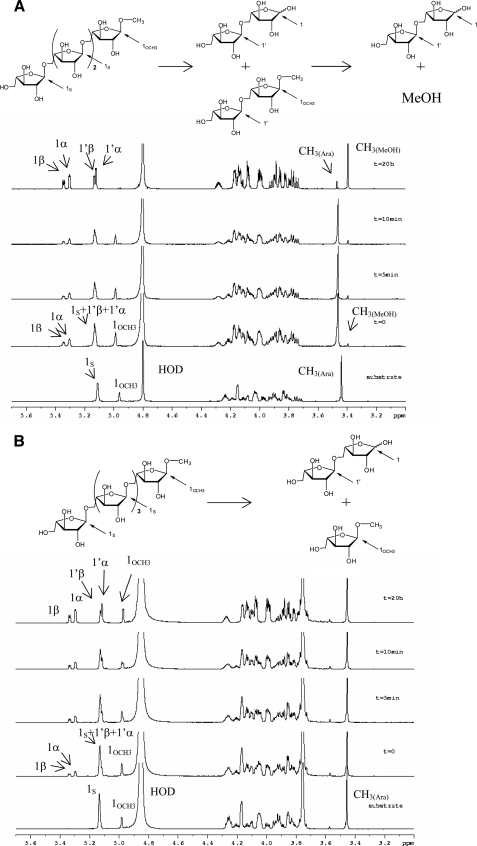
NMR Experiments—Product formation from the cleavage of Me-α-1,5-l-arabinotetraoside by Arb93A was followed in solution by NMR. Within a few minutes, peaks corresponding to arabinobiose (45) could be identified (Fig. 2A). The α/β ratio immediately stabilized at 1.6:1, indicating a very fast mutarotation equilibrium that appears to be particular to this furanose ring. It was therefore not possible to confirm the retention or inversion mechanism of the enzyme by this approach. Interestingly, the peaks corresponding to the methyl group disappeared after a few min, whereas a new peak with the chemical shift of methanol appeared at 3.39 ppm. At the end of the experiments, only arabinobiose and methanol were present in the solution. This demonstrated that the hydrolysis occurs by the nonreducing end and that an aglycon group can be cleaved from arabinobiose-derivative.
FIGURE 2.
Kinetics analysis of the hydrolysis of methylated arabinooligosaccharides by Arb93A followed by 1H NMR. A, hydrolysis of Me-α-1,5-l-arabinotetraoside. B, hydrolysis of Me-α-1,5-l-arabinopentaoside. Spectra were recorded at the indicated times. Labeling of the different anomeric protons of interest is indicated in the schematic representation of the reaction above the spectra.
A second experiment with pentasaccharide substrate was performed at higher pH and lower temperature to slow down the enzyme reaction. As expected, only arabinobiose and α-Me-arabinoside were produced, confirming the requirement of an arabinobiose moiety for cleavage of the aglycon (Fig. 2B). With this substrate and reaction conditions, mutarotation was still too fast to evaluate the configuration of the anomeric carbon immediately after glycosidic bond cleavage. Trials to slow down the reaction and the mutarotation by lowering the temperature failed. Indeed, at temperatures below 20 °C, the solvent peak gets broader and covers an essential area of the spectra.
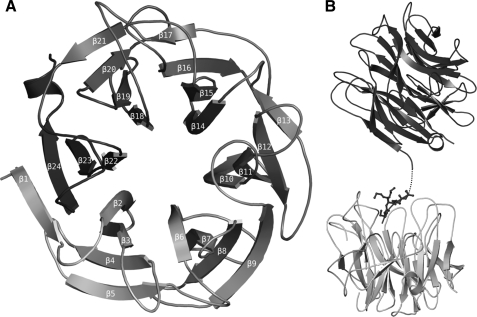
Overall Crystal Structure—The Arb93A crystal structure was solved by the SAD method using a seleno-methionine protein derivative, because molecular replacement trials with different sialidase models failed. The native structure described below was then determined and fully refined to 1.85 Å (Table 1). In the crystal as in solution, Arb93A is a monomeric protein constituted of a single catalytic domain of 366 amino acids (positions 16–381). One molecule is observed in the asymmetric unit, and all residues, apart from Gln20 and Lys21, could be modeled in the electron density. The protein presents a six-bladed β-propeller fold where each blade is constituted of a four-stranded antiparallel sheet. The first β-strand participates in the formation of the last blade and closes the β-propeller by forming the “molecular Velcro” classically observed for this fold (46) (Fig. 3A). The N and C termini extend by some extra residues outside of the β-propeller. The C terminus forms a small α-helix. A five-amino acid coil at the N terminus is stabilized in the active site of a symmetry-related molecule (Fig. 3B).
