Abstract
Evidence is growing to support a functional role for the prion protein (PrP) in copper metabolism. Copper ions appear to bind to the protein in a highly conserved octapeptide repeat region (sequence PHGGGWGQ) near the N terminus. To delineate the site and mode of binding of Cu(II) to the PrP, the copper-binding properties of peptides of varying lengths corresponding to 2-, 3-, and 4-octarepeat sequences have been probed by using various spectroscopic techniques. A two-octarepeat peptide binds a single Cu(II) ion with Kd ≈ 6 μM whereas a four-octarepeat peptide cooperatively binds four Cu(II) ions. Circular dichroism spectra indicate a distinctive structuring of the octarepeat region on Cu(II) binding. Visible absorption, visible circular dichroism, and electron spin resonance spectra suggest that the coordination sphere of the copper is identical for 2, 3, or 4 octarepeats, consisting of a square-planar geometry with three nitrogen ligands and one oxygen ligand. Consistent with the pH dependence of Cu(II) binding, proton NMR spectroscopy indicates that the histidine residues in each octarepeat are coordinated to the Cu(II) ion. Our working model for the structure of the complex shows the histidine residues in successive octarepeats bridged between two copper ions, with both the Nɛ2 and Nδ1 imidazole nitrogen of each histidine residue coordinated and the remaining coordination sites occupied by a backbone amide nitrogen and a water molecule. This arrangement accounts for the cooperative nature of complex formation and for the apparent evolutionary requirement for four octarepeats in the PrP.
Keywords: octarepeat peptides, nuclear magnetic resonance, circular dichroism, electron spin resonance
Prion diseases are a novel class of neurodegenerative diseases, including scrapie in sheep, bovine spongiform encephalopathy in cattle, and Creutzfeldt-Jacob disease in humans (1). A new variant form of Creutzfeldt-Jacob disease has been reported that is thought to be caused by the ingestion of infected beef (2, 3). A variety of biochemical, biophysical, cell biologic, and transgenetic experiments have indicated that the critical pathogenic event in prion disease is the misfolding of a benign cellular prion protein (PrPC) to form the infectious disease-causing isoform, the scrapie isoform of PrP (4–7).
Until recently, little has been known about the normal function of PrPC in the brain. There is now a body of evidence to indicate a role for PrPC in copper metabolism. Mice deficient in PrPC showed a >10-fold reduction of copper in a microsomal fraction from brain relative to wild-type mice and a reduction in activity of Cu/Zn superoxide dismutase (8). It also has been shown that cerebellar cells from mice deficient in PrPC are more sensitive to copper toxicity and oxidative stress (9).
Mature Syrian hamster PrPC is a glycoprotein containing two N-linked carbohydrates and one disulfide bridge. Post-translational processing results in the cleavage of a 22-residue leader sequence and the C-terminal tail after the attachment of a glycosylphosphatidylinositol anchor to serine 231. The solution structures of the mouse prion protein fragment, PrP(121–231) (10, 11), and of Syrian hamster PrP(90–231) (12) have been reported. The sequence of PrP(90–231) corresponds to the protease-resistant core of the scrapie isoform of PrP (PrP27–30), which can mediate prion disease.
The secondary structure of the full length Syrian hamster PrP(29–231) has been determined, and the dynamic properties of the protein backbone have been measured (13). The secondary structural elements of the full length apo PrP(29–231) are identical to those of PrP(90–231). The N-terminal half of the apoprotein, residues 29–124, is unstructured, with considerable backbone flexibility (13). Residues 51–91 contain an unusual glycine-rich repeat every eight residues; this sequence is termed the octarepeat region. Residues 60–91 consist of four octarepeat sequences (PHGGGWGQ)4, and residues 51–59 have a homologous sequence but lack the histidine residue (PQGGGTWGQ). The octarepeat region is among the most conserved parts of PrP in mammals (14), and analogous sequences are present in other prion proteins: for example, the seven hexameric repeats found in chicken PrP (14). Some cases of prion disease have been linked to the presence of additional octarepeat sequences (15).
As early as 1992, it was suggested that the octarepeat region of the prion protein, rich in histidine residues, may have a role in binding transition metal ions (16). In fact, a copper-containing affinity column was essential for the purification of full length PrPC (17). Studies of peptide fragments corresponding to the octarepeat region using x-ray fluorescence indicated Cu(II) binding with a Kd of 6.7 μM (18). The full length protein has a similar affinity for Cu(II), (Kd = 14 μM) (19). However, there is disagreement in the literature as to the number of Cu(II) ions that bind PrPC with high affinity, with stoichiometries varying between 1.8 and 5.6 ± 0.4 (8, 19). In addition, there is little understanding of the structural nature of the copper complex. It is likely that PrPC has a role in copper metabolism, and it is the aim of our work to investigate the manner in which the prion protein binds Cu(II).
MATERIALS AND METHODS
Preparation of Proteins and Peptides.
Peptides were synthesized by using fluorenylmethoxycarbonyl chemistry on a PerSeptive Biosystems (Framingham, MA) Pioneer peptide synthesizer. After removal from the resin and deprotection, the samples were purified by using reverse phase HPLC and were characterized by using mass spectrometry and 1H NMR. All peptides were blocked at the N terminus with an acetyl group and at the C terminus with an amide. Recombinant Syrian hamster PrP(29–231) and PrP(90–231) were expressed and purified as described (13, 20). Peptide and protein concentrations were determined by using the extinction coefficient at 280 nm, which was calculated for the octarepeat peptides by using 5,609 M−1⋅cm−1 multiplied by the number of tryptophan residues (21). The pH was measured before and after each spectrum was recorded.