FIGURE 3.
Schematic diagram of Arb93A fold. A, overall representation with numbering of the strands. B, interaction with a neighboring protein in the crystals displaying the binding of the N-terminal residues (B). Dotted lines, amino acids that could not be modeled in the electron density.
As expected from sequence alignments, the search for structural relatives of Arb93A revealed extensive similarity with the sialidases/neuraminidases from clan GHE. The closest relatives, as determined by MSDFold (47), are sialidases of the family GH33, including the one from M. viridifaciensis (PDB 1EUS) and the human enzyme Neu2 (1VCU), presenting root mean square deviation values of 2.42 and 2.15 Å for 280 and 285 aligned residues, respectively (43, 48). Arb93A also presents two Asp β-hairpin boxes similar to the ones of GH33 sialidases in the first and second blades (49). The role of this motif first identified in sialidases and later in structurally unrelated proteins remains unclear (50). They are located far from the active site and have been proposed to maintain the integrity of the β-propeller (43).
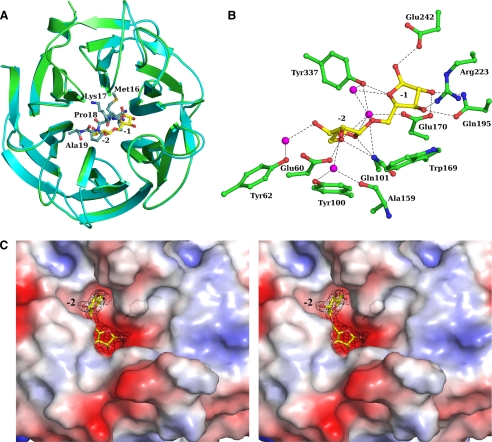
Complex Structure—Crystals of Arb93A crystal soaked with Me-α-1,5-l-arabinotetraoside diffracted to 2.05 Å. The complex structure solved by molecular replacement revealed unambiguously the presence of an α-1,5-l-arabinobiose moiety in the center of the propeller after inspection of the first electron density map. Only residues 22–381 could be modeled from the electron density. Upon sugar binding, the five residues of the N terminus have been displaced from the active site and are disordered. Small rigid body movement of about 1 Å is observed for some surface loops around the active site (loops between strands β11 and β12, β13 and β14, and β15 and β16) in order to accommodate the sugar. The root mean square deviation between the native and complex structure is 0.37 Å (Fig. 4A).
FIGURE 4.
Crystal structure of Arb93A complexed with α-1,5-l-arabinobiose. A, schematic overlay of the native (cyan/marine) and the disaccharide complex (green/yellow) Arb93A structures. B, ball and stick representation of the interactions between Arb93A amino acids and the α-1,5-l-arabinobiose (water molecules are displayed as magenta spheres). C, electrostatic surface representation of Arb93A active site and maximum likelihood weighted 2 mFo - DFc electron density (contoured at 1 σ/0.38 electrons/Å3) for the α-1,5-l-arabinobiose complex in wall-eye stereo.
The active site is located in a deep acidic L-shaped crevice at the center of the β-propeller (Fig. 4C). Two subsites are occupied by arabinofuranose and were identified as -1 and -2. Both arabinofuranoside moieties adopt an E3 ring shape that corresponds to a low energy conformation for this flexible ring (51). The active site extends via a cleft that corresponds to the +1 subsite. It extends further in a more open area where it is difficult to clearly identify additional subsites. The -2 subsite is rather deep and is surrounded by aromatic residues (Tyr44, Tyr62, Tyr100, and Tyr337) and hydrophobic residues (Leu359 and Pro360). The O5 hydroxyl group is deeply buried, with no possible extension of the oligosaccharide at the nonreducing end. In the -1 subsite, no decoration can be tolerated, which could explain the Arb93A preference for linear arabinan.
Sugar Contacts—In the -2 subsite, the O5 hydroxyl group forms hydrogen bonds with Gln101 NE2, Glu60 OE2, and, via water molecules, Glu170 OE2 and Tyr337 OH. The O4 ring oxygen makes direct contact with Gln101 NE2, whereas interactions of the O3 and O2 hydroxyl groups are mediated by water molecules with the Tyr62 OH for the first and the Ala159 main chain oxygen and the Tyr100 OH for the second. In the -1 subsite, the O5 glycosidic bond oxygen presents an indirect hydrogen bond to the Tyr337 OH. The O4 ring oxygen is hydrogen-bonded to the Tyr337 OH, the O3 hydroxyl group is hydrogen-bonded to Gln195 OE1 and Arg223 NH2, the O2 hydroxyl group is hydrogen-bonded to Glu170 OE1 and Gln195 NE2, and finally the anomeric oxygen (O1) is hydrogen-bonded to Glu242 OE2 (Fig. 4B). Stacking interactions have also been identified with Tyr100 in the -2 subsite and Trp169 in the -1 subsite as well as a strong hydrophobic interaction with Pro161 at the level of the glycosidic bond (Table 4). The anomeric oxygen of the reducing ring does not carry a methyl group. The hydrolysis from the nonreducing end is therefore confirmed by structural evidence. The electron density is consistent with a 100% α-configuration, which supports the retention of the configuration of the anomeric carbon proposed for its homologue Abxn (22).
TABLE 4.
List of contacts between arabinobiose and the protein
| Atom | –2 subsite | –1 subsite |
|---|---|---|
| O5 | 3.0 NE2 Gln101 | 3.0 Wat-3.0 OH Tyr337 |
| 2.7 OE2 Glu60 | 3.0 Wat-3.0 O4′ | |
| 3.0 Wat-2.8 OE2 Glu170 | ||
| 3.0 Wat-2.6 OH Tyr337 | ||
| O4 | 3.0 NE2 Gln101 | 3.0 OH Tyr337 |
| O3 | 2.8 Wat-2.7 OH Tyr62 | 2.7 OE1 Gln195 |
| 2.8 NH2 Arg223 | ||
| O2 | 2.9 Wat-2.7 O Ala159 | 2.8 OE1 Glu170 |
| 2.9 Wat-2.8 OH Tyr100 | 2.8 NE2 Gln195 | |
| O1′ | 2.6 OE2 Glu242 | |
| Stacking | Tyr100 | Trp169 |
Catalytic Mechanism and Mutant Analysis—Two essential catalytic residues are required in the classical retention mechanism described by Koshland (3), an acid/base and a nucleophile. Examination of the amino acids close to the anomeric carbon revealed that two carboxylic residues, Glu242 and Glu170, could act as the possible catalytic residues of family GH93. Glu170 is located under the glycosidic C1 in a perfect position for nucleophilic attack. The position of Glu242 is in accordance with a putative acid/base role and is hydrogen-bonded to the anomeric oxygen. The two carboxylate residues are 5.9 Å apart, which is in the range generally observed for other retaining enzymes (52).
Sialidases of the clan GHE use a tyrosine as the nucleophile, and structure overlay between Arb93A and these sialidases reveals that an equivalent tyrosine, Tyr337, is also present in the Arb93A active site. However, contrary to the sialidases, it is not in a proper position for nucleophilic attack. It is not located under the anomeric carbon but on the side and interacts with the ring oxygen. It could, however, play a role in catalysis that should be determined.
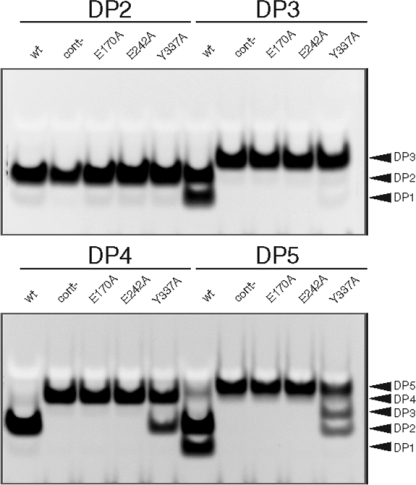
Each of the three hypothetical catalytic residues was mutated to alanine, and activities of the resulting mutants were compared with the wild-type Arb93A. Using linear α-1,5-l-arabinan as the substrate, Arb93A-E170A and Arb93A-E242A show no activity. Arb93A-Y337A displays a residual activity of only 7% compared with that of Arb93A-WT (data not shown). Fig. 5 displays PACE analysis of the digestion products of α-1,5-l-arabino-oligosaccharides by Arb93A and its mutants. α-1,5-l-Arabinobiose (DP2) is not hydrolyzed by any of the four tested proteins. As expected, no hydrolysis products of any tested arabinofuranose-oligosaccharides were observed with Arb93A-E170A and Arb93A-A242A. Arb93A-WT was active on arabinofuranose-oligosaccharides with DPs of 3, 4, and 5, whereas Arb93A-Y337A was only able to cleave polymers with DPs of 4 and 5.