Titration of Metal Ions.
A number of different metal ions were added to the octarepeat peptides by using aliquots from stock aqueous solutions of CuSO4, NiCl2, ZnCl2, CoCl2, and CdCl2. Some buffers affect Cu(II) binding to the prion peptides: it was found that Tris buffer competes successfully for Cu(II). The presence of 20 mM Tris buffer at pH 7.5 will completely remove Cu(II) from PrP(51–75) as judged by the circular dichroism (CD) spectrum. Phosphate, acetate, and N-ethylmorpholine buffers were found not to interfere with the Cu(II) binding to the octarepeats. Sodium azide 0.1% (used to inhibit proteolysis of the full length protein) was found not to affect the CD spectrum of Cu(II)[PrP(51–75)]. The addition of Cu(II) to the octarepeat peptides greatly reduces the solubility of the peptides. Typically, the octarepeat peptides were found to be soluble at 5 mM at pH 7.4; on addition of Cu(II), the peptides became insoluble, but the precipitated material could be redissolved by lowering the concentration to <0.5 mM.
NMR Spectroscopy.
1H NMR experiments were carried out on Bruker (Billerica, MA) AMX spectrometers at 500 MHz. Two-dimensional nuclear Overhauser effect, total correlation, and double quantum filtered correlated spectra were acquired in 90% H2O/10% D2O at 10°C by using the watergate method for solvent suppression (22). Nuclear Overhauser effect spectra were acquired with mixing times of 200 ms. Total correlation spectra were obtained with a DIPSI2 spin lock period (23) of 60-ms duration (7 kHz power).
CD.
CD spectra were recorded at 22°C on an Aviv (Lakewood, NJ) 61DS spectropolarimeter. Typically, a 0.1-cm path-length cell was used for spectra recorded between 185 and 260 nm, with sampling points every 0.5 nm. A 1-cm cell path length was used for data between 290 and 760 nm, with a 1-nm sampling interval.
Absorption Spectroscopy (UV/Visible).
UV/visible electronic absorption spectra were obtained with a Hewlett–Packard 8453 spectrophotometer, using a 1-cm path length.
Electron Spin Resonance (ESR) Spectroscopy.
X-band ESR spectra (9.526 GHz) were measured at liquid nitrogen and liquid helium temperatures by using a Bruker EMX spectrometer.
Binding Curves.
Plots of Y vs. [free Cu2+], 1/Y vs. 1/[free Cu2+], and a Scatchard plot Y/[free Cu2+] vs. Y were used to analyze the binding of Cu(II) to the peptides, where Y = [bound Cu(II)]/[total peptide (free + bound)]. [Bound Cu2+] was calculated by using the intensity of CD bands at 570 or 222 nm.
RESULTS
Design of Peptides.
Octarepeat peptides synthesized containing 2, 3, and 4 histidines included PrP(51–75) (2-His) PQGGGTWGQPHGGGWGQPHGGGWGQ, PrP(73–91) (2-His) WGQPHGGGWGQPHGGGWGQ, PrP(76–86) (2-His) PHGGGWGQPHG, PrP(66–91) (3-His) GQPHGGGWGQPHGGGWGQPHGGGWGQ, and PrP(58–91) (4-His) GQPHGGGWGQPHGGGWGQPHGGGWGQPHGGGWGQ.
Addition of Cu(II) to the Octarepeat Peptides.
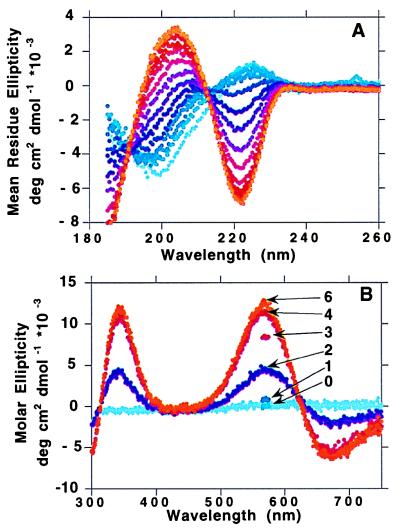
The CD spectrum of PrP(51–75) in the absence of Cu(II) had a strong negative band at 200 nm and a weak positive band at 225 nm. As CuSO4 was titrated into the peptide solution at pH 7.4, the band at 200 nm decreased in intensity, and a new band appeared at 222 nm, reaching a maximum negative per-residue ellipticity of 3,000 deg cm2⋅dmol−1. After the addition of 1 mole-equivalent of Cu(II), further additions of Cu(II) had no effect on the CD spectrum. There was a clear isodichroic point at 212 nm. The octarepeat peptide containing four His residues, PrP(58–91), behaved similarly, but the intensity of the band at 222 nm reached a maximum only after 4 mole-equivalents of Cu(II) were added (Fig. 1A). The maximum negative per-residue ellipticity for Cu4[PrP(58–91)] was 6,850 deg cm2⋅dmol−1 at 222 nm, and the maximum positive ellipticity at 204 nm was 3,400 deg cm2⋅dmol−1.