FIGURE 5.
PACE analysis of hydrolysis products of Arb93A and its mutants on various arabino-oligosaccharides. DP2, DP3, DP4, and DP5 correspond to α-1,5-l-arabinobiose, α-1,5-l-arabinotriose, α-1,5-l-arabinotetraose, and α-1,5-l-arabinopentaose, respectively. cont-, the negative control, corresponding to reactions without enzyme. wt, E170A, E242A, and Y337A, the proteins Arb93A-WT, Arb93A-E170A, Arb93A-E242A, and Arb93A-Y337A, respectively. Hydrolysis reactions were performed at pH 5 and 40 °C.
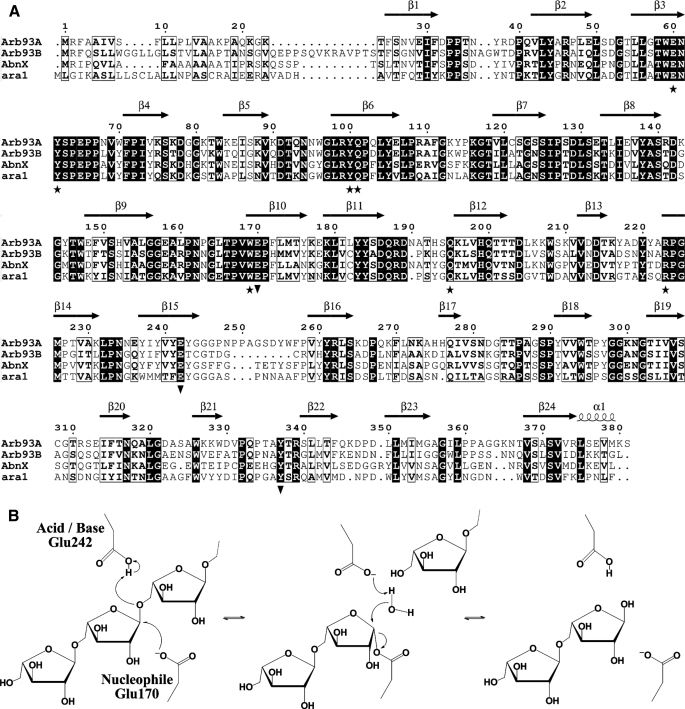
Sequence alignment Arb93A with the other GH93 α-l-arabinanase from F. graminearum (Arb93B) and Abnx from P. chrysogenum (21) and Ara1 from Fusarium oxyporum shows that the three residues are conserved in those enzymes (Fig. 6A). Those results support a role of Glu170 and Glu242 in the catalytic mechanism, whereas the role of Tyr337 during the catalysis remains unclear. We propose, therefore, a classical retaining mechanism for family GH93, with Glu242 functioning as an acid/base and Glu170 as a nucleophile, as depicted in Fig. 6B.
FIGURE 6.
Proposed catalytic mechanism of Arb93A. A, sequence alignment of Arb93A with the second GH93 arabinanase of F. graminearum (Arb93B), Abnx from P. chrysogenum and Ara1 from F. oxyporum (figure drawn with ESPript (60)). B, schematic representation of the proposed Arb93A mechanism.
DISCUSSION
According to the CAZY classification, GHs are distributed in 114 families based on their primary sequence (available on the World Wide Web). For about 30 of them, no three-dimensional structure has been solved to date. Crystal structures of native GHs and their complex with substrates generally provide information about the detailed mechanism, transition states, and specificity. We report here the first structure of a GH93 enzyme: an α-1,5-l-arabinanase from the fungus F. graminearum.
Structural Similarities with Clan GHE—According to its six-bladed β-propeller fold, Arb93A should be included into clan GHE. Until now, this clan consisted of only the sialidase super-family, which comprises family 33 with most bacterial and eukaryotic sialidases and transialidases, and families 34 and 83, which contain viral sialidases and hemagglutinin-neuraminidases, respectively.
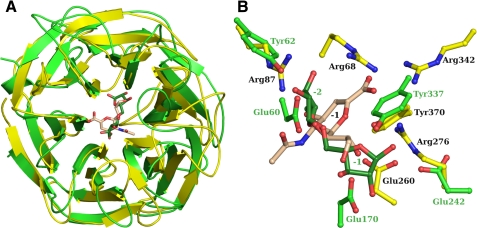
The comparison of the Arb93A structure with its closest related sequence, the family 33 sialidase from M. viridifaciensis (43), is displayed in Fig. 7 and could be extended to other sialidases. Apart from the same canonical six-bladed β-propeller fold, Arb93A presents major differences compared with M. viridifaciensis sialidase. One of them is that Arb93A lacks a carbohydrate binding module. Although carbohydrate binding modules have been proposed to play a role in the increase of the catalytic efficiency (53), the kcat/Km value of the sialidase using p-nitrophenyl α-d-sialoside as substrate is of the same order of magnitude as the values of Arb93A on the different arabino-oligosaccharides (54). For both of these exoenzymes, the active site is located in an acidic crevice at the center of the β-propeller. The Arb93A active site is more extended, since it cleaves off a disaccharide, whereas sialidases cleave only one terminal sialic acid. The active site is also more buried in Arb93A. The part of the active site pocket of sialidases that interacts with the carboxylic group of sialic acid is highly basic, which is not observed in Arb93A as would be expected due to the absence of a charged substituent in its substrate.
FIGURE 7.
Comparison of the three-dimensional structures of GH93 and GH33 glycoside hydrolases. A, schematic diagram of the overlay of Arb93A (green/forest) with the GH33 sialidase of M. viridifaciensis (yellow/wheat). B, ball and stick representation of the superposition of the two enzymes active site.
Active Site and Mechanism—NMR and structural data demonstrate that Arb93A cleaves the nonreducing arabinobiose from linear α-1,5-l-arabinans. From the complex crystal structure, the nonreducing extremity of the disaccharide is enclosed and could not be extended because the hydroxyl group at C5 oxygen is the most buried functionality. In contrast, a polysaccharide substrate can be accommodated from the reducing end glycosidic oxygen of the disaccharide, although with an ∼90° kink in the orientation of the chain. The active site is indeed much more open after the +1 subsite at the reducing end. There is no evidence for a +2 subsite yet, but there is a slightly basic platform where carbohydrate could be accommodated (Fig. 4B).
The present work indicates that arabinanases of the GH93 family act with retention of configuration, a mechanism that is also observed in related sialidases. Moreover, it corroborates the results obtained for Abnx, where retention of configuration was observed in the products of its transglycosylation activity (22). Nevertheless, and in contrast to expectations, the enzymes of family 93 present a catalytic machinery that is different from the sialidases (55). Structural evidence and mutation data clearly indicate that Arb93A acts through a classical retaining mechanism involving two conserved carboxylate residues, Glu170 and Glu242, acting as nucleophile and acid/base, respectively (Figs. 6 and 7). The enzyme could also be classified as a antiprotonator, such as those of the GHA clan (56). Sialidases, on the other hand, use a tyrosine residue as the nucleophile (reviewed in Ref. 55). It is the only residue among the eight that are conserved in sialidases that is also present in members of family 93. Tyr337, the equivalent tyrosine in Arb93A, is, however, not positioned properly for nucleophilic attack. This seems to be a result of the different shape of the active site, which is much more extended in Arb93A. The -1 subsite of sialidases is located at the level of the -2 subsite of Arb93A (Fig. 7B). The anomeric carbon in Arb93A is more than 5 Å away from the position observed in sialidases. This locates Tyr337 on the side of the anomeric carbon and not underneath as expected for a nucleophile. The differences in the nature of the catalytic residues and machinery have probably evolved in response to the unique properties of the different substrates (57).
Role of Tyr337—The mutation of Tyr337 to alanine results in a reduction of activity of 93%, which clearly indicates that this conserved residue is important for activity but not critical. One postulated role is that this residue may facilitates the attack by the nucleophile, as observed for other enzymes of the clan GHA, such as cellulases from family 5 or xylanases of family 11 (58, 59). It could help to maintain the ionization state of the nucleophile in the charged state when the structure is unliganded. The tyrosine is, however, 4 Å from the nucleophile in the present structure, but small movements of the side chain could be envisaged during catalysis. Alternatively, this residue could have a role in the stabilization of the substrate in the binding pocket. The Arb93A-Y337A mutant is indeed weakly active on the tetrasaccharide and longer oligosaccharides, but it cannot hydrolyze a trisaccharide. From the active site shape, upon binding, there should be a strong kink between the -1 and +1 subsites introduced by Tyr337 and Leu359 (Fig. 4C). The tyrosine interacts with the ring oxygen in the -1 subsite and will probably interact with arabinofuranose in the +1 subsite, stabilizing the sugar before cleavage. In the mutant, the trisaccharide would not be correctly bound, whereas for higher oligosaccharides, additional contacts would compensate in part for the absence of tyrosine. Similarly, in the arabinanase Arb43A from Cellvibrio japonicus, an aromatic residue, Phe114, influences the substrate conformation in the active site, and its mutation to alanine leads to a low enzymatic activity (14).