Figure 1.
(A) UV CD spectrum (180–260 nm) of PrP(58–91) (0.032 mM, pH 7.5, no buffer) with the addition of Cu(II) in increments of 0.33 mole-equivalents of CuSO4 from 0 (light blue) up to 7 mole-equivalents (orange). (B) Visible CD spectrum (300–750 nm) of PrP(58–91) (0.033 mM, pH 7.5, no buffer) with the addition of Cu(II) in increments of 1.0 mole-equivalent of CuSO4 from 0 (light blue) up to 6 mole-equivalents (orange). The curves for 1, 3, and 7 mole-equivalents are represented as single points at 570 nm.
On addition of Cu(II) to each peptide, a visible absorption band appeared at 625 nm, with an extinction coefficient of ≈40 M−1⋅cm−1 for PrP(51–75), pH 7.4, 1 mole-equivalent Cu(II). An intrinsic ellipticity is associated with this absorption band, centered at 625 nm, with a positive band at 570 nm and a negative band at 670 nm (Fig. 1B). An additional positive band at 340 nm behaved similarly to the band at 570 nm.
The intensity of the CD band at 570 nm is plotted vs. Cu(II) concentration for octarepeat peptides of different lengths containing 2-, 3-, and 4-His residues in Fig. 2A. The maximum positive molar ellipticity at 570 nm was 3,220 deg cm2⋅dmol−1 for the 2-His peptide and 12,400 deg cm2⋅dmol−1 for the 4-His peptide, ≈4× larger. For the 2-His peptides, the intensity increased with [Cu2+] almost linearly between 0 and 1 mole-equivalent Cu(II) added; further additions of Cu(II) had no effect on the intensity. For the peptide containing three histidines, PrP(66–91), the 570-nm maximum reached a plateau after 3 mole-equivalents of Cu(II) were added whereas, for the four-octarepeat peptide PrP(58–91), the stoichiometry was clearly 4 Cu:1 peptide. A plot of Y = {[bound Cu(II)]/[total peptide (free + bound)]} vs. [free Cu2+] and a double-reciprocal plot [1/Y vs. 1/(free Cu2+)] for binding of Cu(II) to PrP(51–75) at 0.034 mM peptide concentration indicated a Kd of ≈6 μM. The intensities of signals from the three- and four-octarepeat peptides appeared to increase cooperatively with increasing amounts of Cu(II). Scatchard plots gave strong positive deviation for both peptides, indicating a positive cooperativity. For PrP(58–91), 3.3 copper ions bound cooperatively, and, for PrP(66–91), 2.4 Cu(II) bound cooperatively. No cooperativity is seen for any of the 2-His containing peptides. Metal binding to the octarepeat peptides was highly specific for Cu(II). The addition of 10 mole-equivalents of Ni(II), Co(II), Zn(II), or Cd(II) to 0.04 mM solutions of PrP(51–75) at pH 7.4 caused no changes in the far-UV CD spectra.
Figure 2.
(A) Cu(II) binding curves: molar ellipticity at 570 nm with increasing amounts of Cu(II), pH 7.5. ●, 2-His peptide, PrP(51–75) (0.34 mM). ⧫, 3-His peptide, PrP(66–91) (0.021 mM). ▴, 4-His peptide, PrP(58–91) (0.033 mM). (B) pH dependence of the ellipticity at 570 nm for PrP(58–91). The pH dependence curve has been fitted to the following equation: Δɛobs = {Δɛacid[H+]n + Δɛbase[H+] Kan }/ [H+]n + Kan}, where n = Hill coefficient and Ka = acid dissociation constant for the transition. The midpoint of the transition is pH 6.7.
pH Dependence of the Copper–Peptide Interaction.
The CD and absorption spectra of the octarepeat peptides in the presence of Cu(II) were strongly pH-dependent. The CD band at 222 nm was not observed below pH 6; above pH 6, the intensity of the negative ellipticity increased cooperatively, reaching a maximum at pH 7.8 and above. The absorption spectrum of PrP(51–75) below pH 6 in the presence of 1 mole-equivalent of Cu(II) showed only a broad weak band at 800 nm, characteristic of CuSO4 in water. As the pH was raised from 6 to 7.4 the band at 800 nm disappeared and a band at 625 nm increased in intensity. As the pH was raised still further, the absorption band at 625 nm shifted to a slightly lower wavelength, with the maximum absorption shifting to 580 nm at pH 9. The pH dependence of the CD band at 570 nm for PrP(58–91) is shown in Fig. 2B. The midpoint of the pH dependence of the signal at 570 nm was 6.7, with a Hill coefficient of 2.2. For the 2-His peptide PrP(51–75), the midpoint was 7.0, with a Hill coefficient of 1.0.
ESR.
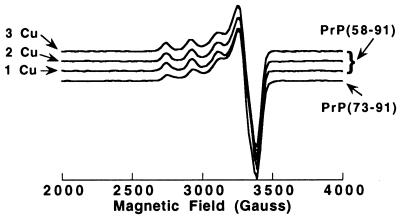
The environment of Cu(II) in the complex with the prion peptides can be deduced from the ESR spectrum. Fig. 3 shows ESR spectra of PrP(58–91) with 1, 2, and 3 mole-equivalents of Cu(II) added at pH 7.4. The three ESR spectra are essentially identical, with g⊥ of 2.05 ± 0.005 and g∥ of 2.26; the hyperfine splitting a∥ is 184 gauss. Fig. 3 also shows the ESR spectrum of PrP(73–91) with 0.8 mole-equivalents of Cu(II) added at pH 7.4; this spectrum is very similar to that of the Cu(II) complex with PrP(58–91), with g⊥ of 2.05 and a slightly smaller g∥ of 2.25. The hyperfine splitting a∥ is 186 gauss. Spectra at pH 6.6 and 7.7 were identical to those at pH 7.4. X-band ESRs (9.536 Hz) at liquid helium and nitrogen temperatures were unsuccessful in resolving ligand super-hyperfine splitting.
Figure 3.
X-band ESR spectra of 1, 2, and 3 mole-equivalents of Cu(II) added to PrP(58–91). A spectrum of 0.8 mole-equivalents of Cu(II) added to PrP(73–91) also is shown. Samples were buffered with 10 mM N-ethylmorpholine HCl, and spectra were acquired at 5 K.
Proton NMR.
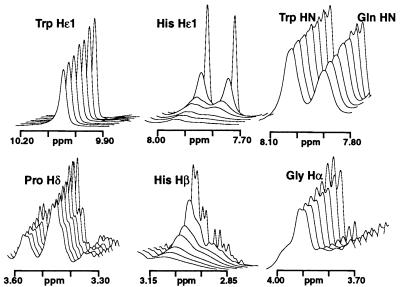
The effects of Cu(II) on the 1H NMR spectrum of a number of peptides [PrP(51–75), PrP(73–91), PrP(76–86), and PrP(58–91)] were studied to determine the nature of the copper binding sites. A combination of two-dimensional total correlation spectroscopy, double quantum filtered correlated spectroscopy, and nuclear Overhauser effect spectroscopy was used to assign the 1H resonances in the absence of Cu(II). The addition of small amounts of CuSO4 to PrP(51–75) caused differential broadening of resonances, which provided an indication of the location of copper binding in the peptides. The effects of Cu(II) addition on the 1H NMR spectrum of PrP(76–86) are shown in Fig. 4. There was a general increase in the linewidth of all resonances with increasing Cu(II) concentration. The Hɛ1, Hδ2, Hβ, and Hα resonances of both His residues were broadened significantly more than other resonances. For example, the two well resolved Hɛ1 His resonances were broadened from an ≈3-Hz line width (at half height) to 13 Hz after the addition of 0.0066 mole-equivalents of Cu(II). The addition of the same amount of Cu(II) caused the linewidths of the Gln and Trp resonances to be increased by only ≈1 Hz; for example, the Trp Hɛ1 resonance increased from 4.7 Hz to 5.5 Hz. The resonances of the two His Hα were overlapped, so it was not possible to determine whether the observed broadening arose from one or both of these resonances. The backbone amide resonances of the two histidines were broadened slightly more than other NH resonances (≈2 Hz). After the addition of 0.15 mole-equivalents Cu(II), the Hɛ1 and Hδ2 resonances were so broad that they were indistinguishable from the baseline whereas other resonances remained relatively sharp. There was no significant broadening of the Gly Hα resonances in any of the PrP peptides containing two His residues. Resonances assigned to proline were relatively unaffected by Cu(II) addition (Fig. 4). By contrast, similar additions of Cu(II) up to 1.5 mole-equivalents to the 4-His peptide, PrP(58–91), resulted in a general broadening of the entire spectrum, with little differential effect on particular resonances.
Figure 4.
1H NMR stack plot showing the effects of addition of Cu(II) to PrP(76–86) (0.5 mM in 90/10% H2O/D2O, pH 7.4, and 10 mM phosphate buffer at 10°C). The CuSO4 was added in aliquots of 0.0033 mole-equivalents up to 0.02 mole-equivalents. The spectral width for each panel is 150 Hz.
Full Length PrP(29–231).
It proved difficult to study the effect of copper addition on the full length prion protein. Although apo PrP(29–231) remained in solution even at quite high concentrations (1 mM) at pH 5.5, raising the pH to 7.4, a pH at which Cu(II) is fully bound to the peptides, caused the protein to precipitate. Binding of Cu(II), as demonstrated by the appearance of the characteristic CD bands centered at 625 nm [as seen in the peptide-Cu(II) complexes] was observed by gradually raising the pH of a Cu(II)-protein solution at pH 5.3. The addition of only 4 molar equivalents of Cu(II) did not give detectable visible CD bands at pH 6.5; additional Cu(II) (10 mole-equivalents) was necessary for these bands to be observed. Fig. 5 shows the visible region of the CD spectra for PrP(29–231) at pH 5.3, 6.2, and 6.5 after the addition of 25 mole-equivalents Cu(II). The CD bands and the pH dependence are very similar to those observed for the octarepeat peptides (Fig. 1B). The molar extinction coefficient at 570 nm of 2,200 deg cm2⋅dmol−1 (at pH 6.5) is very similar to that observed for the PrP(58–91) peptide at a comparable pH. Raising the pH above pH 6.7 resulted in loss of signal because of precipitation of the protein.