Comparison with Sequence-related Arabinanases—It has been demonstrated that F. graminearum expresses two homologous GH93 α-l-arabinanases displaying 52.9% sequence identity (Fig. 6A).3 The reason for the occurrence of two enzymes predicted to catalyze the same reaction is unclear. These two proteins may possibly display slightly divergent substrate specificities, allowing a better adaptation of the fungus to its substrate.
Arb93A displays strict substrate specificity for debranched α-1,5-l-arabino-oligosaccharides with DPs of >2 and cleaves only α-1,5-l-arabinobiose from the backbone. The reducing terminal arabinosyl residue of the commercial arabinan and debranched α-1,5-l-arabinan used for activity measurements is attached through rhamnose to fragments of a rhamnogalacturonan backbone. The observed hydrolysis through the nonreducing end of the molecule is therefore compatible with hydrolysis of such branches. Similar characteristics were observed for Abnx of P. chrysogenum sharing 53.2% sequence identity with Arb93A (15, 22). Enzymes from the GH93 family present the same catalytic machinery, and their mode of action seems quite specific to this family with the release of arabinobiose.
This study provides the first step in the understanding of the catalytic mechanism for the GH93 enzymes. Further experiments with mutants and/or inhibitors will help to confirm the retention mechanism, to determine the requirements both at the enzyme and substrate levels and permit the identification of additional subsites. The understanding of the structural and mechanistic features of the GH93 enzymes as well as the properties of their substrates, is indeed of great importance at the industrial levels. It will help in the comprehension of the degradation mechanisms of the highly diverse polysaccharidic substrates present in nature. Arb93A represents a new step toward the establishment of a defined collection of industrially applicable GHs, which is of primary interest for a better utilization of renewable natural hemicellulosic resources.
Supplementary Material
Acknowledgments
We are very grateful to Cophoudal (France) for supplying the raw hop material. We thank the ESRF (Grenoble, France) for access to synchrotron data collection facilities and Isabelle Jeacomine for help during NMR experiments.
The atomic coordinates and structure factors (codes 2w5n and 2w5o) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by CNRS and the Université de Strasbourg.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: GH, glycoside hydrolase; PACE, polysaccharide analysis using carbohydrate gel electrophoresis; DP, degree of polymerization; WT, wild type.
R. Carapito, J.-M. Jeltsch, and V. Phalip, unpublished data.
References
- 1.Saha, B. C. (2003) J. Ind. Microbiol. Biotechnol. 30 279-291 [DOI] [PubMed] [Google Scholar]
- 2.Ward, O. P., and Moo-Young, M. (1989) Crit. Rev. Biotechnol. 8 237-274 [DOI] [PubMed] [Google Scholar]
- 3.Koshland, D. E. (1953) Biol. Rev. 28 416-436 [Google Scholar]
- 4.Henrissat, B. (1998) Biochem. Soc. Transact. 26 153-156 [DOI] [PubMed] [Google Scholar]
- 5.Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009) Nucleic Acids Res. 37 D233-D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Margolles-Clark, E., Tenkanen, M., Nakari-Setala, T., and Penttila, M. (1996) Appl. Environ. Microbiol. 62 3840-3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Numan, M. T., and Bhosle, N. B. (2006) J. Ind. Microbiol. Biotechnol. 33 247-260 [DOI] [PubMed] [Google Scholar]