Figure 5.
CD spectrum (300–750 nm) of full length PrP(29–231) after the addition of 25 mole-equivalents of Cu(II) at pH 5.3, 6.2, and 6.5 in N-ethylmorpholine buffer.
DISCUSSION
Stoichiometry and Binding Affinity.
The published literature on Cu(II) binding to PrPC (17–19) confirms the specificity of the PrP-Cu(II) interaction. Earlier work showed that full length PrPC but not N-terminally truncated PrPC-II could be purified on a copper affinity column (17) and that the octarepeat region [PrP(58–91)] was the focal point of Cu(II) binding (18). Building on these results, we have studied the binding of Cu(II) to a number of PrP fragments in the octarepeat region, using a variety of spectroscopic techniques. It is clear from the copper binding saturation curves, which monitor the CD band at 570 nm, that the octarepeat peptides containing two His residues all saturate with Cu(II) after 1 mole-equivalent is added. Our measured Kd for the complex of Cu(II) with PrP(51–75) of ≈6 μM is in good agreement with fluorescence and equilibrium dialysis studies that indicate a Kd between 6.7 and 14 μM (18, 19).
The octarepeat peptides PrP(51–75), PrP(73–91), and PrP(76–86) all contain two histidine residues and all bind a single Cu(II) ion. On this basis, one might have expected the PrP(58–91), which contains four histidines, to bind two Cu(II) ions. However, the binding curves indicate that four Cu(II) ions are bound tightly at a peptide concentration of 0.03 mM and pH 7.4 whereas the 3-His-containing peptide, PrP(66–91), binds three Cu(II) ions.
The stoichiometry of Cu(II) binding to the full length PrP is unclear from the literature. Equilibrium dialysis studies indicate saturation of PrP(29–231) by Cu(II) after the addition of ≈1.8 Cu(II) (19) whereas others have reported saturation after 5.6 (±0.4) mole-equivalents of Cu(II) (8) were added to the N-terminal domain. The key factor in these different observations appears to be the pH. The equilibrium dialysis study was performed at pH 6.2, a pH at which our studies indicate that the octarepeat peptides have less than optimal Cu(II) binding. We suggest that four Cu(II) ions bind tightly to the octarepeat region of PrP at pH 7.4 and that protonation of one or more imidazoles at pHs between 6.0 and 7.4 decreases the number of effective Cu(II) binding sites. There may be other binding sites elsewhere in the protein, particularly in the regions that are unstructured in the apoprotein between residues 90–125. No obvious Cu(II) coordination sites are apparent in the structured domain of the protein from the positions of potential Cu(II) binding ligands in the solution structure (10, 12). The high levels of Cu(II) required to observe the visible CD spectrum characteristic of complex formation in the full length protein may be attributable to the necessity for studying PrP(29–231) at pHs below the optimum for Cu(II) binding. Competition from nonspecific Cu(II) binding sites elsewhere on the protein at these pHs also may play a role.
pH as an Important Variable.
The binding of Cu(II), both to the octarepeat peptides and to the full length protein, is strongly pH-dependent. Below pH 6, copper does not bind tightly to the PrP peptides; raising the pH to 6.7 resulted in half the copper sites being bound. A pH dependence centered at 6.7 suggests the involvement of histidine in the coordination of the Cu(II); this was confirmed by the NMR data. The Hill coefficient of 2.2 observed for binding of Cu(II) to the four-histidine peptide PrP(58–91) reflects the sharp cooperative transition between fully bound copper at pH 7.4 and above and no copper binding below pH 6. The binding of Cu(II) to PrP(29–231) has been studied by using tryptophan fluorescence emission and also was found to be pH-dependent (19). However, the center of the transition reported was approximately one pH unit lower than we observed with the octarepeat peptides. This discrepancy probably arose in part from the low solubility of PrP(29–231) at pH >6.5. The study also may have been complicated by the pH dependence of the Trp fluorescence emission itself, irrespective of the presence of Cu(II).
Conformation Change in the Presence of Cu(II).
All of the octarepeat peptides are unstructured in solution in the absence of Cu(II), as expected in view of the highly unstructured nature of this region of apo PrP(29–231) (13). Changes in the far-UV CD spectrum on addition of Cu(II) indicate the formation of structure with a reduction in the negative ellipticity at 200 nm characteristic of unstructured peptides, an increase in a negative CD band at 222 nm, and the development of a positive band at 204 nm. These changes in the CD spectrum are indicative of structure formation but are not consistent with α-helix or β-sheet conformation; they are more characteristic of turns and structured loops. Raman spectroscopic studies on octarepeat peptides suggest copper-dependent α-helix formation (24); our CD results are clearly not consistent with this. A published CD study of the binding of Cu(II) to the octarepeat peptides (18) failed to discern an increase in structure on addition of Cu(II). We suggest that these results are inconsistent with ours because of the use of Tris buffer in the previous study. Our studies (data not shown) indicated that Tris competes successfully with the octarepeat peptides for copper at pH 7.4.
Ligands involved in Cu(II) Coordination.
Cu(II) can adopt a range of coordination geometries in proteins and peptides and will coordinate to nitrogen, oxygen, or sulfur ligands. The ESR spectra of the PrP peptides are typical for the type II class of Cu(II) protein complexes with tetragonal coordination geometry, either square-planar or square-planar with weak axial ligands. The g∥ and a∥ values can give insights into the type of ligands involved. A g∥ of 2.26 and an a∥ of 184 gauss are most typical of a three-nitrogen and one-oxygen [3N, 1O] coordination site, although a [4N] or [2N, 2O] coordination geometry cannot be ruled out (25). For example, the Gly-His-Lys tri-peptide forms a [3N, 1O] square-planar Cu(II) complex (26) (with a weak axial ligand), and the ESR spectrum (27) is very similar to that observed for Cu-PrP. The ESR spectra with increasing mole-equivalents of Cu(II) added to PrP(58–91) have a single set of identical g factors and hyperfine splitting; we therefore conclude that all four of the Cu(II) sites within the Cu4[PrP(58–91)] complex have identical coordination geometry.
The slow electron spin relaxation of the paramagnetic Cu(II) resulted in marked broadening of proton NMR resonances linked to the paramagnetic centers by through-bond (contact) or through-space (pseudocontact) interactions. The low levels of Cu(II) used in the 1H NMR study should enable the observation of significant differential broadening of the resonances of protons close to the copper binding site, as long as the exchange rate of the copper is rapid. The 1H NMR spectra of the 2-His peptides suggest that the imidazole groups of both histidines are involved in coordination of the Cu(II) and that the tryptophan and glutamine side chains also present in each octarepeat do not directly coordinate the metal. The only other possible ligands from the peptide are backbone amide or carbonyl groups. The Hα resonances of the Gln, Trp, and Gly also were relatively unaffected by copper addition, indicating that potential ligands from these residues do not coordinate to the Cu(II). However, the overlapping His Hα resonances were broadened, suggesting that at least one His backbone amide coordinates to the Cu(II) ion. The relative lack of broadening of the His NH resonance is probably attributable to the fact that binding of the Cu(II) to the amide nitrogen displaces the amide proton and thus the His NH signals observed are for the copper-free form of the peptide. The exchange rate of the Cu(II) bonded to PrP(58–91) at low levels of Cu(II) was clearly slower than that observed for the two-His peptides. Differential broadening of the His resonances was not observed; instead, a general broadening of the proton resonances in the peptide takes place, complicating the determination of the copper-binding sites.
An absorption band at 625 nm with an extinction coefficient of 40 M−1⋅cm−1 is typical for a Cu(II) d-d transition (28). The wavelength of the maximum (λmax) can be used as an indication of the number of nitrogen ligands bound to the Cu(II) ion. Increasing numbers of peptide nitrogens ionized for the Cu(II) complex with acetyl-Gly-Gly-His results in a decrease in λmax from 765 nm for 1N to 540 nm for 4N (29). The 3N 1:2 complex of Cu(II) with two acetyl-Gly-His molecules is identical in ligand type to that proposed for the Cu-prion complex (2 His N plus 1 peptide N) and gives an identical λmax of 625 nm and a comparable extinction coefficient of 57 M−1⋅cm−1 (29).
The binding of imidazole groups and backbone amide nitrogens is common in other histidine-containing Cu(II) complexes. Studies with acetyl-Gly-Gly-His-Gly-Gly and other His-containing peptides show that only the backbone amide of the histidine itself or of residues immediately N-terminal to it are normally involved in coordination to the copper (30). This is consistent with our NMR results for the PrP peptides, indicating the possible involvement of the His amide in copper coordination but not any other amide nitrogens (the residue immediately preceding the His is Pro, which cannot be deprotonated to form a ligand). It has been suggested that backbone carbonyl groups could coordinate the Cu(II) (19). Carbonyl coordination can occur in copper–peptide complexes—for example, Cu(Gly-Gly)—but only at a pH < 4; at physiological pH, backbone amide coordination dominates (31). Other examples of amide coordination are seen in the Cu(II) complexes of Gly-Gly-His (32) and Gly-His-Lys (26).
Structure of Copper Complexes with Peptides of Different Lengths.
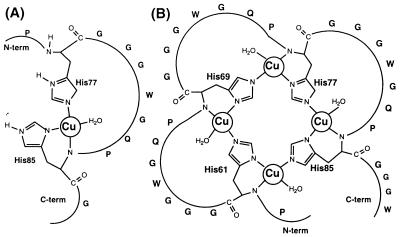
We duduce from the spectroscopic measurements that the Cu(II) complex formed by the 2-His octarepeat peptides [PrP(51-75), PrP(73-91), Prp(76-86)] is square planar, with one Cu coordinating a nitrogen in each of the imidazole rings and with an additional nitrogen ligand from the backbone amide of the more C-terminal His residue. The final ligand to make the copper complex four-coordinate could be a water molecule, as illustrated in Fig. 6A. We consider that coordination of the backbone amide of the more N-terminal His residue to the Cu(II) ion is unlikely from conformational and steric considerations.
Figure 6.
(A) A plausible structure for the complex of Cu(II) with PrP(76–86). (B) A plausible structure for the bridged complex of four Cu(II) ions with PrP(58–91).