- 8.Hovel, K., Shallom, D., Niefind, K., Belakhov, V., Shoham, G., Baasov, T., Shoham, Y., and Schomburg, D. (2003) EMBO J. 22 4922-4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyanaga, A., Koseki, T., Matsuzawa, H., Wakagi, T., Shoun, H., and Fushinobu, S. (2004) J. Biol. Chem. 279 44907-44914 [DOI] [PubMed] [Google Scholar]
- 10.Paes, G., Skov, L. K., O'Donohue, M. J., Remond, C., Kastrup, J. S., Gajhede, M., and Mirza, O. (2008) Biochemistry 47 7441-7451 [DOI] [PubMed] [Google Scholar]
- 11.Taylor, E. J., Smith, N. L., Turkenburg, J. P., D'Souza, S., Gilbert, H. J., and Davies, G. J. (2006) Biochem. J. 395 31-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mai, V., Wiegel, J., and Lorenz, W. W. (2000) Gene (Amst.) 247 137-143 [DOI] [PubMed] [Google Scholar]
- 13.Gielkens, M. M., Visser, J., and de Graaff, L. H. (1997) Curr. Genet. 31 22-29 [DOI] [PubMed] [Google Scholar]
- 14.Nurizzo, D., Turkenburg, J. P., Charnock, S. J., Roberts, S. M., Dodson, E. J., McKie, V. A., Taylor, E. J., Gilbert, H. J., and Davies, G. J. (2002) Nat. Struct. Biol. 9 665-668 [DOI] [PubMed] [Google Scholar]
- 15.Sakamoto, T., and Thibault, J. F. (2001) Appl. Environ. Microbiol. 67 3319-3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunzelle, J. S., Jordan, D. B., McCaslin, D. R., Olczak, A., and Wawrzak, Z. (2008) Arch. Biochem. Biophys. 474 157-166 [DOI] [PubMed] [Google Scholar]
- 17.Proctor, M. R., Taylor, E. J., Nurizzo, D., Turkenburg, J. P., Lloyd, R. M., Vardakou, M., Davies, G. J., and Gilbert, H. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2697-2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandermarliere, E., Bourgois, T. M., Winn, M. D., Van Campenhout, S., Volckaert, G., Delcour, J. A., Strelkov, S. V., Rabijns, A., and Courtin, C. M. (2009) Biochem. J. 418 39-47 [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi, A., Tada, T., Wada, K., Nakaniwa, T., Kitatani, T., Sogabe, Y., Takao, M., Sakai, T., and Nishimura, K. (2005) J. Biochem. (Tokyo) 137 587-592 [DOI] [PubMed] [Google Scholar]
- 20.Varghese, J. N., Hrmova, M., and Fincher, G. B. (1999) Structure 7 179-190 [DOI] [PubMed] [Google Scholar]
- 21.Sakamoto, T., Ihara, H., Shibano, A., Kasai, N., Inui, H., and Kawasaki, H. (2004) FEBS Lett. 560 199-204 [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto, T., Fujita, T., and Kawasaki, H. (2004) Biochim. Biophys. Acta 1674 85-90 [DOI] [PubMed] [Google Scholar]
- 23.Phalip, V., Delalande, F., Carapito, C., Goubet, F., Hatsch, D., Leize-Wagner, E., Dupree, P., Dorsselaer, A. V., and Jeltsch, J. M. (2005) Curr. Genet. 48 366-379 [DOI] [PubMed] [Google Scholar]
- 24.Hatsch, D., Phalip, V., Petkovski, E., and Jeltsch, J. M. (2006) Biochem. Biophys. Res. Commun. 345 959-966 [DOI] [PubMed] [Google Scholar]
- 25.Guldener, U., Mannhaupt, G., Munsterkotter, M., Haase, D., Oesterheld, M., Stumpflen, V., Mewes, H. W., and Adam, G. (2006) Nucleic Acids Res. 34 D456-458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corfield, T. (1992) Glycobiology 2 509-521 [DOI] [PubMed] [Google Scholar]
- 27.Mitchell, D. B., Vogel, K., Weimann, B. J., Pasamontes, L., and van Loon, A. P. (1997) Microbiology 143 245-252 [DOI] [PubMed] [Google Scholar]
- 28.Doublie, S. (1997) Methods Enzymol. 276 523-530 [PubMed] [Google Scholar]
- 29.Lee, R. C., Hrmova, M., Burton, R. A., Lahnstein, J., and Fincher, G. B. (2003) J. Biol. Chem. 278 5377-5387 [DOI] [PubMed] [Google Scholar]
- 30.Miller, G. L. (1959) Anal. Chem. 31 426-428 [Google Scholar]
- 31.Joe, M., Bai, Y., Nacario, R. C., and Lowary, T. L. (2007) J. Am. Chem. Soc. 129 9885-9901 [DOI] [PubMed] [Google Scholar]