The ESR, CD, and visible absorption spectra for the PrP(58–91) peptide in the presence of Cu(II) are strikingly similar to those of the 2-His containing peptides, and a single set of bands is observed at all Cu:peptide ratios. This implies that the copper coordination sphere is the same for octarepeat peptides of different lengths, that the four copper sites within the Cu4[PrP(58–91)] complex are identical, and that a 4-fold locally symmetric structure is formed. Copper binding to PrP(58–91) is cooperative (Fig. 2), which implies that the copper binding sites are not isolated from each other but interact in a specific way.
It is unlikely that a single imidazole binds each copper ion because this would result in less stable complexes than are observed. For example, acetyl-Gly-Gly-His-Gly-Gly binds Cu(II) with a Kd of 10−4 M (30), two orders of magnitude weaker than the observed Cu-PrP binding affinity. A schematic model that can explain all of the data is shown in Fig. 6B. We suggest that the four histidines in successive octarepeats form a bridged copper complex, with the Nɛ2 and Nδ1 imidazole nitrogens from each histidine coordinating two adjacent copper ions. This is reminiscent of the structure of Cu2Zn2 superoxide dismutase, in which the metal ions are 6.3 Å apart, with the imidazolate ring of His63 serving as bridging ligand (33). The crystal structure of the Cu(II) complex with Gly-His-Lys (26) has the His Nδ1 imidazole nitrogen and the His amide nitrogen forming a six-membered ring. We have, therefore, modeled this type of geometry in the proposed Cu-PrP structure. The coordination sphere for the 2-, 3-, and 4-His containing peptides is similar, with a square-planar arrangement of 3N and 1O. A molecular model constructed for the Cu4[PrP(58–91)] complex showed that the four-copper atom cluster could be accommodated without strain at the corners of a distorted tetrahedron, with two imidazole groups bound to each Cu(II) ion at adjacent coordination sites. A model of the 3-His-containing peptide PrP(66–91) also can be built without strain; in this case, the cluster of 3 Cu(II) ions forms a planar triangular arrangement.
The model shown in Fig. 6B can provide an explanation for the cooperativity of Cu(II) binding to PrP(58–91). Because the chelation of a metal to the His Nδ1 nitrogen can lower the pKa of the Nɛ2 NH and can make the nitrogen available for metal chelation (34), we suggest that the chelation of the first Cu(II) lowers the pKa of the second His ring proton enabling, the chelation of the second Cu(II) to the same histidine residue. After the first Cu(II) coordinates two imidazoles, the next two coppers need to trap only one imidazole to gain two ligands, and the fourth site is entirely preorganized after the third copper binds. In addition, the proximity of the Cu(II) may well reduce the pKa of the backbone NH because of a chelate effect (34). Thus, electronic and entropic considerations help to explain the cooperative nature of the binding of the four Cu(II) ions to PrP(58–91).
Copper Binding in the Prion Protein.
Many previous structural studies of PrP have focused on the apoprotein and have speculated on the role of the conformationally flexible regions. Given the nature of the highly specific PrP:Cu(II) interaction, it is incumbent on us to consider the holoprotein, containing the appropriate number of Cu(II) ions bound, as the physiologically relevant structure. Previous studies (17, 19), together with our current work, provide evidence that the binding sites for Cu(II) to the peptides and to the full length protein are the same. This implies that, although residues 29–124 of PrP are highly flexible and unstructured in the absence of Cu(II) (13), the octarepeat region of the holoprotein at physiological Cu(II) concentrations no longer can be considered to be unstructured. If our model of Cu:PrP(58–91) (Fig. 6B) is correct, we suggest that this region will be constrained by the four Cu(II)-coordinating histidines into a rather compact structure. The seven amino acids between each histidine are likely to adopt specific but irregular loop structures. The four glycines in each octarepeat could help to accommodate the tight turns that we suspect are necessary to correctly position the imidazoles for Cu(II) binding. As glycines lack a side chain, this will reduce any problems of steric crowding that the cluster of four copper ions might otherwise experience.
The large number of copper ions per PrP molecule and the cooperative nature of the Cu(II) binding implies PrP has the ability to exchange Cu(II) with other copper binding molecules and suggests a role for PrP in Cu(II) metabolism. PrPC is located on the surface of the cell, fixed to the plasma membrane by the glycosyl-phosphatidylinositol anchor at the C terminus, leaving the apoprotein octarepeat domain free to bind Cu(II). A physiologically important role for Cu(II) binding (9) is supported by studies showing lowered levels of Cu(II) in subcellular fractions from the brains of PrP-deficient mice (8). Proteins that bind to PrPC:Cu(II) are candidates for a copper sensing or transport function.
CONCLUSION
The accumulated evidence suggests that PrP has a functional role in Cu(II) metabolism. Spectroscopic data presented here suggest a bridged arrangement of coordinating histidine imidazole nitrogens ligating four Cu(II) per PrP molecule. The proposed coordination geometry involving four copper ions accounts for the cooperative nature of complex formation and the stoichiometry of copper binding. This cooperative binding may well be a necessary part of the function of PrP and could explain why PrP in the apoprotein (copper-free) form contains a large unstructured domain with an unusual histidine-containing octarepeat sequence, which is highly conserved both in sequence and in number of repeats among mammalian species.
Acknowledgments
We thank Sabine Lahrichi for synthesizing octarepeat peptides, Darlene Groth for expression and purification of PrP(29–231) and PrP(90–231), and John Chung for maintaining NMR facilities. This research was supported by Grant NS14069 from the National Institutes of Health.
ABBREVIATIONS
- PrP
prion protein
- PrPC
cellular isoform of PrP
- CD
circular dichroism
- ESR
electron spin resonance
References
- 1.Prusiner S B. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 2.Chazot G, Broussolle E, Lapras C, Blattler T, Aguzzi A, Kopp N. Lancet. 1996;347:1181. doi: 10.1016/s0140-6736(96)90638-8. [DOI] [PubMed] [Google Scholar]
- 3.Will R G, Ironside J W, Zeidler M, Cousens S N, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofman A, Smith P G. Lancet. 1996;347:921–925. doi: 10.1016/s0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner S B. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 5.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, et al. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horwich A L, Weissman J S. Cell. 1997;89:499–510. doi: 10.1016/s0092-8674(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 7.Kaneko K, Zulianello L, Scott M, Cooper C M, Wallace A C, James T L, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown D R, Qin K F, Herms J W, Madlung A, Manson J, Strome R, Fraser P E, Kruck T, Von Bohlen A, Schulz-Schaeffer W, et al. Nature (London) 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 9.Brown D R, Schmidt B, Kretzschmar H A. J Neurochem. 1998;70:1686–1693. doi: 10.1046/j.1471-4159.1998.70041686.x. [DOI] [PubMed] [Google Scholar]
- 10.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 11.Billeter M, Riek R, Wider G, Hornemann S, Glockshuber R, Wüthrich K. Proc Natl Acad Sci USA. 1997;94:7281–7285. doi: 10.1073/pnas.94.14.7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, et al. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris D A, Falls D L, Johnson F A, Fischbach G D. Proc Natl Acad Sci USA. 1991;88:7664–7668. doi: 10.1073/pnas.88.17.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poulter M, Baker H F, Frith C D, Leach M, Lofthouse R, Ridley R M, Shah T, Owen F, Collinge J, Brown J, et al. Brain. 1998;115:675–685. doi: 10.1093/brain/115.3.675. [DOI] [PubMed] [Google Scholar]
- 16.Sulkowski E. FEBS Lett. 1992;307:129–130. doi: 10.1016/0014-5793(92)80750-b. [DOI] [PubMed] [Google Scholar]
- 17.Pan K M, Stahl N, Prusiner S B. Protein Sci. 1992;1:1343–1352. doi: 10.1002/pro.5560011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornshaw M P, McDermott J R, Candy J M, Lakey J H. Biochem Biophys Res Commun. 1995;214:993–999. doi: 10.1006/bbrc.1995.2384. [DOI] [PubMed] [Google Scholar]
- 19.Stöckel J, Safar J, Wallace A C, Cohen F E, Prusiner S B. Biochemistry. 1998;37:7185–7193. doi: 10.1021/bi972827k. [DOI] [PubMed] [Google Scholar]
- 20.Mehlhorn I, Groth D, Stockel J, Moffat B, Reilly D, Yansura D, Willett W S, Baldwin M, Fletterick R, Cohen F E, et al. Biochemistry. 1996;35:5528–5537. doi: 10.1021/bi952965e. [DOI] [PubMed] [Google Scholar]
- 21.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 22.Piotto M, Saudek V, Sklenár V. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 23.Shaka A J, Lee C J, Pines A. J Magn Reson. 1988;77:274–293. [Google Scholar]
- 24.Miura T, Hori-i A, Takeuchi H. FEBS Lett. 1996;396:248–252. doi: 10.1016/0014-5793(96)01104-0. [DOI] [PubMed] [Google Scholar]
- 25.Peisach J, Blumberg W E. Arch Biochem Biophys. 1974;165:691–708. doi: 10.1016/0003-9861(74)90298-7. [DOI] [PubMed] [Google Scholar]
- 26.Perkins C M, Roes N J, Weinstein B, Stenkamp R E, Jensen L H, Pickart L. Inorg Chim Acta. 1984;82:93–99. [Google Scholar]
- 27.Freedman J H, Pickart L, Weinstein B, Mims W B, Peisach J. Biochemistry. 1982;21:4540–4544. doi: 10.1021/bi00262a004. [DOI] [PubMed] [Google Scholar]
- 28.Solomon E I, Lowery M D, LaCroix L B, Root D E. Methods Enzymol. 1993;226:1–33. doi: 10.1016/0076-6879(93)26003-r. [DOI] [PubMed] [Google Scholar]
- 29.Bryce G F, Gurd F R. J Biol Chem. 1966;241:122–129. [PubMed] [Google Scholar]
- 30.Bryce G F, Roeske R W, Gurd F R. J Biol Chem. 1965;240:3837–3846. [PubMed] [Google Scholar]
- 31.Freeman H C. Adv Protein Chem. 1967;22:257–424. doi: 10.1016/s0065-3233(08)60043-1. [DOI] [PubMed] [Google Scholar]
- 32.Camerman N, Camerman A, Sarkar B. Can J Chem. 1976;54:1309–1316. [Google Scholar]
- 33.Parge H E, Hallewell R A, Tainer J A. Proc Natl Acad Sci USA. 1992;89:6109–6113. doi: 10.1073/pnas.89.13.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundberg R J, Martin R B. Chem Rev. 1974;74:471–517. [Google Scholar]