- 32.Yin, H., D'Souza, F. W., and Lowary, T. L. (2002) J. Org. Chem. 67 892-903 [DOI] [PubMed] [Google Scholar]
- 33.Carapito, R., Carapito, C., Jeltsch, J. M., and Phalip, V. (2009) Bioresourse Technol. 100 845-850 [DOI] [PubMed] [Google Scholar]
- 34.Kabsch, W. (1993) J. Appl. Crystallogr. 26 795-800 [Google Scholar]
- 35.Collaborative Computational Project 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 36.Vonrhein, C., Blanc, E., Roversi, P., and Bricogne, G. (2006) Methods Mol. Biol. 364 215-230 [DOI] [PubMed] [Google Scholar]
- 37.Perrakis, A., Morris, R., and Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458-463 [DOI] [PubMed] [Google Scholar]
- 38.Navaza, J., and Saludjian, P. (1997) Methods Enzymol. 276 581-594 [DOI] [PubMed] [Google Scholar]
- 39.McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C., and Read, R. J. (2005) Acta Crystallogr. Sect. D Biol. Crystallogr. 61 458-464 [DOI] [PubMed] [Google Scholar]
- 40.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 41.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 42.Laskowski, R. A., Macarthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 43.Gaskell, A., Crennell, S., and Taylor, G. (1995) Structure 3 1197-1205 [DOI] [PubMed] [Google Scholar]
- 44.Sakamoto, T. (2006) J. Appl. Glycosci. 53 115-122 [Google Scholar]
- 45.Cros, S., Imberty, A., Bouchemal, N., Du Penhoat, C. H., and Perez, S. (1994) Biopolymers 34 1433-1447 [Google Scholar]
- 46.Jawad, Z., and Paoli, M. (2002) Structure 10 447-454 [DOI] [PubMed] [Google Scholar]
- 47.Krissinel, E., and Henrick, K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60 2256-2268 [DOI] [PubMed] [Google Scholar]
- 48.Chavas, L. M. G., Tringali, C., Fusi, P., Venerando, B., Tettamanti, G., Kato, R., Monti, E., and Wakatsuki, S. (2005) J. Biol. Chem. 280 469-475 [DOI] [PubMed] [Google Scholar]
- 49.Roggentin, P., Rothe, B., Kaper, J. B., Galen, J., Lawrisuk, L., Vimr, E. R., and Schauer, R. (1989) Glycoconj. J. 6 349-353 [DOI] [PubMed] [Google Scholar]
- 50.Copley, R. R., Russell, R. B., and Ponting, C. P. (2001) Protein Sci. 10 285-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cros, S., du Penhoat, C. H., Pérez, S., and Imberty, A. (1993) Carbohydrate Res. 248 81-93 [Google Scholar]
- 52.McCarter, J. D., and Withers, S. G. (1994) Curr. Opin. Struct. Biol. 4 885-892 [DOI] [PubMed] [Google Scholar]
- 53.Thobhani, S., Ember, B., Siriwardena, A., and Boons, G. J. (2003) J. Am. Chem. Soc. 125 7154-7155 [DOI] [PubMed] [Google Scholar]
- 54.Narine, A. A., Watson, J. N., and Bennet, A. J. (2006) Biochemistry 45 9319-9326 [DOI] [PubMed] [Google Scholar]
- 55.Vocadlo, D. J., and Davies, G. J. (2008) Curr. Opin. Chem. Biol. 12 539-555 [DOI] [PubMed] [Google Scholar]
- 56.Heightman, T. D., and Vasella, A. T. (1999) Angew. Chem. Int. Ed. Engl. 38 750-770 [DOI] [PubMed] [Google Scholar]
- 57.Amaya, M. F., Watts, A. G., Damager, I., Wehenkel, A., Nguyen, T., Buschiazzo, A., Paris, G., Frasch, A. C., Withers, S. G., and Alzari, P. M. (2004) Structure 12 775-784 [DOI] [PubMed] [Google Scholar]
- 58.Sabini, E., Sulzenbacher, G., Dauter, M., Dauter, Z., Jorgensen, P. L., Schulein, M., Dupont, C., Davies, G. J., and Wilson, K. S. (1999) Chem. Biol. 6 483-492 [DOI] [PubMed] [Google Scholar]
- 59.Varrot, A., Schulein, M., and Davies, G. J. (2000) J. Mol. Biol. 297 819-828 [DOI] [PubMed] [Google Scholar]
- 60.Gouet, P., Courcelle, E., Stuart, D. I., and Metoz, F. (1999) Bioinformatics 15 305-308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.